Introduction
Polycomb group (PcG) proteins are epigenetic
regulators for gene silencing at the transcription level, acting as
important regulators of DNA repair, proliferation, embryonic
differentiation and cell-fate maintenance during development and in
adult tissue homeostasis (1–3). These proteins exist in at least two
biochemically and functionally distinct PcG core complexes referred
to as polycomb repressive complex (PRC) 1 and 2. PRC2, which is
involved in the initiation of gene repression, mediates the
trimethylation of histone H3 at Lys27 (H3K27me3) and consists of
histone methyltransferase enhancer of zeste homolog 2 (EZH2), the
polycomb repressive complex and embryonic ectoderm development
(4,5).
The mammalian PRC1, which is an ubiquitin E3 ligase complex,
consists of polycomb (PC), polyhomeotic (PH), BMI1, ring finger
protein (Rnf) 1A and 2 (Rnf2) (6),
among which Rnf2 has been identified as the catalytic subunit
(7).
Rnf2, acts as the RING finger E3 ligase responsible
for H2A modification in the PRC1 complex and influences early
development and embryonic stem cell maintenance as well as cancer
development (8,9). Recent evidence showed that Rnf2 is
highly expressed in various types of tumor in comparison to normal
tissue counterparts (10). Knockdown
of Rnf2 in HeLa cells resulted in morphological changes and an
inhibition of cellularproliferation (7), suggesting an oncogenic function of Rnf2.
Additionally, it has been reported that Rnf2 can either directly or
indirectly target a distinct, specific pathway involved in cell
cycle control (11,12). However, in order to acquire a complete
understanding of the mechanisms through which cancer progression is
regulated by Rnf2, additional investigation is required.
Normal cell cycle progression is tightly controlled
by a variety of molecular checkpoints, which supervise the
biological processes that take place in different phases of the
cell cycle (13). Progression through
the cell cycle is governed by a family of cyclin-dependent kinases
(CDKs), whose activity is regulated by phosphorylation, cyclin
binding, the inhibitor of the CDK4 (INK4) family (p16INK4A,
p15INK4B, p18 and p19) and kinase inhibitor protein (KIP) family
(p21, p27 and p57) (14–16). p21 and p27 are the fundamental members
of the KIP family and mediate various biological activities,
primarily by binding to and inhibiting the kinase activity of the
CDKs (17).
As a proliferation inhibitor, p21 performs an
essential role in growth arrest following DNA damage, and
overexpression of p21 leads to G1 and G2 or S-phase arrest
(18,19). PcG proteins can behave as integrators
and/or modulators of cell cycle checkpoints in dividing cells. In
mammals, PRC1 and PRC2 bind to and repress the INK4a/ARF locus,
which encodes several proteins involved in cell cycle regulation
including p14ARF and p16INK4A (20,21). In
melanoma cells, EZH2 depletion induced a decrease in the proportion
of cells in S phase with a concomitant increase of cells in the G1
phase as well as an elevated expression of p21 (22), indicating that PcG has close
associations with p21 in regulating the cell cycle. However, the
precise mechanism behind this remains unclear. The present study
aimed to investigate the expression of Rnf2 and further explore its
effect on cell viability and the cell cycle of gastric cancer (GC)
cells.
Materials and methods
Cell culture
The human stomach cancer SGC-7901 cell line and
normal gastric epithelium GES-1 cell line were obtained from the
Central Laboratory of The Affiliated Hospital of Qingdao University
(Qingdao, China). The cell lines were cultured in RPMI 1640
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, CA, USA)
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin under standard
culture conditions (5% CO2, 37°C and 95% humidity).
Human cell line assays were conducted according to IRB regulations
at The Affiliated Hospital of Qingdao University.
Reverse
transcription-quantitativepolymerase chain reaction (RT-qPCR)
Total RNA was isolated using the TRIzol Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. A random-primed cDNA was generated using a
Superscript II reverse transcription kit (Invitrogen; Thermo Fisher
Scientific, Inc.). Triplicate reactions of cDNA amplification were
performed in SYBR Premix Ex Taq (Takara Biotechnology Co. Ltd.,
Dalian, China) and analyzed using a 7900HT Fast Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Relative expression levels were normalized using GAPDH and
calculated using the 2−∆∆Cq method (23). A negative control without reverse
transcriptase enzyme treatment was used. All experiments were
repeated 3–4 times. Primer sequences are summarized as follows:
p21-sense, 5′-CCATGTGGACCTGTCACTGT-3′ and antisense,
5′-CGGCGTTTGGAGTGGTAGAA-3′; p27-sense, 5′-TAAGGAAGCGACCTGCAACC-3′
and antisense, 5′-TCTGAGGCCAGGCTTCTTG-3′; p16INK4A sense,
5′-CCGAATAGTTACGGTCGGAG-3′ and antisense, 5′-CGGGTCGGGTGAGAGTGG-3′;
p14ARF sense, 5′-CTGTGGCCCTCGTGCTG-3′ and antisense,
5′-CAGCAGCTCCGCCACTC-3′; Rnf2 sense, 5′-TTCAGGCCTCATCCCACACT-3′ and
antisense, 5′-CAGCAGCTCCGCCACTC-3′; GAPDH sense,
5′-TCGACAGTCAGCCGCATCTT-3′ and antisense,
5′-GAGTTAAAAGCAGCCCTGGTG-3′.
Western blotting assay
SGC-7901 and GES-1 cells were washed with 1X PBS and
pelleted at 600 × g for 5 min at 4°C and then lysed in 2% SDS lysis
buffer (2% SDS, 240 mM Tris pH 6.8, and 10% glycerol). Proteins
extracted from cells in log-phase growth were separated by 10%
SDS-PAGE and transferred to polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA) using a semidry transfer system
(Bio-Rad Laboratories, Inc., Hercules, MA, USA). Primary
antibodiesdirected against β-Tubulin (#AT809; 1:3,000; Beyotime
Institute of Biotechnology, Haimen, China), Rnf2 (#5694) p21
(#2946), p27 (#2552) (all 1:3,000; all Cell Signaling Technologies,
Inc., Danvers, MA, USA) were incubated in TBS (pH 7.4) with 0.1%
Tween 20 (TBST) and 5% nonfat milk (Bio-Rad Laboratories, Inc.)
with gentle agitation overnight at 4°C according to the
manufacturer's protocol. Subsequent to washing 3 times with TBST,
the membranes were incubated with horseradish peroxidase-conjugated
secondary antibodies (#SC-2357 and #SC-516102; 1:10,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) for 1 h at room temperature.
The membranes were then washed again in TBST and visualized using
an enhanced chemiluminescence kit (EMD Millipore). β-tubulinwas
used as an internal reference. The experiments were repeated 3
times.
Cell counting kit 8 (CCK-8) assay
Cell proliferation was quantified with a CCK-8 assay
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Cells were
seeded in 96-well culture plates at a density of 1×104
per well in 100 µl fresh RPMI 1640 supplemented with 10% fetal
bovine serum (both Invitrogen; Thermo Fisher Scientific, Inc.).
Subsequent to a 48 h culture at 37°C, 10 µl of CCK-8 were added in
each well and incubated at 37°C for 4 h. Absorbance was measured at
450 nm using a microplate reader (Bio-Rad Laboratories, Inc.). The
measurement for each sample was conducted in triplicate to certify
the accuracy.
Flow cytometric analysis of cell
cycle
Cells were seeded in 6-well plates at a density of
5×105 cells/well and then transfected with Rnf2 small
hairpin RNA (shRNA; sequence, CGAAGTCTACACAGTGAATTA) using
Lipofectamine 3000 following the manufacturer's protocol
(#L3000015; Thermo Fisher Scientific, Inc.) and cultured in RPMI
1640 supplemented with 10% fetal bovine serum at 37°C for 48 h. The
cells were harvested with trypsin, fixed with 70% ice-cold ethanol,
and stained with propidium oxide using a Cell Cycle Analysis kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's protocol. Cell cycle distributions were then
analyzed by FACScan (BD Biosciences, Franklin Lakes, NJ, USA) and
the percentage of cells in G1, S, and G2/M phase was calculated and
compared. Data represent the mean value derived from triplicate
experiments.
Statistical analyses
All statistical analyses were carried out using
SPSSversion 17.0 (SPSS, Inc., Chicago, IL, USA). All of the data
are presented as the mean ± standard deviation for at least 3
independent experiments. The significant differences between any of
two groups were evaluated by one-way analysis of variance.
P<0.05 was considered to indicate a statistically significant
difference.
Results
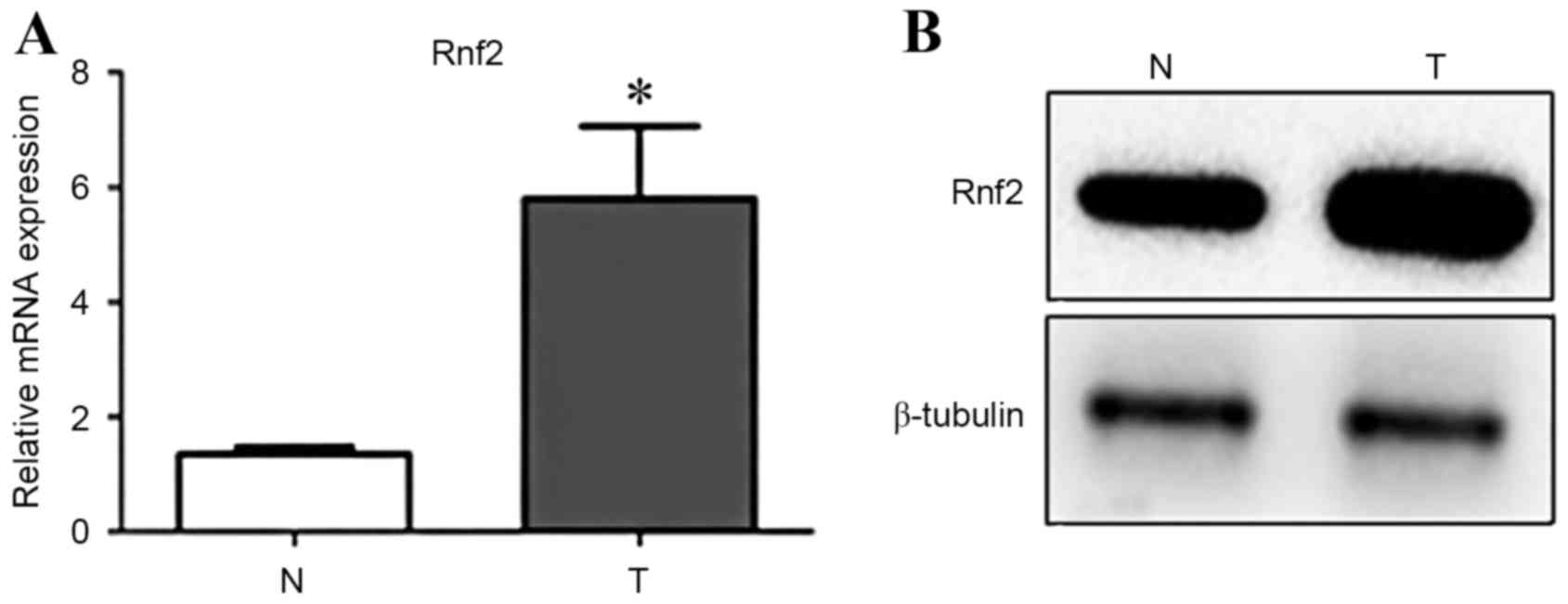
Rnf2 is overexpressed in GC cells
The present study first examined Rnf2 expression in
GC cells and normal gastric cells by RT-qPCR and western blot
analysis. Results demonstrated that mRNA and protein levels of Rnf2
were significantly higher in GC cells compared with the normal
gastric cells (P<0.05) (Fig. 1A and
B).
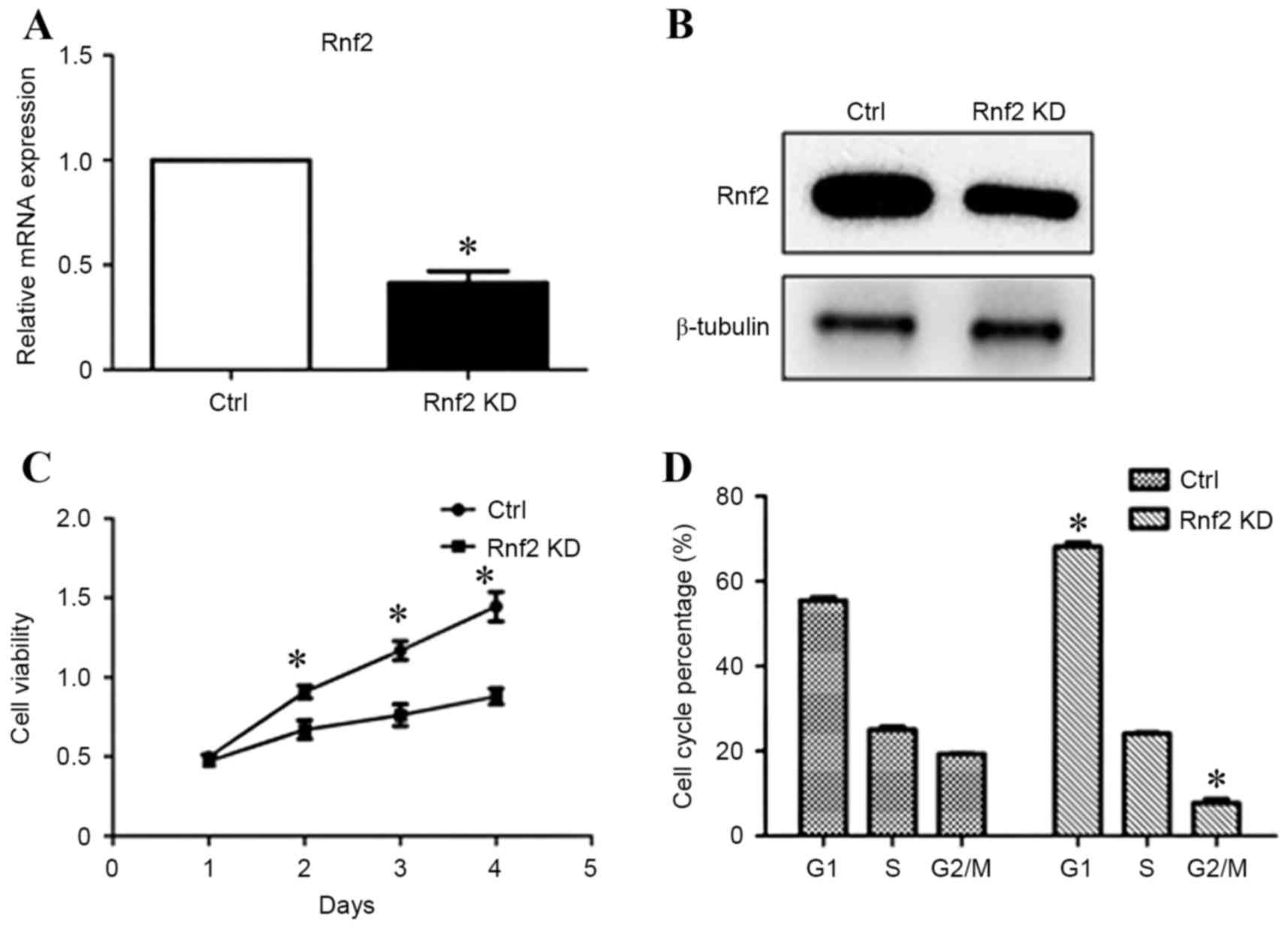
Knockdown of Rnf2 inhibits viability
and induces G1 cell cycle arrest in GC cells
To study the role of Rnf2 in the progression of GC
cells, the present study then investigated the knockdown of Rnf2 in
SGC-7901 and whether Rnf2 influenced cell survival. RT-qPCR and
western blot analysis demonstrated efficient knockdown of Rnf2
following transfection for 48 h (Fig. 2A
and B). Accordingly, the viability of GC cells was suppressed
upon silencing of Rnf2 (P<0.05; Fig.
2C). Furthermore, knockdown of Rnf2 resulted in cell cycle
arrest illustrated by increased percentage of cells in G1 phase and
decreased percentage of cells in G2/M phase (P<0.05; Fig. 2D).
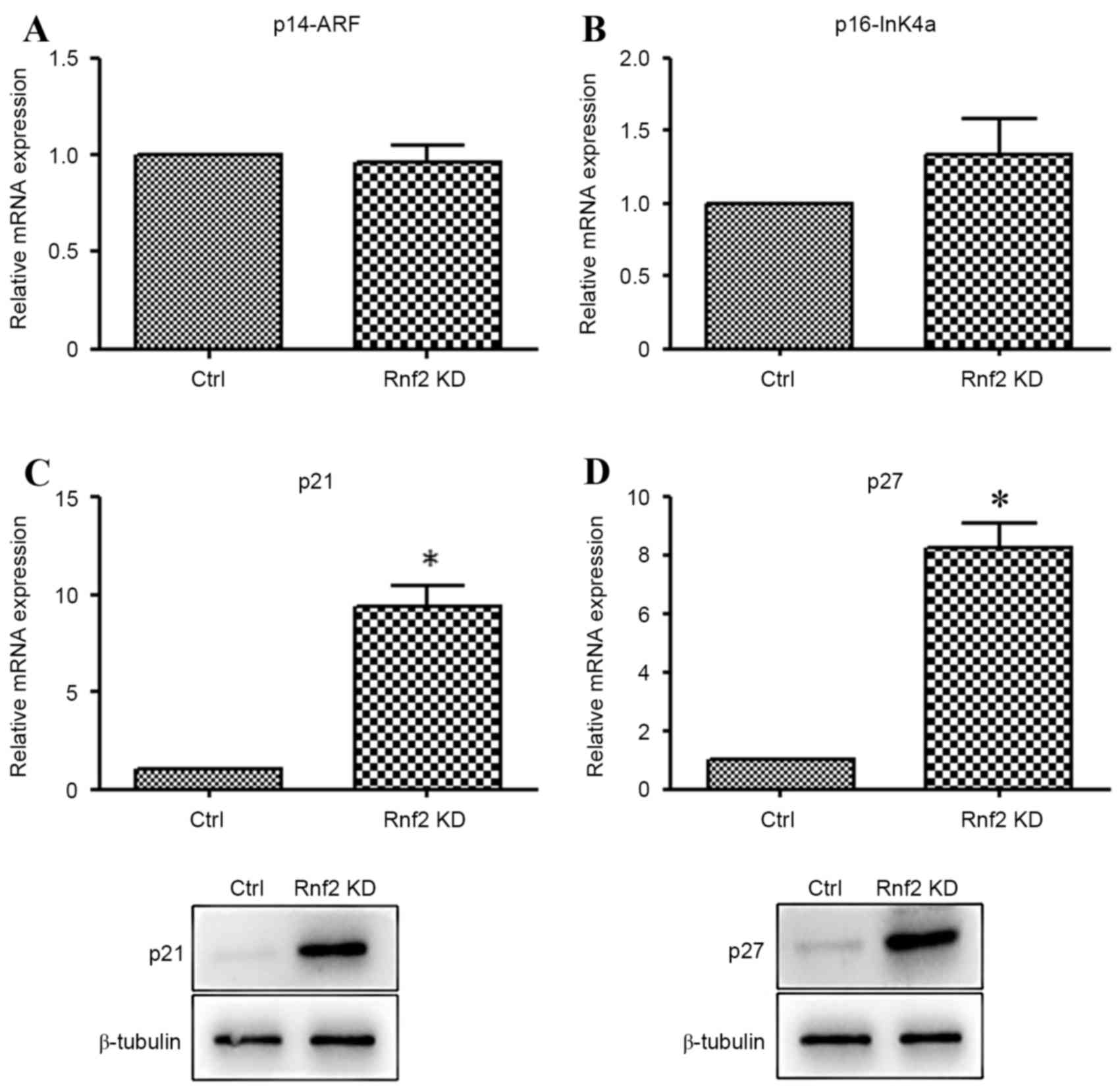
Knockdown of Rnf2 increased the
expression of p21 and p27
In order to further explore the probable mechanism
of Rnf2 in the GC cells, the expression of p14, p16, p21 and p27
was detected by RT-qPCR and western blot analysis. As is
demonstrated in Fig. 3A and B, there
were no significant differences between the expression of p14 and
p16. By contrast, the mRNA and protein levels of p21 and p27 were
markedly increased in the Rnf2 silenced GC cells in comparison with
the control cells (P<0.05) (Fig. 3C
and D).
Discussion
The present study revealed that expression levels of
Rnf2 are significantly increased in GC cells compared with their
counterpart. Following the knockdown of Rnf2, GC cell viability was
inhibited. Additionally, downregulation of Rnf2 blocked cell cycle
progression by arresting cells in the G1 phase, which illustrates a
unique activity of Rnf2 in the proliferation of GC. To further
investigate this mechanism, the present study examined the mRNA
expression level of several cell cycle related genes. The results
demonstrated that Rnf2 knockdown could notably induce the
upregulation of p21 and p27, but the expression of p14 and p16 did
not show the same tendency. Thus, the present study was a
preliminary investigation into the function of Rnf2 in GC.
A major cause of cancer is through the dysregulation
of proper transcriptional control, a process that is directly
regulated by transcription factors, co-activators and co-repressors
(24). Accumulating evidence
indicates that the PC family of epigenetic proteins comprises
transcriptional repressors that are often misregulated in various
types of human cancer, and the expression level is associated with
cancer progression (25), indicating
that PcG proteins may exhibit oncogenic functions. Studies have
identified that EZH2, which functions as a histone H3
methyltransferase, is involved in the progression of prostate
cancer and in neoplastic transformation of breast epithelial cells
(26,27). Human BMI1 has been identified to be
overexpressed in colorectal carcinoma (28), Hodgkin's lymphoma and diffuse large
B-cell lymphoma (29). As for Rnf2,
Sánchez-Beato et al (10)
revealed that Rnf2 expression is higher in gastric and colonic
tumors compared with the stomach or colon surface-epithelial cells.
GC-derived, Burkitt's and Hodgkin's lymphoma also exhibited a
higher level of Rnf2 expression (10). Results from the present study
demonstrate that the expression of Rnf2 is higher in the GC cells
compared with the normal gastric epithelial cells, which is in
accordance with a previous study (10).
Rnf2, a core number of the PRC1 complex, is
upregulated in several types of tumor and performs a critical role
in tumor proliferation, immortalization and metastasis (30). A recent study has shown that the
knockdown of Rnf2 significantly inhibits cell proliferation and
colony formation in the colon cancer HCT116 cell line (31). In addition, it was also associated
with induction of apoptosis by regulating MDM2 and p53 stability
(31). Similarly, Su et al
(9) reported that Rnf2 is essential
for the proliferation of ovarian and testicular cancer cells by
functioning as an E3 ligase for p53 degradation. However, to the
best of our knowledge the role of Rnf2 in the viability of GC has
not been previously investigated. The current study revealed that
downregulation of Rnf2 dramatically reduced the viability of GC
cells. Additionally, downregulation of Rnf2 in GC cells induced
increased G1 phase followed by a substantial reduction of G2/M
(Fig. 2D), suggesting that Rnf2 is of
vital important in regulating the growth of GC.
Considering the evidence that cellular proliferation
is activated by cyclin-dependent kinases and inhibited in response
to various stresses by corresponding inhibitors, including INK
family and KIP family (32), the
present study supposed that several tumor suppressors maybe
involved in this process. The proteins p14 and p16 are alternative
transcript variants of the INK4A-ARF (CDKN2A in humans) gene
located on chromosome 9p21 and function as inhibitors of cell cycle
progression, which possess complementary function as regulators of
two major cell cycle control pathways, p53 and pRB, respectively
(33,34). The p16/pRB and p14/p53 cell cycle
pathways are activated in the majority of human cancers, which
suggests the importance of these pathways in repressing the process
of carcinogenesis (35). p21 is a
common element of the two pathways and may serve as a bridge
between them (36,37).
Previous studies demonstrated that p21 was regulated
by p53-dependent and independent pathways (38). Rnf2 knockdown cells exhibited
increased p21 expression and G1 phase in colon cancer HCT116 cell
lines in an p53-dependent pathway, followed by marked accumulation
of these cells at sub-G1 (39).
Similar results have also been reported in hepatic cancer HepG2
cells (40). Additionally, p21 can
act as a master effector of multiple tumor suppressor pathways on
promoting anti-proliferative activities that are independent of the
classical p53 tumor suppressor pathway. Subsequent tothe depletion
of Ezh2 (a main component of PRC2), marked inhibition of cellular
proliferation, G1 cell cycle arrest and accompanying upregulation
of key cell-cycle regulators p16, p21 and p27 has been identified
in lymphoma and tongue cancer (41,42).
However, the role of these proteins in regulating the GC cell
proliferation and cell cycle has not previously been investigated.
The present study demonstrated for the first time that p21 and p27
were elevated following the knockdown of Rnf2, while similar
results were not shown in the mRNA level of p14 and p16. The reason
of this discrepancy remains unclear and requires further
exploration.
In conclusion, the present results established that
Rnf2 is overexpressed in GC cells and contributes to cell
viability. Furthermore, knockdown of Rnf2 induced G1 phase cell
cycle arrest and upregulation of p21 and p27. Therefore, the
present findings may provide novel insights in the molecular
mechanisms behind GC and provide new information that may be used
in the prognosis and therapy of GC.
Acknowledgements
The study was supported by Shandong Provincial
Education Department Program (grant no. J14LK02), the Natural
Science Foundation of China (grant nos. 81672662, 81170688 and
81470973) and the Shandong Province Natural Science Foundation
(grant no. ZR2014CM040).
Glossary
Abbreviations
Abbreviations:
|
Rnf2
|
ring finger protein 2
|
|
H3K27me3
|
trimethylation of histone H3 at
Lys27
|
|
PC
|
polycomb
|
|
PcG
|
polycomb group
|
|
PH
|
polyhomeotic
|
|
PRC1
|
polycomb repressive complex 1
|
References
|
1
|
Simon JA and Kingston RE: Mechanisms of
polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell
Biol. 10:697–708. 2009.PubMed/NCBI
|
|
2
|
Sparmann A and van Lohuizen M: Polycomb
silencers control cell fate, development and cancer. Nat Rev
Cancer. 6:846–856. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bracken AP and Helin K: Polycomb group
proteins: Navigators of lineage pathways led astray in cancer. Nat
Rev Cancer. 9:773–784. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Russell IF: Auditory perception under
anaesthesia. Anaesthesia. 34:2111979. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanders LL: Gold therapy for rheumatoid
arthritis. J Ark Med Soc. 73:135–137. 1976.PubMed/NCBI
|
|
7
|
Wang H, Wang L, Erdjument-Bromage H, Vidal
M, Tempst P, Jones RS and Zhang Y: Role of histone H2A
ubiquitination in Polycomb silencing. Nature. 431:873–878. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vidal M: Role of polycomb proteins Ring1A
and Ring1B in the epigenetic regulation of gene expression. Int J
Dev Biol. 53:355–370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Su WJ, Fang JS, Cheng F, Liu C, Zhou F and
Zhang J: RNF2/Ring1b negatively regulates p53 expression in
selective cancer cell types to promote tumor development. Proc Natl
Acad Sci USA. 110:1720–1725. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sánchez-Beato M, Sánchez E,
González-Carreró J, Morente M, Díez A, Sánchez-Verde L, Martín MC,
Cigudosa JC, Vidal M and Piris MA: Variability in the expression of
polycomb proteins in different normal and tumoral tissues. A pilot
study using tissue microarrays. Mod Pathol. 19:684–694. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei M, Jiao D, Han D, Wu J, Wei F, Zheng
G, Guo Z, Xi W, Yang F, Xie P, et al: Knockdown of RNF2 induces
cell cycle arrest and apoptosis in prostate cancer cells through
the upregulation of TXNIP. Oncotarget. 8:5323–5338. 2017.PubMed/NCBI
|
|
12
|
Voncken JW, Roelen BA, Roefs M, de Vries
S, Verhoeven E, Marino S, Deschamps J and van Lohuizen M: Rnf2
(Ring1b) deficiency causes gastrulation arrest and cell cycle
inhibition. Proc Natl Acad Sci USA. 100:2468–2473. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yagi A, Machii K, Nishimura H, Shida T and
Nishioka I: Effect of aloe lectin on deoxyribonucleic acid
synthesis in baby hamster kidney cells. Experientia. 41:669–671.
1985. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Morgan DO: Principles of CDK regulation.
Nature. 374:131–134. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sherr CJ: G1 phase progression: Cycling on
cue. Cell. 79:551–555. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reed SI, Bailly E, Dulic V, Hengst L,
Resnitzky D and Slingerland J: G1 control in mammalian cells. J
Cell Sci Suppl. 18:69–73. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abbas T and Dutta A: p21 in cancer:
Intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Niculescu AB III, Chen X, Smeets M, Hengst
L, Prives C and Reed SI: Effects of p21(Cip1/Waf1) at both the G1/S
and the G2/M cell cycle transitions: pRb is a critical determinant
in blocking DNA replication and in preventing endoreduplication.
Mol Cell Biol. 18:629–643. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dulić V, Kaufmann WK, Wilson SJ, Tlsty TD,
Lees E, Harper JW, Elledge SJ and Reed SI: p53-dependent inhibition
of cyclin-dependent kinase activities in human fibroblasts during
radiation-induced G1 arrest. Cell. 76:1013–1023. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maertens GN, El Messaoudi-Aubert S, Racek
T, Stock JK, Nicholls J, Rodriguez-Niedenführ M, Gil J and Peters
G: Several distinct polycomb complexes regulate and co-localize on
the INK4a tumor suppressor locus. PLoS One. 4:e63802009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jacobs JJ, Kieboom K, Marino S, DePinho RA
and van Lohuizen M: The oncogene and Polycomb-group gene bmi-1
regulates cell proliferation and senescence through the ink4a
locus. Nature. 397:164–168. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan T, Jiang S, Chung N, Alikhan A, Ni C,
Lee CC and Hornyak TJ: EZH2-dependent suppression of a cellular
senescence phenotype in melanoma cells by inhibition of p21/CDKN1A
expression. Mol Cancer Res. 9:418–429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamamoto Y, Abe A and Emi N: Clarifying
the impact of polycomb complex component disruption in human
cancers. Mol Cancer Res. 12:479–484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang W, Qin JJ, Voruganti S, Nag S, Zhou J
and Zhang R: Polycomb group (PcG) proteins and human cancers:
Multifaceted functions and therapeutic implications. Med Res Rev.
35:1220–1267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette sTR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, et al: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:624–629. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ,
Li SX, Wang XL and Peng ZH: Expression level of Bmi-1 oncoprotein
is associated with progression and prognosis in colon cancer. J
Cancer Res Clin Oncol. 136:997–1006. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Garcia JF, Camacho FI, Morente M, Fraga M,
Montalbán C, Alvaro T, Bellas C, Castaño A, Díez A, Flores T, et
al: Hodgkin and reed-sternberg cells harbor alterations in the
major tumor suppressor pathways and cell-cycle checkpoints:
Analyses using tissue microarrays. Blood. 101:681–689. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bosch A, Panoutsopoulou K, Corominas JM,
Gimeno R, Moreno-Bueno G, Martín-Caballero J, Morales S, Lobato T,
Martínez-Romero C, Farias EF, et al: The polycomb group protein
RING1B is overexpressed in ductal breast carcinoma and is required
to sustain FAK steady state levels in breast cancer epithelial
cells. Oncotarget. 5:2065–2076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wen W, Peng C, Kim MO, Jeong Ho C, Zhu F,
Yao K, Zykova T, Ma W, Carper A, Langfald A, et al: Knockdown of
RNF2 induces apoptosis by regulating MDM2 and p53 stability.
Oncogene. 33:421–428. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ortega S, Malumbres M and Barbacid M:
Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim
Biophys Acta. 1602:73–87. 2002.PubMed/NCBI
|
|
33
|
Pérez-Sayáns M, Suárez-Peñaranda JM,
Gayoso-Diz P, Barros-Angueira F, Gándara-Rey JM and Garcia-Garcia
A: p16 (INK4a)/CDKN2 expression and its relationship with oral
squamous cell carcinoma is our current knowledge enough? Cancer
Lett. 306:134–141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Llanos S, Clark PA, Rowe J and Peters G:
Stabilization of p53 by p14ARF without relocation of MDM2 to the
nucleolus. Nat Cell Biol. 3:445–452. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dominguez G, Silva J, Garcia JM, Silva JM,
Rodriguez R, Muñoz C, Chacón I, Sanchez R, Carballido J, Colás A,
et al: Prevalence of aberrant methylation of p14ARF over p16INK4a
in some human primary tumors. Mutat Res. 530:9–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng M, Olivier P, Diehl JA, Fero M,
Roussel MF, Roberts JM and Sherr CJ: The p21(Cip1) and p27(Kip1)
CDK ‘inhibitors’ are essential activators of cyclin D-dependent
kinases in murine fibroblasts. EMBO J. 18:1571–1583. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Carnero A, Hudson JD, Price CM and Beach
DH: p16INK4A and p19ARF act in overlapping pathways in cellular
immortalization. Nat Cell Biol. 2:148–155. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kerkhoff E and Rapp UR: Cell cycle targets
of Ras/Raf signalling. Oncogene. 17:(11 Reviews). 1457–1462. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Villabona C, Novials A, Soler J, Morató J,
Gómez JM and Navarro MA: Hormonal evaluation of 27 patients with
acromegaly. Med Clin (Barc). 84:219–222. 1985.(In Spanish).
PubMed/NCBI
|
|
40
|
Sen N, Satija YK and Das S: PGC-1α, a key
modulator of p53, promotes cell survival upon metabolic stress. Mol
Cell. 44:621–634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fiskus W, Wang Y, Sreekumar A, Buckley KM,
Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, et al:
Combined epigenetic therapy with the histone methyltransferase EZH2
inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor
panobinostat against human AML cells. Blood. 114:2733–2743. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Z, Wang Y, Qiu J, Li Q, Yuan C, Zhang
W, Wang D, Ye J, Jiang H, Yang J and Cheng J: The polycomb group
protein EZH2 is a novel therapeutic target in tongue cancer.
Oncotarget. 4:2532–2549. 2013. View Article : Google Scholar : PubMed/NCBI
|