Introduction
Each year, ~480,000 new cases of oral cancer are
diagnosed worldwide, the majority of which are squamous cell
carcinoma (1,2). Globally, oral squamous cell carcinoma
(OSCC) has become the most commonly diagnosed type of head and neck
neoplasm (3). Despite improvements in
tumor treatments, the 5-year survival rate for OSCC has remained
~50% during the last decades (4). In
the treatment of OSCC, surgery and radiation can cause severe
functional impairment, while the efficacy of chemotherapeutic
agents is limited by acquired resistance and drug side effects
(5). As a result, the development of
new molecular targets for the prevention and treatment of OSCC has
become an urgent matter.
WW domain-containing oxidoreductase (WWOX) has been
identified as a tumor-suppressor gene that spans the chromosomal
fragile site FRA16D. Lost or reduced expression of the WWOX gene
commonly presents in numerous types of neoplasms, including breast,
prostate, ovary, lung and oral cancer (6–8). A reduced
expression of WWOX has been observed in cases of OSCC and oral
leukoplakia, which are prevalent precancerous lesions (8,9). By
contrast, normal transcriptomes and proteins of WWOX were observed
to be expressed in normal mucosa (9,10).
Therefore, the present study speculated that WWOX deficiency in
oral squamous carcinoma cells may contribute to oral
carcinogenesis. However, the function and precise molecular
mechanism of WWOX in OSCC remain unclear.
The present study aimed to detect, in vitro,
the effect of WWOX overexpression on cell growth, apoptosis and
cell cycle distribution in oral squamous carcinoma cells. The
present study also used a microarray assay, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis and western blotting to investigate the expression of
genes that are modulated by WWOX, in order to elucidate the
underlying molecular mechanisms of the antitumor effect of
WWOX.
Materials and methods
Cell lines and cell culture
The oral cancer cell lines, Tca8113, CAL27 and FaDu,
were provided by Dr Zhiyong Li (Stomatology Hospital Affiliated to
Zhejiang University College of Medicine, Hangzhou, China). These
cells and 293T cells [Type Culture Collection of Chinese Academy of
Sciences (TCCCAS), Beijing, China] were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (GE
Healthcare Life Sciences, Chalfont, UK). The cells were grown in an
incubator at 37°C with a humidified atmosphere containing 5%
CO2.
Plasmid construction, virus packaging
and cell transfection
The WWOX gene, NM_016373, open reading frame was
amplified from human breast carcinoma MCF-7 cells (TCCCAS, Beijing,
China) using the following specific primers; forward,
5′-GAGGATCCCCGGGTACCGGTCGCCACCATGGCAGCGCTGCGCTAC-3′ and reverse,
5′-TCCTTGTAGTCCATACCGCCGGACTGGCTGCCAAG-3′. The WWOX complementary
DNA (cDNA) was subcloned into a pGV287-LV lentivirus vector,
purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). The
positive clone, pGV287-LV-WWOX, was selected through sequencing.
The reconstructed lentivirus vector, and two helper vectors,
pHelper1.0 and pHelper2.0 (Jikai Gene Biochemical Co., Ltd.,
Shanghai, China), were produced in the 293T packaging cell line.
The cells were placed in 6-well plates and grown to 30% confluency
prior to being infected with the virus (~109 TU/ml, multiplicity of
infection=20). The medium was replaced at 16 h post-infection and
the cells were cultured as normal. RT-qPCR and western blotting
were then used to confirm the expression of WWOX, according to the
protocol described below.
Cell proliferation assay
An MTT assay was used to assess cell proliferation.
The Tca8113 cells, the Tca8113 cells transfected with an empty
vector and the Tca8113 cells transfected with the reconstructed
vector were separately plated in 96-well plates with a density of
2,000 cells/well and incubated at 37°C for 1–5 days. A total of 20
µl MTT (5 mg/ml) was added to each well and incubated for 4 h on
days 1 to 5. Each day, prior to incubation, a total of 20 µl MTT (5
mg/ml) was added to each well and the plates were incubated for 4
h. The crystals formed were subsequently dissolved in dimethyl
sulfoxide. The optical density was assessed at 490 nm
(OD490). Cell proliferation activity was analyzed using
the mean OD490 values for each well.
Colony formation assay
Subsequent to infection, the cells were seeded in
6-well plates at a density of 800 cells/plate and cultured at 37°C
for 14 days until visible colonies appeared. The cells were stained
with methyl violet prior to end of the incubation and the number of
colonies was counted under a light microscope (Olympus Corporation,
Tokyo, Japan).
Apoptosis and cell cycle analysis
The cells were harvested, then washed and
resuspended in PBS 72 h following transfection, then stained with
propidium iodide for cell cycle analysis. Apoptosis analysis was
performed using an Annexin V-APC Apoptosis Detection kit
(eBioscience, Inc., San Diego, CA, USA), according to the protocol
of the manufacturer.
Microarray analysis
Total RNA from six samples, consisting of three
samples from Tca8113 cells with WWOX overexpression and three
samples from Tca8113 cells infected with the empty lentivirus
vector, was isolated. TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to lyse the cells, then chloroform and
isopropanol were used to isolate the RNA from the cell lysate, and
alcohol treated with diethyl pyrocarbonate was used to inactivate
RNases. The RNA samples were tested using NanoDrop 2000 (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA) and a 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). The
RNA samples that met the following criteria: NanoDrop 2000, 1.7<
Absorbance (A)260/A280 <2.2; and 2100
Bioanalyzer, RNA integrity number ≥7.0 and 28S/18S >0.7, were
analyzed by microarray expression profiling using
GeneChip® PrimeView™ Human Gene Expression Array
(Affymetrix, Inc., Santa Clara, CA, USA) according to standard
protocol (11). Genes that were
significantly differentially expressed in cells, with or without
WWOX overexpression, were selected based on exhibiting P<0.05
and a 1.5-fold-change (FC). The Reactome FI Cytoscape Plugin
(www.reactome.org) was used to perform function
pathway analysis according to the protocol of the manufacturer.
RT-qPCR
Each reaction was carried out in triplicate. Total
RNA separately from the Tca8113 cells with empty vectors and the
Tca8113 cells from with WWOX overexpression was isolated from the
cultured cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) for cDNA synthesis. Reverse transcription was
performed for one cycle under the following conditions: 70°C for 10
min; 42°C for 1 h; then 70°C for 10 mißn using the Reverse
Transcription System (Promega Corporation, Madison, WI, USA). qPCR
was performed using a StepOneä Real-Time PCR System (Applied
Biosystems, Carlsbad, CA, USA) with the One Step SYBR PrimeScript
RT-PCR kit II (Takara Bio, Inc., Otsu, Japan). The relative
expression of all investigated genes was calculated using the
2−ΔΔCq (12) method
subsequent to the normalization of the reference gene GAPDH. The
primer sequences were as follows: GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′; WWOX forward,
5′-CCAACCACCCGGCAAAGATA-3′ and reverse, 5′-AATGCTGCACGCTACGGAG-3′;
dual specificity phosphatase (DUSP) 5 forward,
5′-TCCTCACCTCGCTACTCG-3′ and reverse, 5′-ACATCCACGCAACACTCAG-3′;
DUSP6 forward, 5′-GAACTGTGGTGTCTTGGTACATT-3′ and reverse,
5′-GTTCATCGACAGATTGAGCTTCT-3′; nuclear receptor subfamily 4 group A
member 1 (NR4A1) forward, 5′-TCATGGACGGCTACACAG-3′ and reverse,
5′-GTGGCTGAGGACGAGGATG-3′; mitogen-activated protein kinase kinase
(MAP2 K) 5 forward, 5′-ACTGACGAGCGGTGGAC-3′ and reverse,
5′-GTCGGAAGGTTCTGGA-3′; and fibroblast growth factor receptor 2
(FGFR2) forward, 5′-GGTGGCTGAAAAACGGGAAG-3′ and reverse,
5′-AGATGGGACCACACTTTCCATA-3′. PCR was performed under the following
conditions: 95°C for 30 sec; 40 cycles of 95°C for 5 sec and 60°C
for 30 sec.
Western blot analysis
Cell proteins were separated using 10% SDS-PAGE and
transferred to a polyvinylidene fluoride membrane subsequent to
cell protein concentration being quantified with a Bio-Rad assay
kit (cat. no. 5000002; Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The membranes were blocked in 5% non-fat milk at room
temperature for 1 h and incubated with the primary antibodies (all
antibodies were purchased from Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) anti-WWOX (dilution, 1:200; cat. no. sc-390175),
anti-DUSP5 (dilution, 1:500; cat. no. sc-393801), anti-DUSP6
(dilution, 1:500; cat. no. sc-137245), anti-NR4A1 (dilution, 1:500;
cat. no. sc-365113), anti-MAP2K5 (dilution, 1:500; cat. no.
sc-135986) and anti-FGFR2 (dilution, 1:500; cat. no. sc-6930), at
4°C overnight. Subsequent to 3 washes in TBS/0.1% Tween-20, the
membranes underwent hybridization with a peroxidase-conjugated
secondary antibody (dilution, 1:2,000; cat. no. sc-2031; Santa Cruz
Biotechnology, Inc.) for 1 h at room temperature. Signals were then
visualized with the BeyoECL Plus detection kit (Beyotime Institute
of Biotechnology, Haimen, China).
Statistical analysis
All quantitative data were presented as the mean ±
standard deviation. One-way analysis of variance and Student's
t-test were used to compare the normally distributed continuous
variables. The statistical significance of the microarray results
was analyzed using FC values and a Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analysis was performed with SPSS 16.0 (SPSS, Inc.,
Chicago, IL, USA).
Results
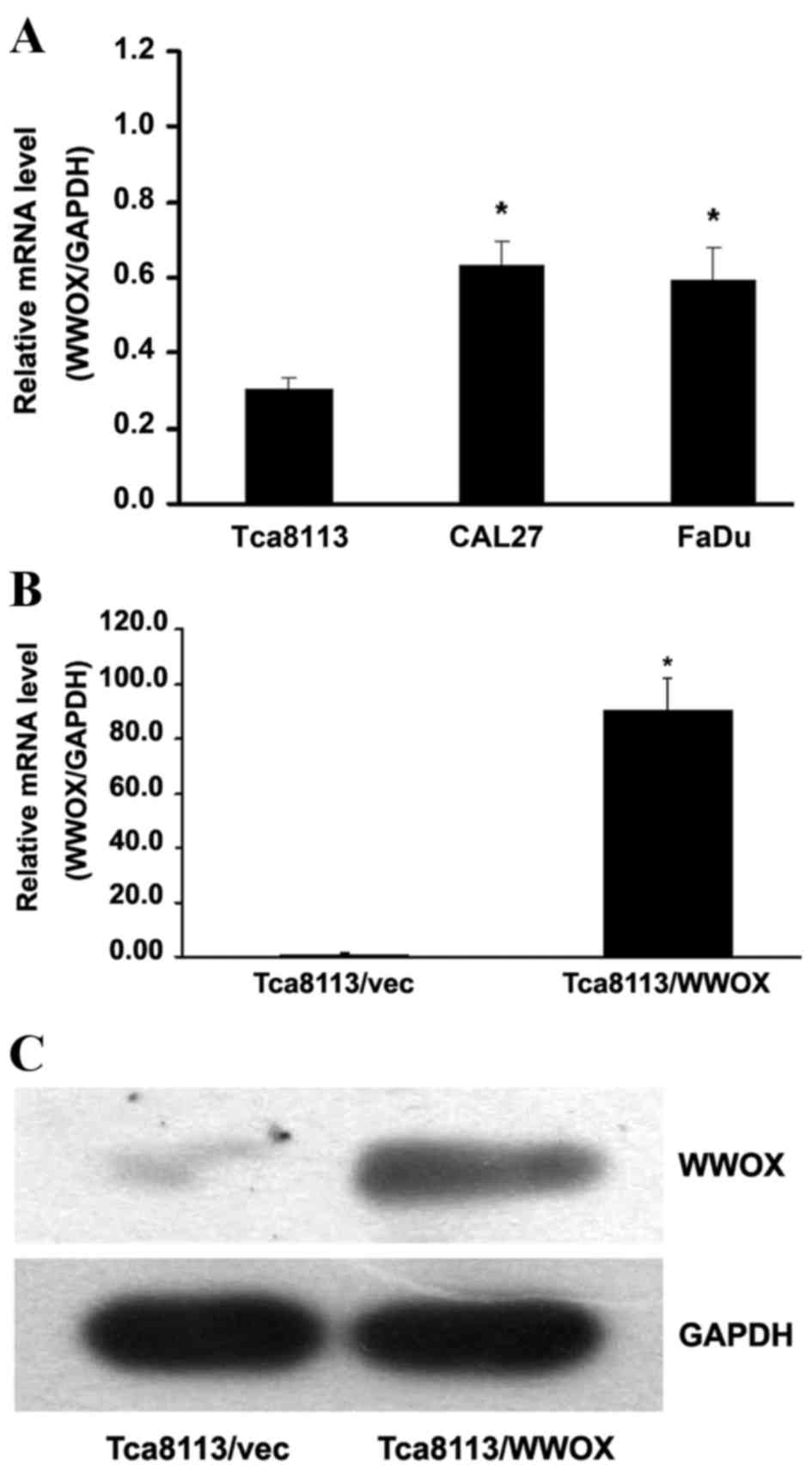
Lentivirus infection enables the
overexpression of the WWOX gene in Tca8113 cells
By comparing the relative WWOX messenger RNA (mRNA)
levels in the oral cancer Tca8113, CAL27 and FaDu cell lines, the
present study observed that Tca8113 cells exhibited the lowest
expression of the WWOX gene (P<0.05; Fig. 1A). Subsequent to the Tca8113 cells
being infected with the lentivirus plasmid pGV287-LV-WWOX, the mRNA
expression of the WWOX gene was >90.593-fold, significantly
greater than that detected in the cells infected with the empty
vector pGV287-LV (P<0.05; Fig.
1B). Western blotting detected elevated levels of WWOX protein
expression in Tca8113 cells with WWOX overexpression (Fig. 1C).
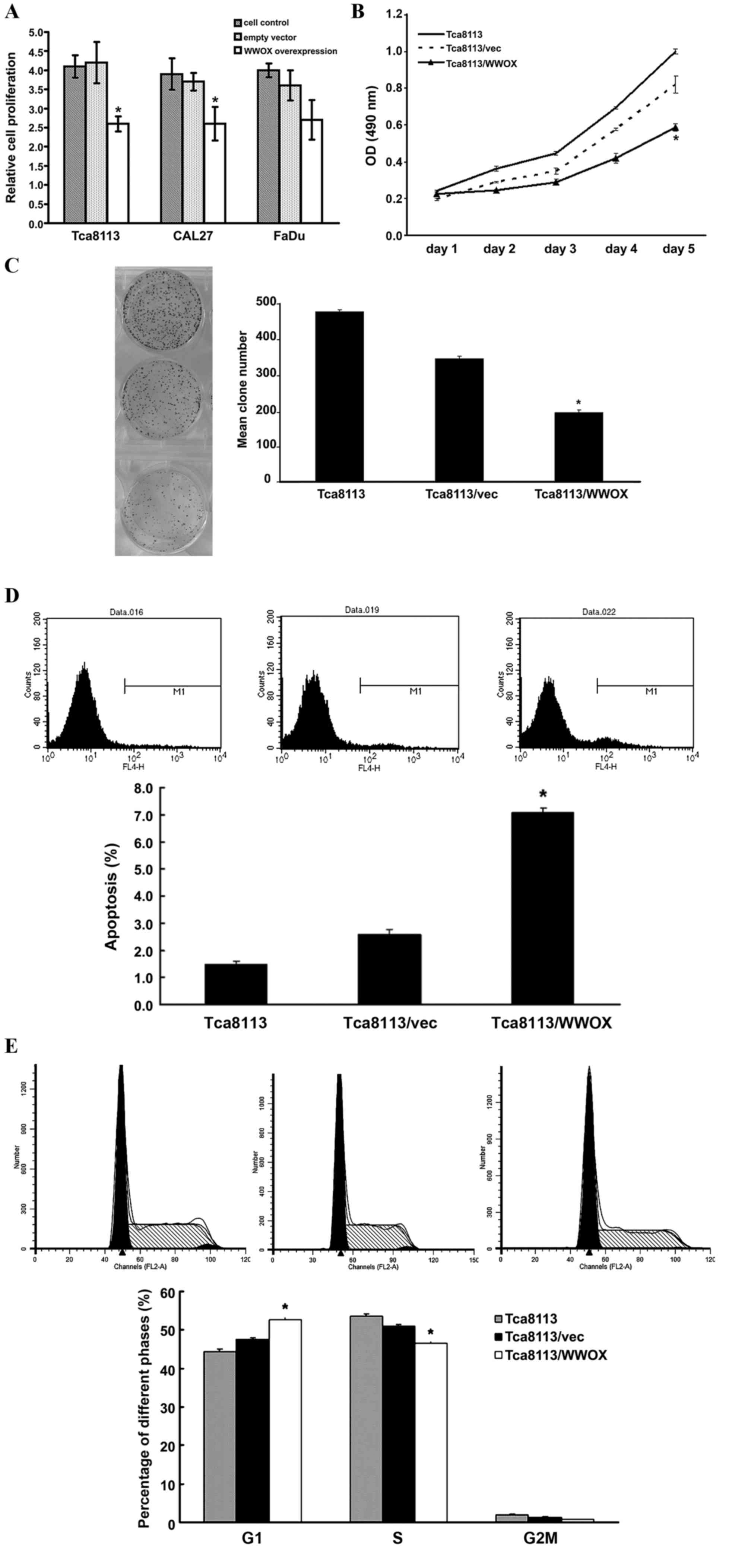
Overexpression of the WWOX gene
inhibits cell growth in vitro
To investigate the effect of WWOX on cell growth,
WWOX overexpression plasmids were transfected into Tca8113, CAL27
and FaDu cells. WWOX overexpression inhibited cell proliferation as
determined by the MTT method. This inhibition was most prevalent in
the Tca8113 cells (P<0.05; Fig. 2A and
B). The colony formation assay also revealed similar inhibition
in the Tca8113 cells (P<0.05; Fig.
2C). Furthermore, flow cytometry assays revealed a significant
increase in apoptosis and G0/G1-phase population subsequent to
transfection with the WWOX overexpression plasmids (P<0.05;
Fig. 2D and E).
Overexpression of the WWOX gene
modulates several key pathways in Tca8113 cells, as determined by
global gene expression analysis
To investigate the mechanism of WWOX
tumor-suppressor function, the present study compared the
transcriptomes of cells transfected with the WWOX gene with those
transfected with an empty vector. The present study used the
Affimetrix Human Gene 1.0 ST Array to identify 347 transcriptomes
that were significantly differentially expressed, of which, 171
genes were upregulated and 176 genes were downregulated, based on
an FC >1.5 and P<0.05 threshold in Tca8113 cells with WWOX
overexpression compared with the findings in the associated control
(Fig. 3A). Through a functional
analysis of genes using the Reactome Functional Interaction
network, the present study demonstrated that WWOX overexpression
modulated the activation of certain key signaling pathways,
including the mitogen-activated protein kinase (MAPK) signaling
pathway (Fig. 3B and C).
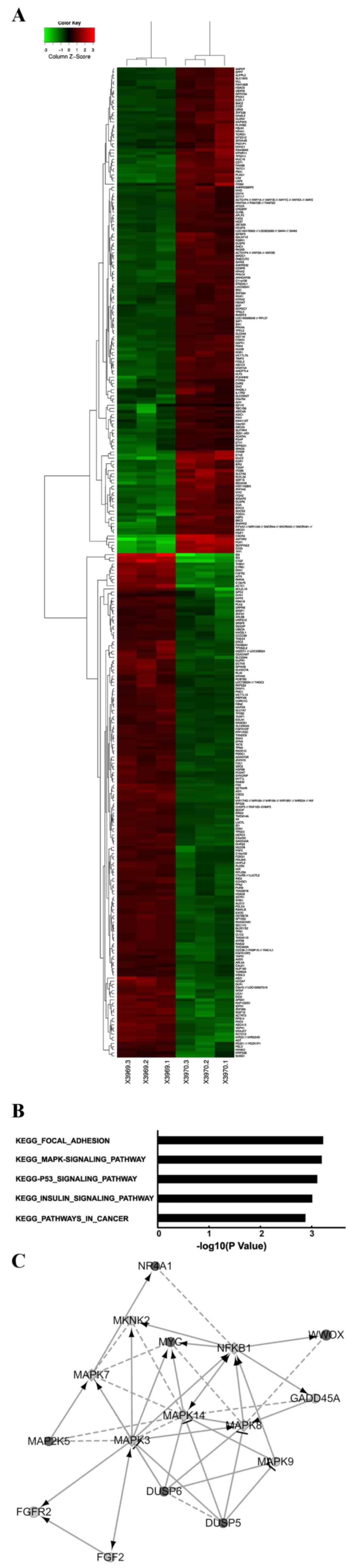 | Figure 3.Global changes in the Tca8113 cell
transcriptome with WWOX overexpression. (A) Heat map depicting 347
transcripts, which were significantly differentially expressed in
Tca8113 cells with WWOX overexpression (P<0.05). The rows and
columns represent samples and transcriptomes, respectively. The red
and green colors represent upregulated and downregulated genes,
respectively. X3969.1-X3969.3 represents the samples from the
Tca8113 cells carrying empty vectors and X3970.1-X3970.3 represent
the samples from the Tca8113 cells overexpressing the WWOX gene.
(B) Functional pathway analysis of the differentially expressed
genes was conducted by Reactome Functional Interaction network. In
the Tca8113 cells with WWOX overexpression, the five pathways
modulated significantly by WWOX overexpression are depicted,
indicated by the inverse logarithm of P-values. (C) Network between
the WWOX gene and the MAPK signaling pathway. In the map, black
circles, grey circles and grey squares are used to indicate changes
in gene expression: Upregulation, downregulation and predicted
genes, respectively. The larger the diameter of the circle is, the
more significant the difference between the two groups. Solid lines
indicate confirmed interactions and dashed lines represent
predicted interactions. WWOX, WW domain-containing oxidoreductase;
MAPK, mitogen-activated protein kinase; vec, empty vector; KEGG,
Kyoto Encyclopedia of Genes and Genomes. |
Overexpression of the WWOX gene
regulates the expression of certain genes involved in modulating
MAPK signaling
The present study screened selected genes associated
with MAPK signaling. Utilizing global gene expression analysis to
determine absolute FC subsequent to the overexpression of WWOX, the
present study identified that the absolute FC of DUSP5, DUSP6,
NR4A1 and MAP2K5 increased, while the absolute FC of FGFR2
decreased, compared with that of the Tca8113/vector control group
(all P<0.05; Fig. 4A).
Subsequently, the mRNA and protein expression levels of the genes
were examined by RT-qPCR and western blotting, yielding results
that were for the majority of genes consistent with the initial
findings of the present study (P<0.05; Fig. 4B and C), indicating the upregulation
of DUSP5, DUSP6, NR4A1 and MAP2K5, and the downregulation of FGFR2,
with respect to mRNA and protein levels.
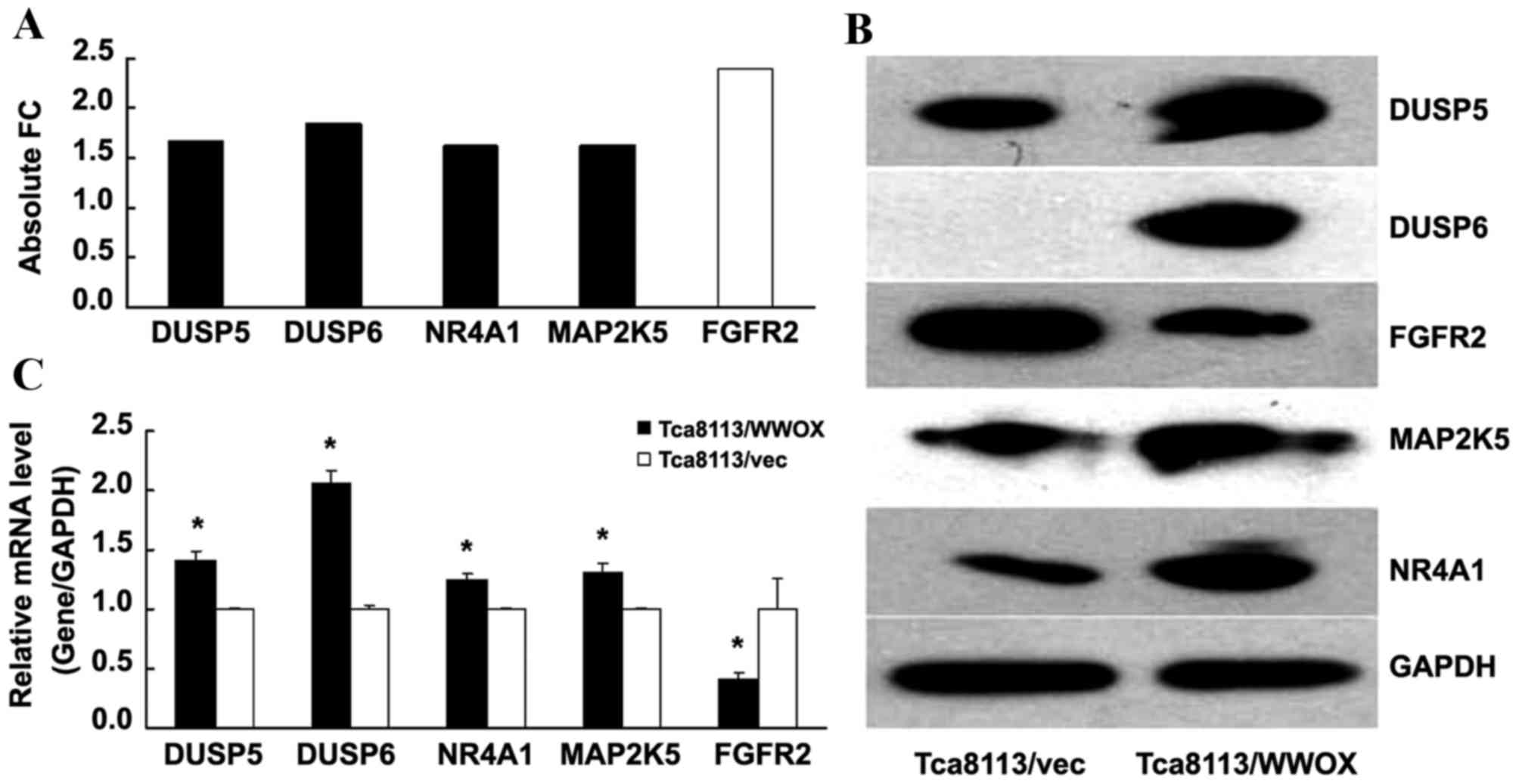 | Figure 4.WWOX modulates the expression of genes
involved in mediating MAPK signaling. (A) Absolute FC indicates the
expression level of genes, compared with that of the Tca8113/vec
control (which was equal to 1). Black columns represent upregulated
genes and white columns represent downregulated genes. (B) Western
blot analysis of the proteins was conducted, including the cells
with and without WWOX overexpression. (C) The relative mRNA level
of DUSP5, DUSP6, NR4A1, MAP2K5 and FGFR2 was detected by reverse
transcription-quantitative polymerase chain reaction, normalized to
GAPDH and compared with the control (which was equal to 1).
*P<0.05 compared with the Tca8113/vec cells. WWOX, WW
domain-containing oxidoreductase; DUSP, dual-specificity
phosphatase; NR4A1, nuclear receptor subfamily 4 group A member 1;
MAP2K5, mitogen-activated protein kinase kinase 5; FGFR2,
fibroblast growth factor receptor 2; FC, fold-change; vec, empty
vector; mRNA, messenger RNA. |
Discussion
WWOX has been identified as a gene located on a
chromosomal fragile site, and deficiency/deletion with respect to
the expression of the gene is common in various types of carcinoma
(6). Reduced expression was also
demonstrated in OSCC and oral precancerous lesions subsequent to
examination with PCR and immunohistochemistry (8–10). Reduced
expression of WWOX contributed to tumor development and
progression, and exogenous WWOX expression significantly suppressed
tumor growth (13,14). Qu et al (15) revealed that the reconstitution of WWOX
inhibited cell proliferation and induced apoptosis, while the
knockdown of WWOX resulted in the opposite effect in cervical
cancer cells. Lin et al (16)
concluded that WWOX suppressed prostate cancer cell progression by
inducing cell cycle arrest in the G1 phase. The present study
investigated the effect of WWOX overexpression on cell growth in
oral squamous carcinoma cells, and the results are consistent with
the findings of previous studies with respect to the WWOX gene
inhibiting cell proliferation, and promoting apoptosis and cell
cycle arrest. To investigate the underlying tumor-suppression
mechanism of the WWOX gene, the present study used microarray
analysis to evaluate the genetic changes exhibited in Tca8113 cells
subsequent to WWOX overexpression.
To investigate the underlying tumor-suppression
mechanism of the WWOX gene, the present study analyzed the genetic
change of Tca8113 cells following WWOX overexpression by microarray
analysis, and noticed an increase in the expression of DUSP5,
DUSP6, NR4A1 and MAP2K5, and a decrease in the expression of FGFR2.
These genes are closely associated with the extracellular-signal
regulated protein kinase (ERK)/MAPK signaling pathway, and mediate
various biological events involved in cell proliferation,
differentiation and survival (17).
DUSP5 and DUSP6 are members of the MAPK phosphatase
family (18). Okudela et al
(19), Li et al (20) and Nunes-Xavier et al (21) observed that DUSP5 and DUSP6 act as
negative mediators in the regulation of ERK1/2 phosphorylation and
cell growth in tumor cells. Wang et al (18) indicated that, in corneal epithelial
cells, DUSP6 overexpression specifically prevented the formation of
phosphorylated ERK1/2 and slowed cell growth, whereas DUSP5
knockdown was observed to enhance ERK1/2 phosphorylation and cell
growth. The authors therefore concluded that DUSP5 and DUSP6 serve
a role in the negative feedback regulation of ERK/MAPK signaling
when their expression is upregulated through the activation of the
ERK/MAPK signaling pathway. The present study demonstrated that,
subsequent to WWOX overexpression, the increased expression of
DUSP5 and DUSP6 is accompanied by the inhibition of Tca8113 cell
growth. Therefore, the present study hypothesizes that WWOX
overexpression activates the ERK/MAPK signaling pathway, and
upregulates the expression of DUSP5 and DUSP6. Conversely, DUSP5
and DUSP6 reduce ERK phosphorylation, and suppress the growth of
Tca8113 cells.
NR4A1, also referred to as Nur77, is a member of the
nuclear receptor subfamily 4, group A, and can be activated via a
cascade involving MAP2K5, MAPK7 and NR4A1, which is also dependent
on the ERK/MAPK signaling pathway (22). In OSCC, NR4A1 activated through the
MAPK signaling pathway can induce apoptosis (23). The present study demonstrated that the
combination of the upregulation of NR4A1 and MAP2K5 increased the
level of apoptosis subsequent to WWOX overexpression in Tca8113
cells. Previous studies identified that NR4A1 induces apoptosis by
associating with B-cell lymphoma 2 and initiating the release of
cytochrome c (23,24). Zhang et al (25) reported that the ectopic overexpression
of WWOX also induces a release of cytochrome c from the
mitochondria. As a result, the present study hypothesizes that the
overexpression of WWOX upregulates the expression of MAP2K5 and
NR4A1 by activating the ERK/MAPK signaling pathway, and induces
apoptosis in Tca8113 cells through the release of cytochrome
c.
FGFR2 is a tyrosine kinase receptor that is crucial
with respect to controlling tumor proliferation, angiogenesis,
migration and survival (26). Katoh
and Nakagama (27) demonstrated that
the expression of FGFR2 was amplified in breast and gastric cancer.
In the colorectal cancer NCI-H716 cell line, which exhibits a high
expression of FGFR2, the inhibition of FGFR2 by small molecule
inhibitors or FGFR2 short hairpin (sh)RNA was shown to decrease
cell viability (28). In pancreatic
cancer, tumor cells with FGFR2-shRNA transfection exhibited
attenuated proliferation rates, migration and invasion levels, and
a reduced level of phosphorylation of ERK compared with that of the
control cells (29). These findings
demonstrate that the inhibition of FGFR2 contributes to the
suppression of cell proliferation and ERK phosphorylation. In the
present study, a reduced expression of FGFR2 and the inhibition of
growth in Tca8113 cells were also observed when WWOX was
overexpressed.
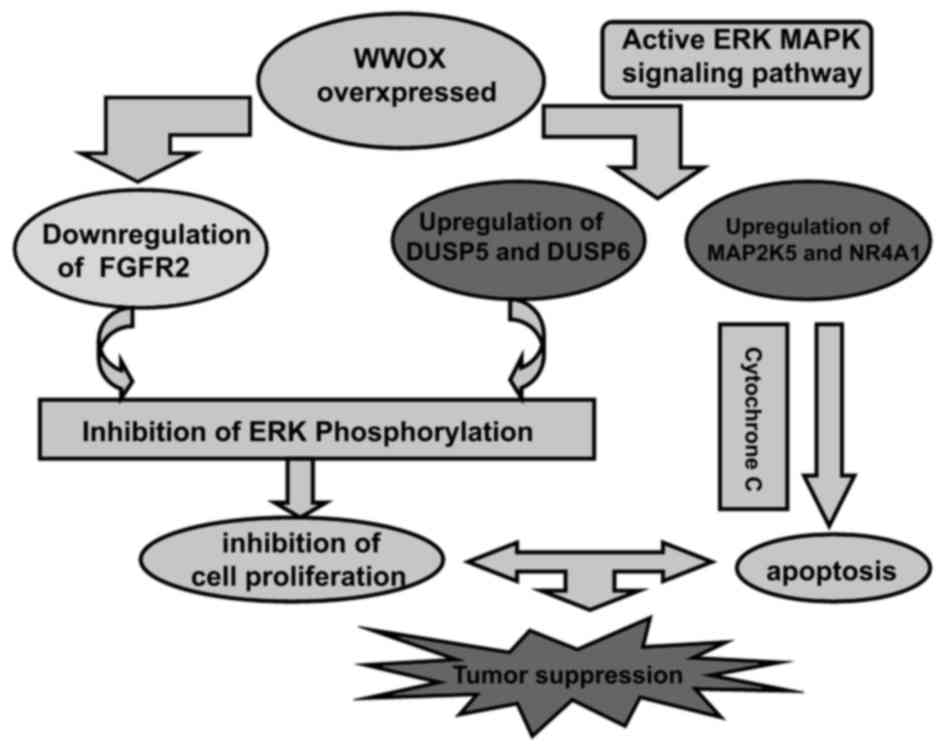
In summary, the overexpression of the WWOX gene in
Tca8113 cells suppressed cell growth, and induced apoptosis and
cell cycle arrest. This tumor suppression is associated with a
modulation of the expression of genes that mediate the ERK/MAPK
signaling pathway. The conclusions of the present study regarding
the tumor-suppressor functions of the WWOX gene in a diagrammatic
sketch are presented in Fig. 5.
Targeting WWOX may be an effective method for the treatment of oral
cancer.
Acknowledgements
The present study was supported by grants from the
Public Welfare Technology and Application Research Projects (grant
no. 2013C37019), the Science and Technology Plans of Taizhou City
(grant no. 131ky17) and the Science and Technology Development Plan
of Jilin City (grant no. 20163066).
References
|
1
|
Minicucci EM, da Silva GN and Salvadori
DM: Relationship between head and neck cancer therapy and some
genetic endpoints. World J Clin Oncol. 5:93–102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ratajczak-Wrona W, Jablonska E, Antonowicz
B, Dziemianczyk D and Grabowska SZ: Levels of biological markers of
nitric oxide in serum of patients with squamous cell carcinoma of
the oral cavity. Int J Oral Sci. 5:141–145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rivera C and Venegas B: Histological and
molecular aspects of oral squamous cell carcinoma (Review). Oncol
Lett. 8:7–11. 2014.PubMed/NCBI
|
|
4
|
Warnakulasuriya S: Living with oral
cancer: Epidemiology with particular reference to prevalence and
life-style changes that influence survival. Oral Oncol. 46:407–410.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Giannola LI, De Caro V, Giandalia G,
Siragusa MG, Paderni C, Campisi G and Florena AM: 5-Fluorouracil
buccal tablets for locoregional chemotherapy of oral squamous cell
carcinoma: Formulation, drug release and histological effects on
reconstituted human oral epithelium and porcine buccal mucosa. Curr
Drug Deliv. 7:109–117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lewandowska U, Zelazowski M, Seta K,
Byczewska M, Pluciennik E and Bednarek AK: WWOX, the tumour
suppressor gene affected in multiple cancers. J Physiol Pharmacol.
60:(Supp l). S47–S56. 2009.
|
|
7
|
Ekizoglu S, Muslumanoglu M, Dalay N and
Buyru N: Genetic alterations of the WWOX gene in breast cancer. Med
Oncol. 29:1529–1535. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pimenta FJ, Cordeiro GT, Pimenta LG, Viana
MB, Lopes J, Gomez MV, Aldaz CM, De-Marco L and Gomez RS: Molecular
alterations in the tumor suppressor gene WWOX in oral leukoplakias.
Oral Oncol. 44:753–758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pimenta FJ, Gomes DA, Perdigão PF, Barbosa
AA, Romano-Silva MA, Gomez MV, Aldaz CM, De Marco L and Gomez RS:
Characterization of the tumor suppressor gene WWOX in primary human
oral squamous cell carcinomas. Int J Cance. 118:1154–1158. 2006.
View Article : Google Scholar
|
|
10
|
Pimenta FJ, Cordeiro GT, Pimenta LG, Viana
MB, Lopes J, Gomez MV, Aldaz CM, De Marco L and Gomez RS: Molecular
alterations in the tumor suppressor gene WWOX in oral leukoplakias.
Oral Onco. 44:753–758. 2008. View Article : Google Scholar
|
|
11
|
Kabbout M, Garcia MM, Fujimoto J, Liu DD,
Woods D, Chow CW, Mendoza G, Momin AA, James BP, Solis L, et al:
ETS2 mediated tumor suppressive function and MET oncogene
inhibition in human non-small cell lung cancer. Clin Cancer Res.
19:3383–3395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aqeilan RI, Abu-Remaileh M and Abu-Odeh M:
The common fragile site FRA16D gene product WWOX: Roles in tumor
suppression and genomic stability. Cell Mol Life Sci. 71:4589–4599.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang H, Kong L, Cui Z, Du W, He Y, Yang
Z, Wang L and Chen X: The WWOX gene inhibits the growth of U266
multiple myeloma cells by triggering the intrinsic apoptotic
pathway. Int J Mol Med. 34:804–809. 2014.PubMed/NCBI
|
|
15
|
Qu J, Lu W, Li B, Lu C and Wan X: WWOX
induces apoptosis and inhibits proliferation in cervical cancer and
cell lines. Int J Mol Med. 31:1139–1147. 2013.PubMed/NCBI
|
|
16
|
Lin JT, Li HY, Chang NS, Lin CH, Chen YC
and Lu PJ: WWOX suppresses prostate cancer cell progression through
cyclin D1-mediated cell cycle arrest in the G1 phase. Cell Cycle.
14:408–416. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Whelan JT, Hollis SE, Cha DS, Asch AS and
Lee MH: Post-transcriptional regulation of the Ras-ERK/MAPK
signaling pathway. J Cell Physiol. 227:1235–1241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Z, Reinach PS, Zhang F, Vellonen KS,
Urtti A, Turner H and Wolosin JM: DUSP5 and DUSP6 modulate corneal
epithelial cell proliferation. Mol Vis. 16:1696–1704.
2010.PubMed/NCBI
|
|
19
|
Okudela K, Yazawa T, Woo T, Sakaeda M,
Ishii J, Mitsui H, Shimoyamada H, Sato H, Tajiri M, Ogawa N, et al:
Down-regulation of DUSP6 expression in lung cancer: Its mechanism
and potential role in carcinogenesis. Am J Pathol. 175:867–881.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Song L, Ritchie AM and Melton DW:
Increased levels of DUSP6 phosphatase stimulate tumourigenesis in a
molecularly distinct melanoma subtype. Pigment Cell Melanoma Res.
25:188–199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nunes-Xavier CE, Tárrega C, Cejudo-Marín
R, Frijhoff J, Sandin A, Ostman A and Pulido R: Differential
up-regulation of MAP kinase phosphatases MKP3/DUSP6 and DUSP5 by
Ets2 and c-Jun converge in the control of the growth arrest versus
proliferation response of MCF-7 breast cancer cells to phorbol
ester. J Biol Chem. 285:26417–26430. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mori Sequeiros Garcia M, Gorostizaga A,
Brion L, González-Calvar SI and Paz C: cAMP-activated Nr4a1
expression requires ERK activity and is modulated by MAPK
phosphatase-1 in MA-10 Leydig cells. Mol Cell Endocrinol.
408:45–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu PY, Sheu JJ, Lin PC, Lin CT, Liu YJ,
Ho LI, Chang LF, Wu WC, Chen SR, Chen J, et al: Expression of Nur77
induced by an n-butylidenephthalide derivative promotes apoptosis
and inhibits cell growth in oral squamous cell carcinoma. Invest
New Drugs. 30:79–89. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thompson J and Winoto A: During negative
selection, Nur77 family proteins translocate to mitochondria where
they associate with Bcl-2 and expose its proapoptotic BH3 domain. J
Exp Med. 205:1029–1036. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang P, Jia R, Ying L, Liu B, Qian G, Fan
X and Ge S: WWOX-mediated apoptosis in A549 cells mainly involves
the mitochondrial pathway. Mol Med Rep. 6:121–124. 2012.PubMed/NCBI
|
|
26
|
Feng S, Zhou L, Nice EC and Huang C:
Fibroblast growth factor receptors: Multifactorial-contributors to
tumor initiation and progression. Histol Histopathol. 30:13–31.
2015.PubMed/NCBI
|
|
27
|
Katoh M and Nakagama H: FGF receptors:
Cancer biology and therapeutics. Med Res Rev. 34:280–300. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mathur A, Ware C, Davis L, Gazdar A, Pan
BS and Lutterbach B: FGFR2 is amplified in the NCI-H716 colorectal
cancer cell line and is required for growth and survival. PLoS One.
9:e985152014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsuda Y, Yoshimura H, Suzuki T, Uchida
E, Naito Z and Ishiwata T: Inhibition of fibroblast growth factor
receptor 2 attenuates proliferation and invasion of pancreatic
cancer. Cancer Sci. 105:1212–1219. 2014. View Article : Google Scholar : PubMed/NCBI
|