Introduction
Lung cancer is one of the leading causes of
cancer-associated death, and non-small cell lung cancer (NSCLC)
accounts for ~80–85% of all lung cancer cases (1,2). Gefitinib
is an epidermal growth factor receptor tyrosine kinase inhibitor
(EGFR-TKI) approved for first-line treatment of locally advanced or
metastatic NSCLC with activating mutations of epidermal growth
factor receptor (EGFR) tyrosine kinase (3). NSCLC initially exhibits an excellent
response to gefitinib treatment (4).
However, acquired resistance has been observed in many NSCLC
patients after 6–12 months of treatment (5). Investigating the mechanism of resistance
to EGFR-TKIs and identifying strategies capable of overcoming this
resistance is thus an important clinical goal.
EGFR is overexpressed in NSCLC by 40–89% (6) and is one of the most important signaling
components involved in cell growth and survival. As well as
activating the phosphatidylinositol-3 kinase/protein kinase B
(PI3K/AKT) signaling pathway and promoting tumor cell
proliferation, EGFR overexpression may also decrease cell apoptosis
by activating anti-apoptotic factors such as B-cell lymphoma 2-like
protein 4 (Bax), Bcl-2-associated death promoter (Bad), and
caspase-9, as well as inactivating pro-apoptotic transcription
factors such as p53 (7).
The human transcriptome comprises large numbers of
protein-coding messenger RNAs (mRNAs), and a large set of
nonprotein coding transcripts, including long noncoding RNA
(lncRNA) (8). lncRNAs are >200
nucleotides long, and their dysregulation appears to contribute to
the growth and progression of human tumors (9,10). Some
studies have reported that dysregulated lncRNAs are related to lung
cancer with lymph node metastasis, advanced stage lung cancer,
metastasis development and poor patient prognosis (11–16).
However, the mechanism by which changes in lncRNA levels affect the
expression of gene products that may contribute to gefitinib
resistance remains largely unknown.
The aim of the present study was to investigate gene
expression profiling in the PC9 (formerly known as PC14) human
non-small cell lung adenocarcinoma cell line. Microarray expression
profiling of lncRNAs was undertaken in both PC9 cells and PC9 cells
resistant to gefitinib (PC9-R). Expression levels of different
lncRNAs in the two cell lines were examined, with the aim of
revealing the mechanism by which PC9-R cells acquire resistance to
gefitinib.
Materials and methods
Cell culture
The PC9 human non-small cell lung adenocarcinoma
cell lines (formerly known as PC14) were obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China) and PC9
cells resistant to gefitinib (PC9-R) were routinely cultured in
RPMI-1640 medium (Gibco-BRL; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin, 100
mg/ml streptomycin, and 1% L-glutamine, and were maintained in a 5%
CO2 incubator at 37°C with saturated humidity.
Subcultures were produced by trypsinization and were reseeded for
experiments.
Cell proliferation assay
Cells were plated in 96-well flat-bottomed culture
plates at a density of 5×103 cells/well. Various
concentrations (0.01, 0.1, 1, 5, 10, 20, 40 and 80 µM) of gefitinib
were then added to the plates. Following this, the media were
replaced with RPMI-1640 and 1% FBS (HyClone; GE Healthcare Life
Sciences). The proliferative activity of cells after treatment with
gefitinib was assessed using the Cell Counting Kit-8 (CCK-8;
Dojindo Molecular Technologies, Inc., Kumamoto, Japan). Subsequent
to incubation for 48 h in a 5% CO2 humidified atmosphere
at 37°C, 10 µl CCK-8 reagent was added to each well, and the plates
were incubated for a further 4 h. Absorbance at 450 nm was
determined spectrophotometrically using a microplate reader. Data
were analyzed by the median-effect method to evaluate the drug
concentrations that resulted in 50% growth inhibition
(IC50). The combination effect was evaluated by the
CCK-8 assay. Confidence interval values of <1, 1 and >1
indicated synergism, additive effect and antagonism, respectively.
Each treatment was assayed in triplicate during the same
experiment.
Cell apoptosis assay
Apoptosis was detected using an Annexin V-FITC/PI
double staining kit (BD Biosciences, Franklin Lakes, NJ, USA)
according to the manufacturer's protocol. Cells were seeded at a
concentration of 5×103 cells/100 µl/well in 96-well
culture plates, then 0.1 µM gefitinib was added 48 h prior to
detection. Cells were harvested and washed twice with cold PBS by
gentle shaking. Cells were then re-suspended and added to Binding
buffer (1X), and cell density was adjusted to
2–5×105/ml. In the dark, 5 µl Annexin V-FITC was added
to the cell suspension volume of 195 µl and incubated for 10 min at
room temperature prior to the addition of 190 µl binding buffer
(1X) and 10 µl propidium iodide (PI). A total of 10,000 events per
sample were acquired using a FACS-scan flow cytometer (BD
Biosciences, San Jose, CA, USA), and the percentage of cells
undergoing apoptosis was analyzed using BD CellQuest™
Pro Software Analysis Tutorial (Version 5.1; BD Biosciences).
Cell cycle analysis
PC9-R cells seeded in 6-well plates
(3×105/well) were treated either with transfection of
MIR31HG siRNA or a siRNA scramble control in an RPMI-1640 medium
with 10% fetal bovine serum for cell cycle analyses, and 0.1 µM
gefitinib was added to the plates for 48 h. The cells were then
harvested and fixed in cold 70% ethanol. The samples were incubated
with RNase A (60 µg/ml) and PI (25 µg/ml) for 20 min in the dark at
room temperature. Samples were measured on a FACSCalibur flow
cytometer (BD Biosciences), and cell cycle stages were analyzed
using ModFit Software (version 3.2; Verity Software House Inc.,
Topsham, ME, USA).
lncRNA microarray
PC9 and PC9-R cells were used to synthesize
double-stranded cDNA. Double-stranded cDNA was labeled and
hybridized to the 8660 K LncRNA Expression Microarray (Array Star
Inc., Rockville, MD, USA). After hybridization and washing,
processed slides were scanned with the Agilent DNA Microarray
Scanner (G2505B; Agilent Technologies, Inc., Santa Clara, CA, USA).
Agilent Feature Extraction software ver. 10.7.3.1 (Agilent
Technologies, Inc.) was used to analyze acquired array images.
Quantile normalization and subsequent data processing were
performed using the Gene Spring GX software package version 11.5.1
(Agilent Technologies, Inc.). Differentially expressed genes were
identified through the random variance model. A P-value was
calculated using the paired t-test. The threshold set
for up and downregulated genes was a fold-change ≥2.0 and a P-value
≤0.05. Each cell line performed lncRNA microarray in
triplicate.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The differential genes were verified by using
RT-qPCR. The specific operation steps are as follows: Total
cellular RNA was isolated from the cultured PC9 and PC9-R cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the protocol of the manufacturer. Total RNA was used
to synthesize cDNA with PrimeScript™ RT reagent kit
(cat. no. RR037A; Takara Bio, Inc., Otsu, Japan), and subjected to
qPCR. The relative expression levels of lncRNA were measured using
a SYBR® Premix Ex Taq™ (cat. no. RR420A;
Takara Bio, Inc.) following the protocol of the manufacturer with
GADPH as an internal control. RT-qPCR primers were synthesized by
Sangon Biotech Co., (Shanghai, China). The 5′-3′ primer sequences
used for RT-qPCR were as follows: PVT1 forward, GGACGGACTTGAGAACTGT
and reverse, GGCTTGTGAATCTGGGAG; H19 forward, GCACTAAGTCGATTGCACTGG
and reverse, GCCTCAAGCAGACGGCCACA; MIR31HG forward,
TCCCAGTTTCAGACCACC and reverse, CCAGGCTATGTCTTTCCTCTAT; CBR3-AS1
forward, ACAGCACGCATTCACCAG and reverse, TTGTAGCCGCCAAGTTTC;
lincRNA-p21 forward, GGGTGGCTCACTCTTCTGGC and reverse,
TGGCCTTGCCCGGGCTTGTC; GAPDH forward, GACTCATGACCACAGTCCATGC and
reverse, AGAGGCAGGGATGATGTTCTG. For RT-qPCR, the reaction was
performed on ABI 7500 Sequence Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The following cycles
were used: 95°C for 30 sec, 40 cycles of 95°C for 5 sec and 60°C
for 31 sec. The dissociation stage was 95°C for 15 sec, 60°C for 1
min and 95°C for 15 sec. The relative expression levels of target
genes were calculated as 2−(Cq of target genes)-(Cq of
GAPDH) (17) subsequent to
normalization with reference to the quantification of GAPDH. The
experiment was repeated three times.
Western blot analysis
PC9-R cells seeded in 6-well plates
(3×105/well) were treated either with transfection
MIR31HG siRNA or a si-scramble control for 48 h. For the RNA
interference-mediated knockdown of MIR31HG, three siRNAs for
depletion of MIR31HG were synthesized and generated by GenePharma,
Inc. (Sunnyvale, CA, USA). The sequence for MIR31HG siRNA was:
siRNA-1, 5′-GCAAAGAAGUCCGAGGC-3′; siRNA-2,
5′-UCAAAGGACACGCCAAGUG-3′; siRNA-3, 5′-GAGAAGAAAGAAGUCACC-3′. Cells
were harvested and lysed with immunoprecipitation buffer
supplemented with 1 mM sodium vanadate, 0.5 mM dithiothreitol, 1 mM
phenylmethylsulfonyl fluoride, 2 mM leupeptin, 2 mM aprotinin, and
2 mM pepstatin on ice for 30 min. The total cellular protein
content was determined using a Bio-Rad protein assay reagent
(Bio-Rad Laboratories Inc., Hercules, CA, USA). Aliquots of the
protein extracts were resolved by 10% SDS-PAGE and transferred to
nitrocellulose (cat. no. HATF00010; EMD Millipore, Billerica, MA,
USA) membranes. Membranes were blocked with 5% nonfat dry milk in
Tris-buffered saline and incubated with antibodies against p-EGFR
(cat. no. PL-0302648; dilution, 1/1,000; PLLABS, Nanaimo, BC,
Canada), total EGFR (cat. no. AB36836; dilution, 1:2,000; AbSci,
Vancouver, BC, Canada), p-PI3K (cat. no. BS4605; dilution, 1:2,000;
Bioworld Technology, Inc., St. Louis Park, MN, USA), total PI3K
(cat. no. NB100-75198; dilution, 1:1,500; Novus Biologicals, Inc.,
Abingdon, UK), p-AKT (cat. no. xyP001a; dilution, 1:3,000; Abcam,
Cambridge, UK), total AKT (cat. no. AB27174; dilution, 1:10,000;
AbSci), phosphorylated mouse minute 2 homolog (p-Mdm2; cat. no.
US1506136; dilution, 1:500; Merck & Co., Inc., Whitehouse
Station, NJ, USA), P53 (cat. no. 1026-1; dilution, 1:2,000:
Epitomics, Burlingame, CA, USA), GAPDH (cat. no. PA116777;
dilution, 1:2,000: Thermo Fisher Scientific, Inc.), Caspase-3 (cat.
no. 1087-1; dilution, 1:1,000; Epitomics), Caspase-9 (cat. no.
DB081; dilution, 1:5,000; Acris Antibodies GmbH; OriGene, Herford,
Germany), Bax (cat. no. PA112602; dilution, 1:1,000; Thermo Fisher
Scientific, Inc.) and Bcl-2 (cat. no. MA126233; dilution, 1:1,000;
Thermo Fisher Scientific, Inc.) at 4°C overnight. The membranes
were washed and subsequently probed with secondary antibody, goat
anti-mouse IgG conjugated to horseradish peroxidase (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) at a dilution of 1:4,000 for
1 h at room temperature. Proteins were visualized with a
chemiluminescence reagent (Pierce; Thermo Fisher Scientific, Inc.).
GAPDH was used as the internal control.
Statistical analysis
SPSS version 19.0 for Windows (IBM SPSS, Armonk, NY,
USA) was used for all analyses. Student's t-test was used to
compare the differences between groups. P-values were based on the
two-sided statistical analysis and P<0.05 was considered to
indicate a statistically significant difference.
Results
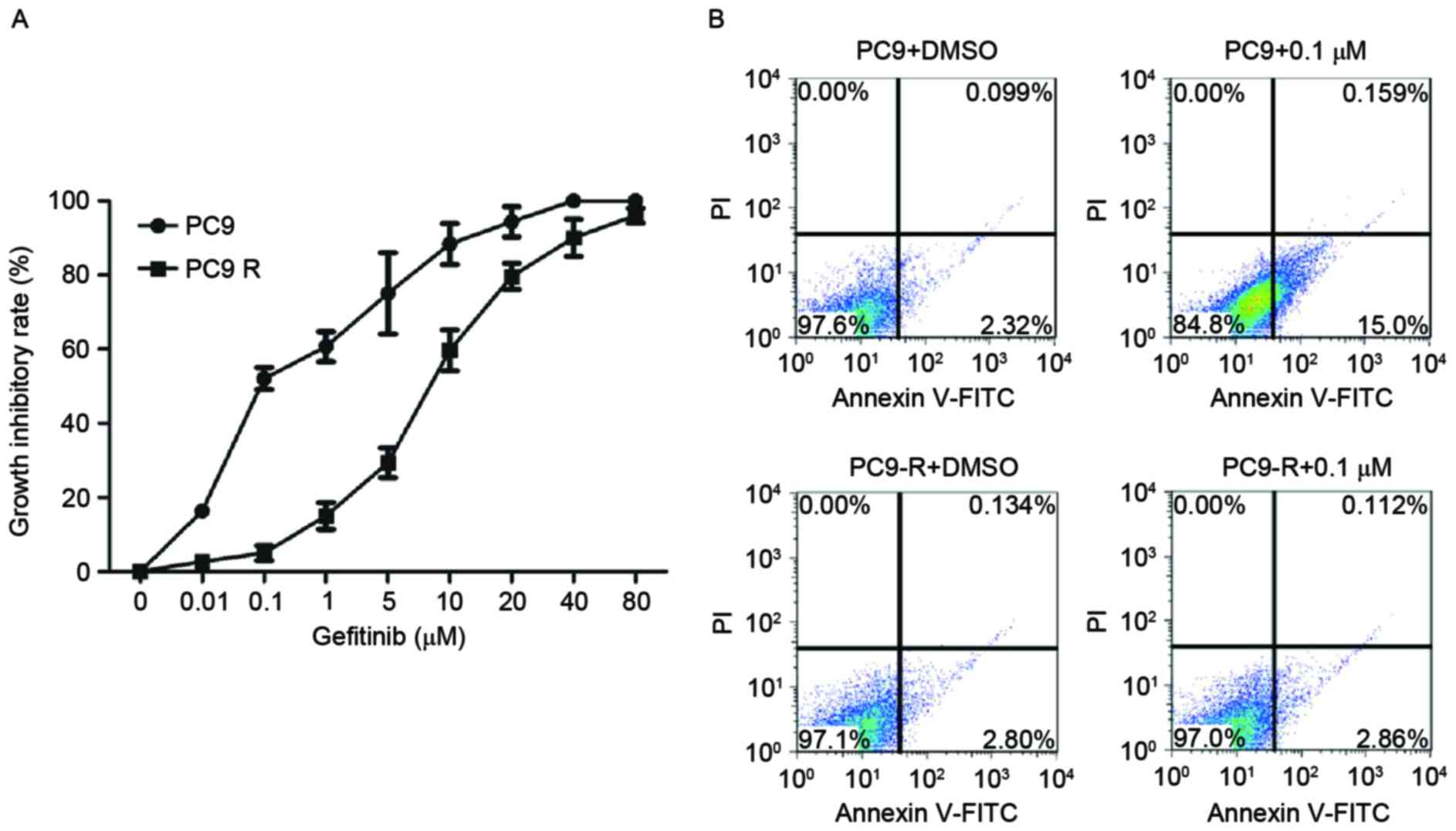
Sensitivity of PC9 and PC9-R cells to
gefitinib
The IC50 values of gefitinib in PC9 and
PC9-R cells treated with various concentration of the drug ranging
from 0.01–80 µM for 48 h were assessed. As expected, PC9-R cells
were resistant to gefitinib (IC50=8.1 µM), and PC9 cells
were sensitive to gefitinib (IC50=0.115 µM; Fig. 1A).
Rates of apoptosis of PC9 and PC9-R
cells following gefitinib treatment
The rates of apoptosis of PC9 and PC9-R cells
treated for 48 h in 0.1 µM gefitinib were analyzed by flow
cytometry. The rate of apoptosis in PC9 cells markedly increased
(P<0.001), but in PC9-R cells it did not (Fig. 1B).
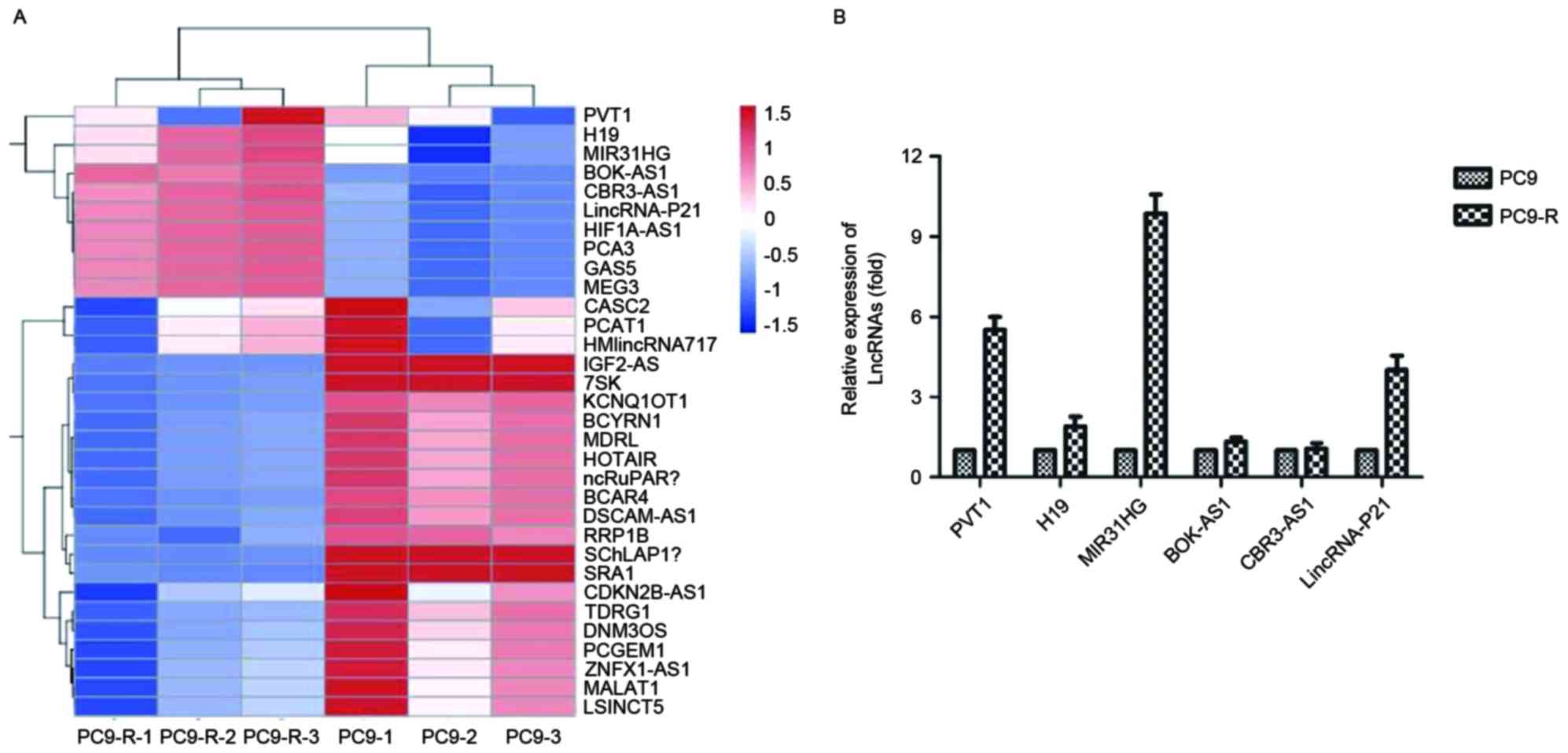
Microarray of PC9 and PC9-R cell
lines
A gene chip study was performed using the Array Star
probe dataset (Array Star, Inc.) to investigate possible lncRNA
expression changes that may cause gefitinib resistance.
Hierarchical clustering showed systematic variations in the
expression of lncRNAs between PC9 and PC9-R (Fig. 2A).
Validation of the microarray data
using RT-qPCR
To validate the microarray analysis findings, the
expression levels of lncRNAs in both PC9 and PC9-R cell lines were
analyzed using RT-qPCR. The expression of PVT1, H19, MIR31HG,
BOK-AS1, CBR3-AS1 and LincRNA-p21 differed, and PVT1 (P=0.0058),
MIR31HG (P<0.001) and LincRNA-p21 (P=0.0036) were significantly
decreased, especially expression of MIR31HG, which was ten times
higher in PC9-R than in PC9 (Fig.
2B).
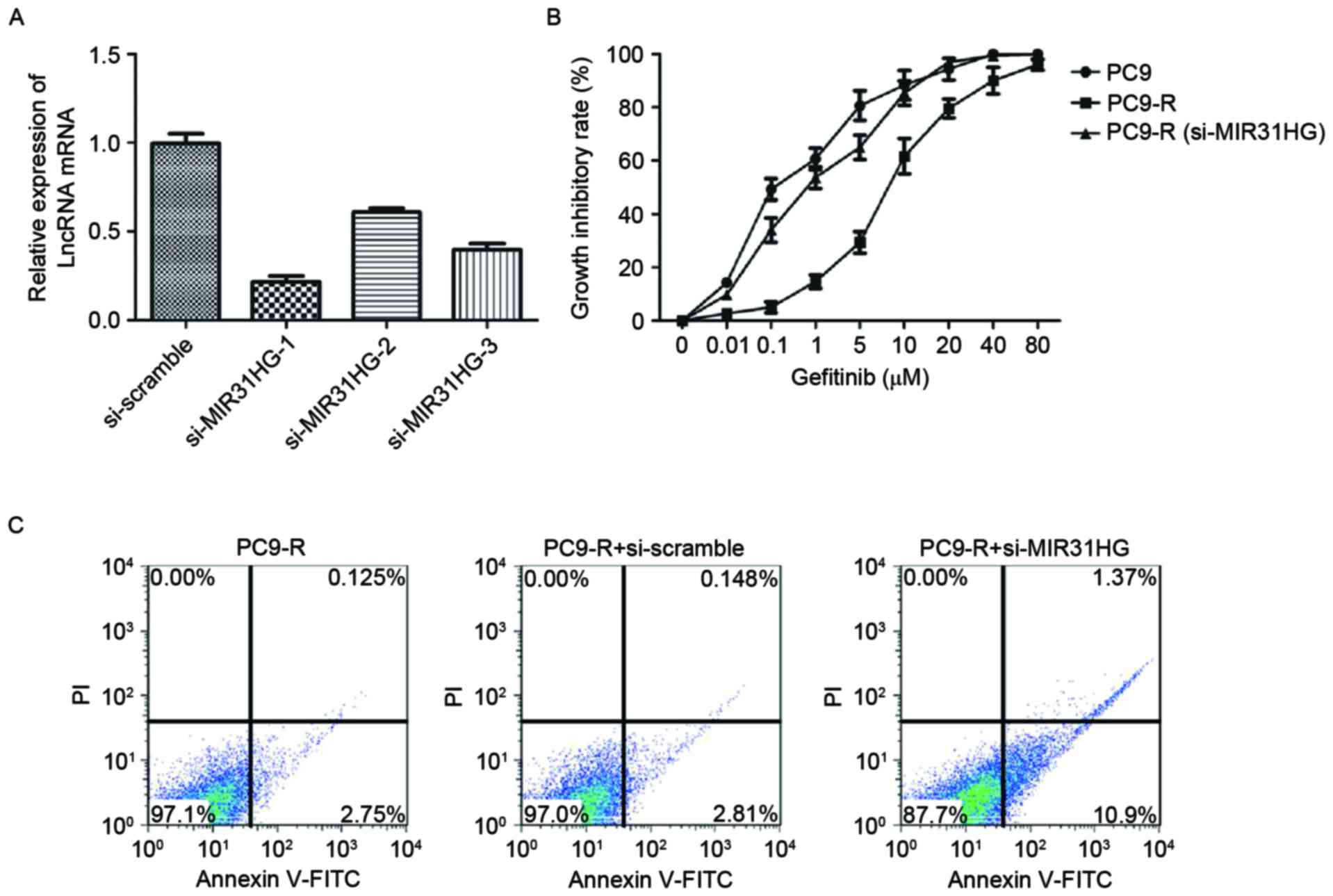
MIR31HG lncRNAs knockdown sensitizes
PC9-R cells to gefitinib
The effect of MIR31HG lncRNA knockdown on PC9-R
sensitivity to gefitinib was investigated. Knockdown was carried
out by designing and then transfecting siRNA targeting MIR31HG into
PC9-R cells. MIR31HG lncRNA expression in the transfected cells was
measured by RT-qPCR. As expected, MIR31HG lncRNA expression levels
were decreased (P<0.001) in the cells containing si-MIR31HG,
compared to those transfected with a si-scramble control (Fig. 3A). To determine the sensitivity of
si-MIR31HG of PC9-R cells to gefitinib, the CCK-8 cell viability
assay (Dojindo Molecular Technologies, Inc.) was used to measure
the effect of gefitinib on the cells transfected with si-MIR31HG,
and both PC9 and PC9-R cells. The results of the cell viability
assay demonstrated that knockdown of MIR31HG lncRNA in PC9-R cells
significantly increases their sensitivity to gefitinib
(IC50, 0.93 µM), compared with PC9-R cells
(IC50, 8.1 µM), P<0.001. Indeed, they exhibit
sensitivity to gefitinib comparable with PC9 cells
(IC50, 0.115 µM; Fig. 3B).
In addition, the rate of apoptosis in PC9-R cells transfected with
si-MIR31HG lncRNA was 12.3% and significantly increased, compared
with PC9-R cells and those transfected with a si-scramble control
(Fig. 3C), respectively were 2.9 and
3.0% (P<0.001).
MIR31HG lncRNAs knockdown sensitizes
PC9-R cells to gefitinib
To delineate the molecular mechanism involved in the
MIR31HG lncRNA knockdown sensitization of PC9-R cells to gefitinib,
western blotting was used to analyze the changes in protein levels
of the key components of the EGFR/PI3K/AKT signaling pathways in
PC9-R cells transfected with si-MIR31HG. The western blot analysis
indicated that knockdown of MIR31HG lncRNA significantly reduces
the expression of p-EGFR (P=0.0027), p-PI3K (P=0.0016), p-AKT
(P=0.0003) and p-Mdm-2 (P=0.0003) proteins in PC9-R cells, compared
to untreated and si-scramble cells, but does not alter total EGFR,
PI3K or AKT levels. It also activates expression of p53 (Fig. 4A). This indicates that transfection of
PC9-R with si-MIR31HG turns off the EGFR/PI3K/AKT signaling
pathway. Furthermore, the western blot analysis showed that PC9-R
cells transfected with si-MIR31HG increased the expression of
proteins involved in the cell mitochondrial apoptosis pathway.
Compared with expression levels in the control group, Caspase-3
(P=0.0031), Caspase-9 (P=0.0047) and Bax (P<0.001) were
significantly increased, however Bcl-2 (P<0.001) expression was
repressed (Fig. 4B), demonstrating
that transfection of PC9-R cells with si-MIR31HG induces cell
apoptosis by activating the mitochondrial apoptosis pathway.
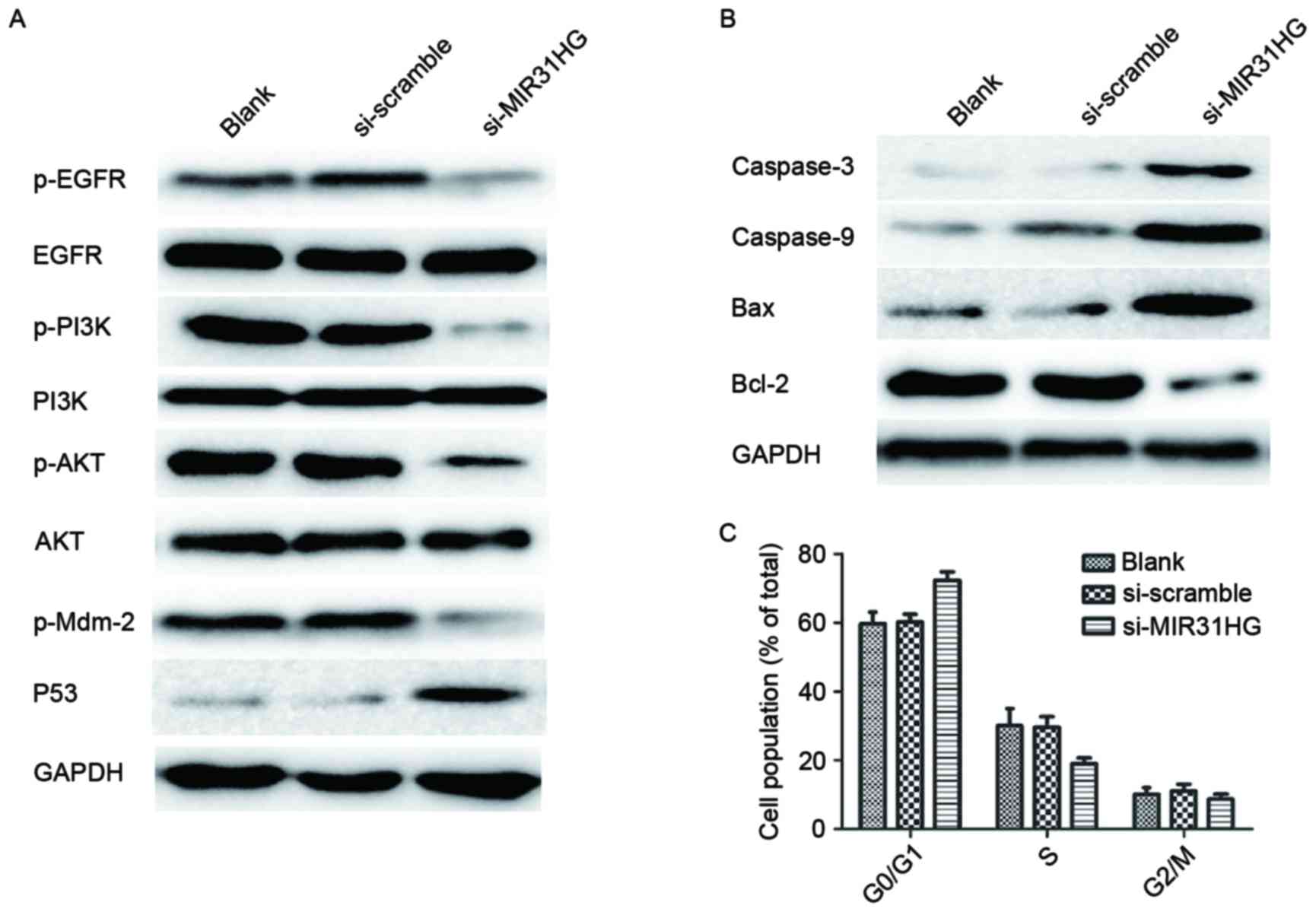 | Figure 4.Knockdown of MIR31HG alters protein
expression and cell cycle distribution in PC9-R cells. (A) Western
blot analysis revealed that PC9-R cells transfected with si-MIR31HG
repressed p-EGFR, p-PI3K, p-AKT and p-Mdm-2 expression, but did not
alter total EGFR, PI3K or AKT levels. It also stimulated expression
of p53. (B) The result showed that PC9-R cells containing
si-MIR31HG increased expression of the proteins Caspase-3,
Caspase-9 and Bax, but repressed Bcl-2, compared to levels in the
control group. (C) The effect of si-MIR31HG on cell cycle was
analyzed by flow cytometry. This showed that PC9-R cells
transfected with si-MIR31HG were able to arrest the cell cycle at
the G0/G1 phase. EGFR, epidermal growth factor receptor; PI3K,
phosphatidylinositol-3 kinase; AKT, protein kinase B; p-EGFR,
phosphorylated epidermal growth factor receptor; p-I3K,
phosphorylated phosphatidylinositol-3 kinase; p-AKT, phosphorylated
protein kinase B; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase. |
Cell cycle distribution of PC9-R cells
following knockdown by MIR31HG lncRNAs
The effect of si-MIR31HG on the cell cycle was
analyzed by a FACS Calibur flow cytometer (BD Biosciences), which
showed that a higher percentage of cells in the si-MIR31HG group
were in the G0/G1 phase than in the PC9-R cells group and those
transfected with si-scramble group, the P-values were 0.0189 and
0.016, respectively (Fig. 4C). The
si-MIR31HG group also contained a lower percentage of cells in the
S phase compared with other groups, P-values were 0.0264 and 0.0307
respectively (Fig. 4C). These results
indicate that PC9-R cells transfected with si-MIR31HG are able to
arrest the cell cycle in the G0/G1 phase.
Discussion
Gefitinib has proven to be highly effective at
treating advanced or metastatic NSCLC in patients harboring an
activating EGFR mutation, which competes with ATP for binding to
the tyrosine kinase pocket of the receptor (18). Compared with conventional chemotherapy
and radiotherapy, EGFR-TKI drugs have fewer side effects and
significantly longer progression free survival rates for patients
with activating EGFR mutations (19,20).
However, many patients treated with gefitinib later develop
resistance to the drug. The T790M mutation and MET amplification
are well-studied mechanisms for acquired gefitinib resistance
(21,22).
lncRNAs are long non-protein coding RNAs whose
dysregulation is related to prognosis and chemotherapy resistance
in human cancers (23–26). Various studies have suggested that the
dysregulation of lncRNA contributes to lung cancer with lymph node
metastasis, advanced stage, metastasis development and poor patient
prognosis (11–16,27,28). Cheng
et al (29) have analyzed
EGFR-TKI-sensitive and EGFR-TKI-resistant human lung cancer cells
by lncRNA microarray. Their results suggested that numerous lncRNAs
were differentially expressed in gefitinib-sensitive and
gefitinib-resistant PC9 cells. However, the exact mechanism by
which differentially expressed lncRNAs are correlated with EGFR-TKI
resistance remained unknown.
The present study identified differentially
expressed lncRNAs in PC9 and PC9-R cells by microarray and RT-qPCR.
The results indicated that levels of expression of PVT1, H19,
MIR31HG, BOK-AS1, CBR3-AS1 and LincRNA-P21 differed significantly
between the two cell lines, in particular the expression of
MIR31HG. Following this, the molecular mechanism involved in
EGFR-TKIs resistance in NSCLC was delineated using a CCK-8 cell
viability assay to determine the sensitivity of PC9-R cells
transfected with si-MIR31HG to gefitinib. Western blotting was
carried out to monitor the changes in protein levels of key
components of EGFR/PI3K/AKT signaling pathways involved.
A number of previous studies have demonstrated that
the activation of PI3K/AKT and MEK/ERK cell signaling pathways is
associated with EGFR TKI resistance in NSCLC (30,31). Kang
et al (32) have reported that
bufalin inhibits cell proliferation and induces cell apoptosis by
inhibiting the MET/PI3K/AKT pathway and activating death-signaling
pathways. PI3K/AKT is an important downstream signaling cascade of
EGFR, which is overexpressed in NSCLC (33). Dysregulation of PI3K/AKT signaling
pathways is related to reduced rates of apoptosis and the phenotype
of multidrug resistance (34).
In the present study, PC9-R cells transfected with
si-MIR31HG lncRNA exhibited an increased sensitivity to gefitinib
and a higher rate of apoptosis. The si-MIR31HG PC9-R cells also had
a reduced expression of p-EGFR, p-PI3K, p-AKT and p-Mdm-2 proteins,
and increased expression of p53. Total levels of EGFR, PI3K and AKT
remained the same. Mdm-2 has been identified as a protein that
represses p53 transcriptional activity and so its reduced
expression in si-MIR31HG PC9-R cells may increase p53 expression,
which initiates cell apoptosis and regulates the cell cycle
(35). Therefore, inhibition of
EGFR/PI3K/AKT pathway could decrease cell proliferation and promote
apoptosis by increasing levels of p53.
Mitochondrial integrity is central to both
caspase-dependent and -independent cell death. Regulation of the
mitochondrial pathway is under the control of the Bcl-2 family,
which includes pro-apoptotic proteins such as Bax, Bad, and Bak,
and anti-apoptotic proteins, such as Bcl-2, Bcl-XL, and Bcl-W
(36). The mitochondrial pathway is
activated by the release of cytochrome c, which is followed by
caspase-9 and caspase-3 activation (7,37). The
current study demonstrated that PC9-R cells transfected with
si-MIR31HG lncRNAs expressed significantly higher levels of
Caspase-3, Caspase-9 and Bax proteins, but reduced levels of Bcl-2.
This has demonstrated for the first time that PC9-R cells
transfected with si-MIR31HG exert pro-apoptotic function via the
mitochondrial pathway by inhibiting the EGFR/PI3K/AKT pathway.
Furthermore, regulation of the cell cycle is vital to regulate cell
growth, and some proteins or chemical compounds could trigger
apoptosis in tumor cells accompanied by cell arrest (38). The present study demonstrated that
PC9-R cells transfected with si-MIR31HG were able to arrest the
cell cycle in G0/G1 phase, thus regulating the cell cycle.
In conclusion, PC9-R cells transfected with
si-MIR31HG developed enhanced sensitivity to gefitinib by
inhibiting the EGFR/PI3K/AKT pathway and activating p53. They also
induced cell apoptosis via activation of the mitochondrial
apoptosis pathway leading to arrest of the cell cycle in the G2/M
phase. Therefore, over-expression of MIR31HG lncRNA contributes to
gefitinib resistance in the PC9-R cell, by affecting cell
proliferation, apoptosis and the cell cycle through activation of
the EGFR/PI3K/AKT pathway.
Acknowledgements
The present study was supported by grants from the
National Nature Science Foundation of China (grant no. 81301927),
Zhejiang Provincial Nature Science Foundation (grant no.
LY13H160023) and Hangzhou Medical Major Disease Project (grant no.
20130733Q03).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cruz CS Dela, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hotta K, Kiura K, Ueoka H, Tabata M,
Fujiwara K, Kozuki T, Okada T, Hisamoto A and Tanimoto M: Effect of
gefitinib (‘Iressa’, ZD1839) on brain metastases in patients with
advanced non-small-cell lung cancer. Lung Cancer. 46:255–261. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Burotto M, Manasanch EE, Wilkerson J and
Fojo T: Gefitinib and erlotinib in metastatic non-small cell lung
cancer: A meta-analysis of toxicity and efficacy of randomized
clinical trials. Oncologist. 20:400–410. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Engelman JA and Settleman J: Acquired
resistance to tyrosine kinase inhibitors during cancer therapy.
Curr Opin Genet Dev. 18:73–79. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prabhakar CN: Epidermal growth factor
receptor in non-small cell lung cancer. Transl Lung Cancer Res.
4:110–118. 2015.PubMed/NCBI
|
|
7
|
Chen J, Wang W, Wang H, Liu X and Guo X:
Combination treatment of ligustrazine piperazine derivate DLJ14 and
adriamycin inhibits progression of resistant breast cancer through
inhibition of the EGFR/PI3K/Akt survival pathway and induction of
apoptosis. Drug Discov Ther. 8:33–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y,
Wei M, Chen J, Gao X, Xu C, et al: The prostate cancer-up-regulated
long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation
through reciprocal regulation of androgen receptor. Urol Oncol.
31:1117–1123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun M, Liu XH, Wang KM, Nie FQ, Kong R,
Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al: Down regulation of
BRAF activated non-coding RNA is associated with poor prognosis for
non-small cell lung cancer and promotes metastasis by affecting
epithelialmesenchymal transition. Mol Cancer. 13:682014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30:6942013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Zhou XF, Pan GF and Zhao JP:
Enhanced expression of long non-coding RNA ZXF1 promoted the
invasion and metastasis in lung adenocarcinoma. Biomed
Pharmacother. 68:401–407. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ricciuti B, Mecca C, Crinò L, Baglivo S,
Cenci M and Metro G: Non-coding RNAs in lung cancer. Oncoscience.
1:674–705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thomas D and Livak KJ: Analyzing real-time
PCR data by the comparative C(T) method. Nat Protoc. 3:1101–1108.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rahman AF, Korashy HM and Kassem MG:
Gefitinib. Profiles Drug Subst Excip Relat Methodol. 39:239–264.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pilkington G, Boland A, Brown T, Oyee J,
Bagust A and Dickson R: A systematic review of the clinical
effectiveness of first-line chemotherapy for adult patients with
locally advanced or metastatic non-small cell lung cancer. Thorax.
70:359–367. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan Y, Li XF, Chen JQ, Dong CX, Weng SS
and Huang JJ: Critical appraisal of the role of gefitinib in the
management of locally advanced or metastatic non-small cell lung
cancer. Onco Targets Ther. 7:841–852. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Engelman JA and Jänne PA: Mechanisms of
acquired resistance to epidermal growth factor receptor tyrosine
kinase inhibitors in non-small cell lung cancer. Clin Cancer Res.
14:2895–2899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Engelman JA, Zejnullahu K, Mitsudomi T,
Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen
J, et al: MET amplification leads to gefitinib resistance in lung
cancer by activating ERBB3 signaling. Science. 316:1039–1043. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang J, Zhu N and Chen X: A novel long
noncoding RNA LINC01133 is upregulated in lung squamous cell cancer
and predicts survival. Tumour Biol. 36:7465–7471. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cai H, Chen J, He B, Li Q, Li Y and Gao Y:
A FOXM1 related long non-coding RNA contributes to gastric cancer
cell migration. Mol Cell Biochem. 406:31–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Y, Chen G, Zeng Y, Zeng J, Lin M, Liu
X and Liu J: Invasion and metastasis-related long noncoding RNA
expression profiles in hepatocellular carcinoma. Tumour Biol.
36:7409–7422. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J,
Wei M, Xu C, Wu C, Zhang Z, et al: Long non-coding RNA metastasis
associated in lung adenocarcinoma transcript1 derived miniRNA as a
novel plasma-based biomarker for diagnosing prostate cancer. Eur J
Cancer. 49:2949–2959. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng N, Li X, Zhao C, Ren S, Chen X, Cai
W, Zhao M, Zhang Y, Li J, Wang Q and Zhou C: Microarray expression
profile of long non-coding RNAs in EGFR-TKIs resistance of human
non-small cell lung cancer. Oncol Rep. 33:833–839. 2015.PubMed/NCBI
|
|
30
|
Nguyen KS, Kobayashi S and Costa DB:
Acquired resistance to epidermal growth factor receptor tyrosine
kinase inhibitorsin non-small-cell lung cancers dependent on the
epidermal growth factor receptor pathway. Clin Lung Cancer.
10:281–289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li H, Schmid-Bindert G, Wang D, Zhao Y,
Yang X, Su B and Zhou C: Blocking the PI3K/AKT and MEK/ERK
signaling pathways can overcome gefitinib-resistance in non-small
cell lung cancer cell lines. Adv Med Sci. 56:275–284. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kang XH, Xu ZY, Gong YB, Wang LF, Wang ZQ,
Xu L, Cao F and Liao MJ: Bufalin reverses HGF-induced resistance to
EGFR-TKIs in EGFR mutant lung cancer cells via blockage of
Met/PI3k/Akt pathway and induction of apoptosis. Evid Based
Complement Alternat Med. 2013:2438592013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sordella R, Bell DW, Haber DA and
Settleman J: Gefitinib-sensitizing EGFR mutations in lung cancer
activate anti-apoptotic pathways. Science. 305:1163–1167. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qiao M, Sheng S and Pardee AB: Metastasis
and AKT activation. Cell Cycle. 7:2991–2996. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Slack A, Chen Z, Tonelli R, Pule M, Hunt
L, Pession A and Shohet JM: The p53 regulatory gene MDM2 is a
direct transcriptional target of MYCN in neuroblastoma. Proc Natl
Acad Sci USA. 18:731–736. 2005. View Article : Google Scholar
|
|
36
|
Harris MH and Thompson CB: The role of the
Bcl-2 family in the regulation of outer mitochondrial membrane
permeability. Cell Death Differ. 7:1182–1191. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Elkholi R, Renault TT, Serasinghe MN and
Chipuk JE: Putting the pieces together: How is the mitochondrial
pathway of apoptosis regulated in cancer and chemotherapy? Cancer
Metab. 2:162014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Muthu M, Cheriyan VT and Rishi AK:
CARP-1/CCAR1: A biphasic regulator of cancer cell growth and
apoptosis. Oncotarget. 6:6499–6510. 2015. View Article : Google Scholar : PubMed/NCBI
|