Introduction
Gastric cancer (GC) is the third most common cancer
in China, with the third highest mortality rate (1). Patients are frequently diagnosed at an
advanced stage, resulting in a poor prognosis. It is estimated that
~50% of patients with GC exhibit metastasis at diagnosis (2). Tumor metastasis is a common cause of
mortality in GC, therefore it is important to identify novel
biomarkers that may contribute to the early diagnosis of tumor
invasion and metastasis.
MicroRNAs (miRNAs/miRs) were initially described in
Caenorhabditis elegans in 1993 (3). They are endogenous non-coding small
molecule RNAs, between 19 and 24 ribonucleotides in length, which
post-transcriptionally regulate gene expression in plants and
animals (2,4). miRNAs have important roles in regulating
a number of cell processes, including cell differentiation,
apoptosis and cell cycle progression (5–9). miRNAs
serve as biomarkers in various types of cancer, including GC and
prostate cancer (10,11). Although a number of miRNAs have been
identified to be dysregulated in GC tissues, the role of miR-1258
in GC has not, to the best of our knowledge, previously been
investigated.
Heparanase (HPSE) is a 65-kDa inactive precursor
that cleaves heparan sulfate and participates in the degradation
and remodeling of the extracellular matrix (ECM) (12). It is widely accepted that tumor
metastasis begins with the degradation of the ECM and breakdown of
the basement membrane (13). Our
previous studies demonstrated an increased expression of HPSE in GC
tissues and identified that HPSE facilitates invasion and
metastasis of GC cells (13,14). However, the underlying molecular
mechanism of the upregulation of HPSE in GC tissues remains
unclear. Zhang et al (15)
identified that miR-1258 suppressed breast cancer brain metastasis
by inhibiting the expression and activity of HPSE, by directly
targeting the HPSE 3′-untranslated region. Furthermore, Liu et
al (16) identified that miR-1258
influenced the morbidity and metastasis of non-small cell lung
cancer by regulating the expression of HPSE. However, the potential
association between miR-1258 and HPSE in GC remains unclear. The
aim of the present study was to investigate the role of miR-1258 in
GC and to determine the regulation of HPSE by miR-1258 in GC.
Materials and methods
Patients and tissue specimens
The present study was approved by the First Hospital
of China Medical University (Shenyang, China) and all specimens
were obtained from patients who provided informed consent. The
present study complied with the principles of The Declaration of
Helsinki, and received approval from the Research Ethics Committee
of China Medical University. GC was diagnosed according to
histopathological evaluation, and the histological grade was staged
according to the seventh tumor-node-metastasis (TNM) staging
(17). None of the patients in the
present study received preoperative therapy, and all received
surgery at the time of hospitalization between January 2007 and
December 2009. A total of 116 pairs of GC tissue were obtained, and
the tissues adjacent to the proximal excision margin were regarded
as matched non-tumor adjacent tissues (NATs). The tumor size was
represented by the maximum diameter and this measurement was
obtained from the pathological reports of the patients. The paired
and GC tissues were preserved in liquid nitrogen and stored at
−80°C immediately.
Cell culture
Three GC cell lines (MGC-803, BGC-823 and SGC-7901)
were obtained from the Institute of Biochemistry and Cell Biology
at the Chinese Academy of Sciences (Shanghai, China). For all
assays, the cells were cultured at 37°C and 5% CO2 using
RPMI-1640 medium (HyClone; GE Healthcare Life Sciences, Logan City,
UT, USA) supplemented with 10% fetal bovine serum (FBS).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA of the paired specimens and cultured cells
was isolated using the TRIzol® reagent (Ambion; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. A Poly (A) Tailing kit (Ambion; Thermo
Fisher Scientific, Inc.) was used to add a poly (A) tail, according
to the manufacturer's protocol. RNA was reverse-transcribed into
cDNA using a PrimeScript™ RT Reagent kit (Takara Biotechnology,
Co., Ltd., Shiga, Japan), according to the manufacturer's protocol.
Subsequently, qPCR was employed using 2 µl diluted cDNA products
were added to 12.5 µl SYBR Premix Ex Taq II (Takara Biotechnology,
Co., Ltd.), 0.5 µl forward and reverse primers (10 µM) and 9.5 µl
nuclease-free water in a final volume of 25 µl on a Light Cycler
480 II Real-Time PCR system (Roche Diagnostics, Basel, Switzerland)
with a thermocycling protocol of 94°C for 5 sec, 58°C for 20 sec
and 72°C for 30 sec, for 45 cycles. U6 small nuclear RNA (U6)
expression was used as an internal control and the
2−ΔΔCq method was used to determine expression levels
(18). The primers used were:
RT-primer1,
5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTTA-3′;
RT-primer2,
5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTTC-3′;
RT-primer3,
5′-GCTGTCAACGATACGCTACGTAACGGCATGACAGTGTTTTTTTTTTTTTTTTTTTTTTTTG-3′;
miR-1258-F, 5′-AGTTAGGATTAGGTCGTGGAAAA-3′ miR-1258-R,
5′-GCTGTCAACGATACGCTACGT-3′; U6 primer-F,
5′-CGCTTCGGCAGCACATATAC-3′; and U6 primer-R,
5′-TTCACGAATTTGCGTGTCAT-3′.
Transfection of GC cells
miR-1258 mimic was purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). The cells were seeded in
6-well plates. SGC-7901 cells at a concentration of
2×105 cells/well were transfected with ~50 nM miR-1258
mimic or negative control miRNA (NC) using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
sequence of miR-1258 mimic was 5′-AGUUAGGAUUAGGUCGUGGAA-3′ and the
sequence of the NC was 5′-UUCUCCGAACGUGUCACGUdTdT-3′.
Cell proliferation assay
The proliferation ability of untransfected or
transfected SGC-7901 cells was detected using an MTT assay. A total
of ~8×103 cells were seeded in 96-well plates, and
cultured for 24, 48, 72 or 96 h. Following culture, cells were
incubated with 20 µl 5 mg/ml MTT at 37°C for 4 h prior to
dissolution of the formazan crystals generated with 150 µl
dimethylsulfoxide for 20 min at room temperature. The optical
density was determined at a wavelength of 490 nm using a Model 680
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
In vitro cell invasion assay
A Transwell invasion assay was used to determine the
invasion ability of the GC cells. Untransfected or transfected
SGC-7901 cells were cultured for 24 h. A 50 µl volume of Matrigel
(BD Biosciences, San Jose, CA, USA) diluted 1:12 was added to the
upper chamber of a 24-well tissue culture plate (Corning
Incorporated, Corning, NY, USA), and allowed to solidify for 4 h at
37°C. Subsequently, 200 µl medium containing 5×104 cells
was added on top of the solidified Matrigel. Simultaneously, ~600
µl medium with 10% FBS was added to the lower chamber, to act as a
chemoattractant. Following incubation at 37°C and 5% CO2
for 24 h, the cells remaining on the upper surface were removed
using a wet cotton swab. Cells attached to the lower surface were
fixed using methanol for 1 min and stained with 0.4% hematoxylin
and 0.5% eosin (H&E) separately for 3 min and 30 sec. These
steps were performed at room temperature. The cells were examined
and counted under a light microscope. All the experiments were
performed independently and in triplicate.
In vivo cancer cell metastasis
assay
In order to investigate the effect of miR-1258
expression on GC cell metastasis, SGC-7901 cells transfected with
miR-1258 mimic or NC, and untransfected SGC-7901 cells were
separately injected into the lateral tail vein of 3 groups of
four-week-old female BALB/c mice with an average weight of 15 g,
which were purchased from HFK Bio-Technology Co., LTD (Beijing,
China) and housed in the animal care facility of the China Medical
University under specific pathogen-free conditions. Each group
included 8 mice that were injected with 1×106 cells
resuspended in 0.1 ml PBS. After 5 weeks, the mice were sacrificed.
The lungs were isolated and fixed in 4% paraformaldehyde in PBS at
room temperature for >24 h. Following embedding in paraffin and
sectioning, 4 µm sections were stained with H&E. The H&E
staining is performed using separate stains of 0.4% hematoxylin and
0.5% eosin, for 5 min and 2 min, respectively, at room temperature.
The visible lung metastases were counted under a light microscope.
All procedures involving animals were performed in accordance with
the institutional animal welfare guidelines of the Guide for the
Care and Use of Laboratory Animals (National Institutes of Health
publication no. 80-23, revised 1996).
Protein extraction and western blot
analysis
To extract the total protein from cells, a Total
Protein Extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China) was used according to the manufacturer's protocol. Protein
(50 µg/lane) was separated by SDS-PAGE (12% gels) prepared using an
SDS-PAGE Gel Preparation kit (Nanjing KeyGen Biotech Co., Ltd.),
according to the manufacturer's protocol, then transferred onto 0.2
µm pore size polyvinyl difluoride membranes (EMD Millipore,
Billerica, MA, USA). The membranes were blocked using the 5%
skimmed milk powder at room temperature for 2 h. Then the membranes
were incubated overnight at 4°C with rabbit anti-HPSE polyclonal
antibody (1:200; sc-25825; Santa Cruz Biotechnology, Dallas, TX,
USA) or rabbit anti-β-actin monoclonal antibody (1:5,000; A5441;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) as a control.
Following washing three times for 20 min in Tris-buffered saline
containing Tween 20 (TBST), membranes were incubated with the
horseradish peroxidase-conjugated AffiniPure goat anti-mouse or
anti-rabbit IgG secondary antibody (1:5,000; ZB-2305 and ZB-2301,
respectively; OriGene Technologies, Inc., Beijing, China) for 2 h
at room temperature, then washed again in TBST three times for 20
min. SuperSignal Chemiluminescent substrates made by the mixture of
the Stable Peroxide Solution and Luminol/Enhancer Solution (Thermo
Fisher Scientific, Inc.) were used, according to the manufacturer's
protocol, to detect protein bands, which were observed using
GelCapture software (version 2.0.0.0; DNR Bio-Imaging Systems,
Ltd., Jerusalem, Israel), and the relative protein expression was
analyzed using FluorChem software (version 2.01; ProteinSimple, San
Jose, CA, USA).
Statistical analysis
All results are presented as the mean ± standard
deviation and were analyzed using SPSS software (version 18.0;
SPSS, Chicago, IL, USA). Student's t test and non-parametric tests
(Mann-Whitney U and Kruskal-Wallis tests) were used for statistical
analysis. All the experiments were performed at least in
triplicate. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-1258 expression is downregulated
in human GC cells and patient tissues
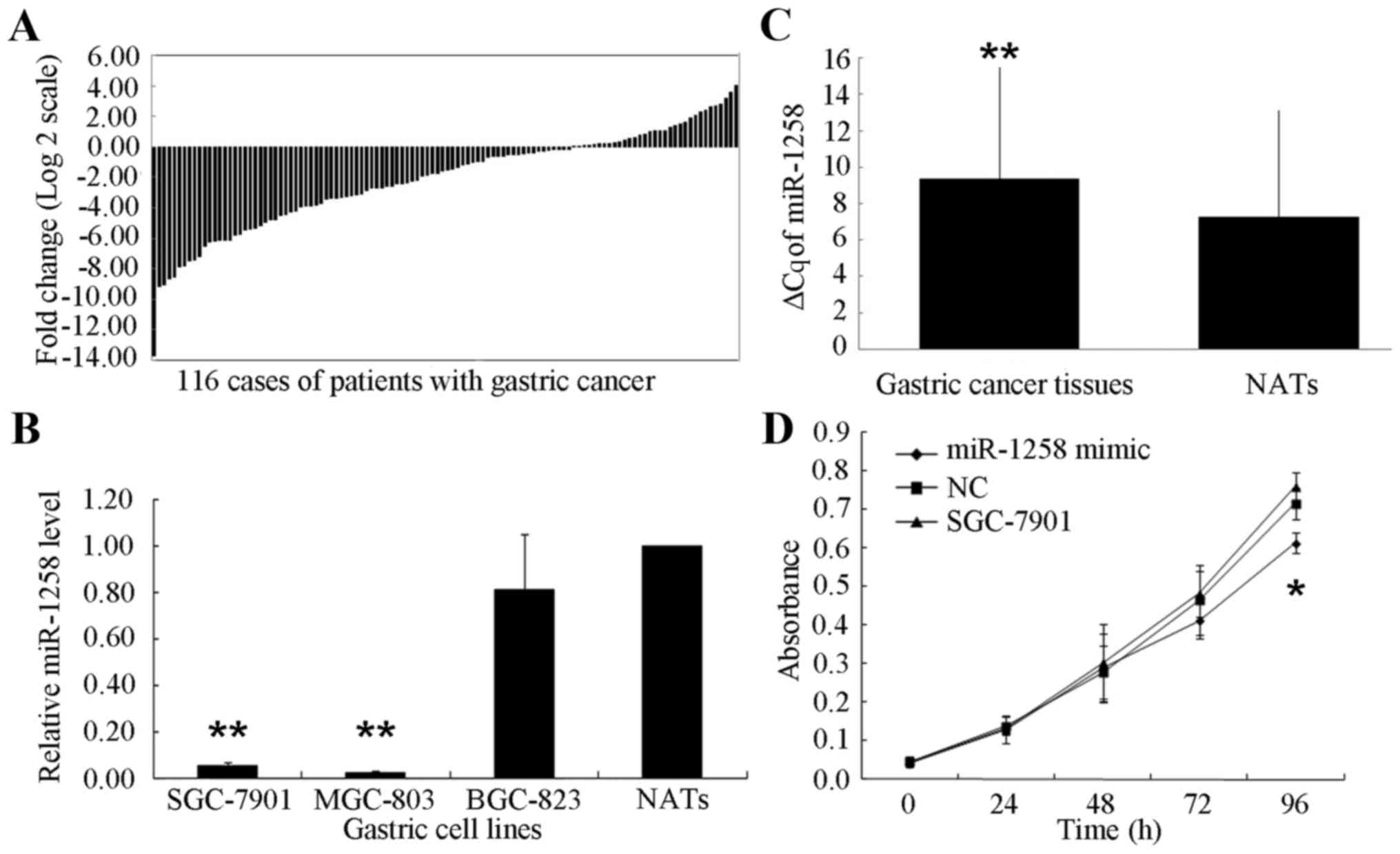
Among the 116 patients with GC, a decreased
expression of miR-1258 was exhibited in 83 cases (71.55%) compared
with their NATs (Fig. 1A). The ΔCq
for miR-1258 was significantly increased in GC tissues compared
with NATs (9.3957±6.1351 and 7.2786±5.8543, respectively;
P<0.001; Fig. 1B). Furthermore,
compared with the NATs from the patients with GC, a significantly
decreased expression of miR-1258 was identified in SGC-7901 cells
(0.05±0.04-fold; P<0.001) and MGC-803 cells (0.02±0.01-fold,
P<0.001) (Fig. 1C).
As shown in Table I,
according to results of the Mann-Whitney U test and the
Kruskal-Wallis test concerning the relative miR-1258 expression
levels and the clinicopathological characteristics, older patients
exhibited a decreased miR-1258 expression (P=0.042), and patients
with the decreased miR-1258 expression tended to be classified in
an advanced pathological tumor (pT) category (P=0.027).
Furthermore, miR-1258 expression was significantly decreased in
cases in which lymphatic vessel invasion was positive (P=0.044).
However, no significant association between the expression level
and gender, tumor size, macroscopic type, histological grade,
pathological node (pN) category or TNM stage was identified
(Table I). Although no significant
association between miR-1258 expression and pN classification was
identified, a decreased miR-1258 expression was observed in 76.74%
of the 86 cases with metastatic lymph nodes. By contrast, of the 30
cases with no metastatic lymph nodes, only 56.67% exhibited a
decreased expression of miR-1258.
 | Table I.Associations between the expression of
miR-1258 with clinicopathological characteristics in 116 patients
with gastric cancer. |
Table I.
Associations between the expression of
miR-1258 with clinicopathological characteristics in 116 patients
with gastric cancer.
| Characteristic | n | miR-1258
T/Na | P-value |
|---|
| Gender |
|
| 0.410 |
|
Male | 90 | 1.048
(0.068–1.068) |
|
|
Female | 26 | 1.577
(0.044–2.044) |
|
| Age, years |
|
| 0.042c |
|
<62 | 57 | 1.568
(0.083–1.083) |
|
|
≥62 | 59 | 0.778
(0.032–0.032) |
|
| Tumor size, cm |
|
| 0.579 |
|
<5 | 53 | 1.342
(0.081–1.081) |
|
| ≥5 | 63 | 1.018
(0.051–1.051) |
|
| Macroscopic
typeb |
|
| 0.560 |
|
Borrmann I+II | 12 | 1.061
(0.035–1.035) |
|
|
Borrmann III+IV | 100 | 1.181
(0.057–1.057) |
|
| Histological
grade |
|
| 0.920 |
| Well
and moderately differentiated | 28 | 1.582
(0.028–1.028) |
|
| Poorly
differentiated | 88 | 1.034
(0.057–1.057) |
|
| pT stage |
|
| 0.027c |
|
T1+T2 | 23 | 2.312
(0.138–2.138) |
|
|
T3+T4 | 93 | 0.883
(0.045–0.045) |
|
| pN stage |
|
| 0.343 |
|
Negative | 30 | 1.821
(0.086–1.086) |
|
|
Positive | 86 | 0.938
(0.050–0.050) |
|
| pN stage |
|
| 0.350 |
| N0 | 28 | 1.914
(0.092–2.092) |
|
| N1 | 17 | 1.401
(0.086–2.086) |
|
| N2 | 28 | 0.486
(0.046–0.046) |
|
| N3 | 43 | 1.030
(0.027–0.027) |
|
| pTNM stage |
|
| 0.199 |
| I | 13 | 2.077
(0.055–1.055) |
|
| II | 27 | 1.586
(0.103–2.103) |
|
|
III | 76 | 0.862
(0.038–0.038) |
|
| Invasion into
lymphatic vessels |
|
| 0.044c |
|
Negative | 75 | 1.395
(0.103–1.103) |
|
|
Positive | 41 | 0.748
(0.030–0.030) |
|
Association between miR-1258 and cell
proliferation in GC
An MTT assay of the effect of miR-1258 on SGC-7901
cell proliferation following transfection with miR-1258 mimic or NC
revealed that the miR-1258 mimic exhibited a limited effect on the
cell growth. An MTT assay was performed at 24, 48, 72 and 96 h. No
significant differences were identified in the first 72 h. Only at
96 h was a significant decrease in the number of SGC-7901 cells
identified in the miR-1258-transfected cells compared with the
NC-transfected and untransfected cells (P<0.05; Fig. 1D).
Upregulation of the miR-1258
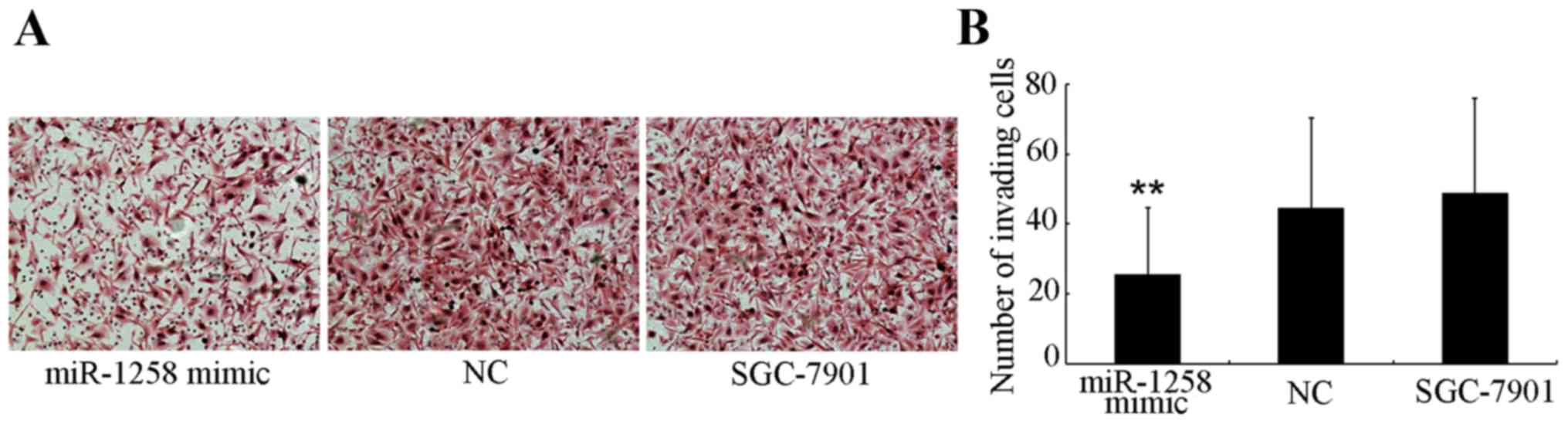
expression level suppresses SGC-7901 cell invasion in vitro
According to Mann-Whitney U test analysis, a
significant association between the expression of miR-1258 and
advanced pT category was identified, therefore the influence of
miR-1258 on the invasive ability of GC cells was investigated.
Cells that were able to invade to the lower side of the membrane in
the Transwell assays after a 24-h incubation were harvested
(Fig. 2A) and quantified (Fig. 2B). The number of miR-1258-transfected
SGC-7901 cells (25.46±19.12) that invaded through the Matrigel was
significantly decreased (P<0.001) compared with the number of
NC-transfected (44.44±25.77) and untransfected (48.75±27.09)
SGC-7901 cells.
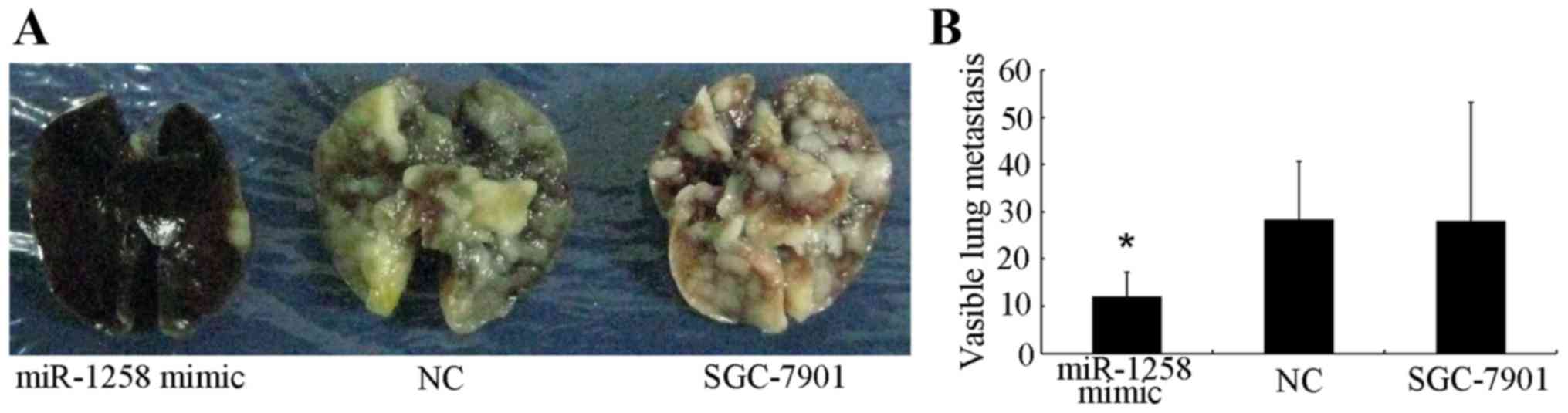
Upregulation of miR-1258 expression
level inhibits SGC-7901 cell metastasis in vivo
In order to investigate the function of miR-1258 in
GC metastasis, a metastasis formation assay was established using
nude mice. After 5 weeks, microscopic histological analyses were
performed by dissecting the lungs of the sacrificed mice. The
number of lung metastases in first group (miR-1258 mimic) was
12.00±5.2, whereas the number of metastases in the second (NC) and
third (untransfected SGC-7901) were 28.25±12.40 and 28±25.19,
respectively (Fig. 3A and B). These
results demonstrate that upregulation of miR-1258 expression
significantly inhibits cell metastasis in vivo.
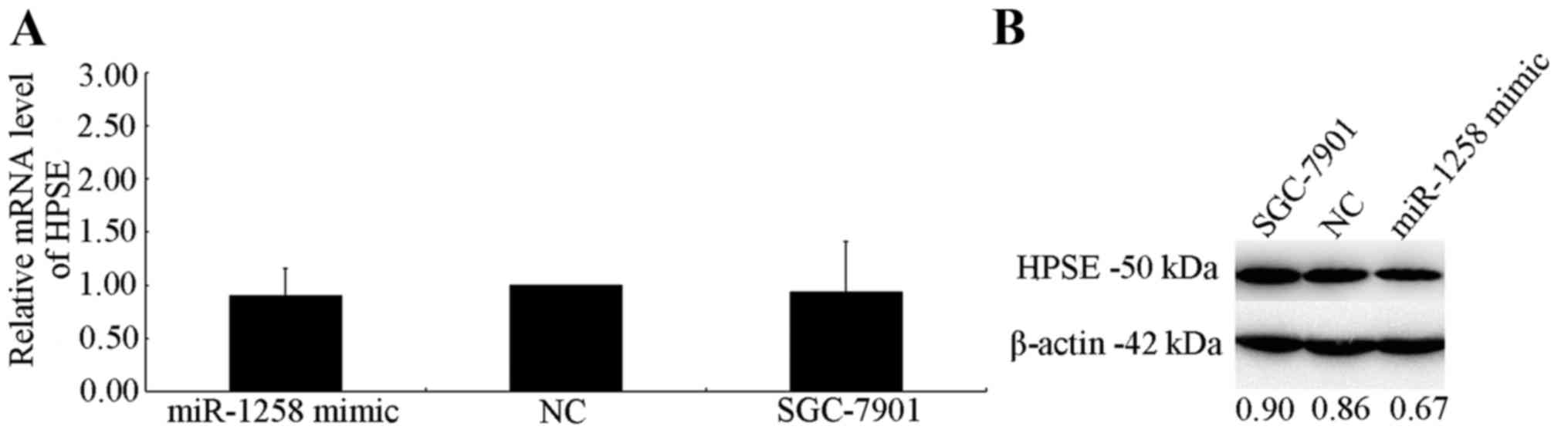
miR-1258 may target HPSE in SGC-7901
cells
RT-qPCR and western blot analysis were used to
investigate the effect of HPSE expression on mRNA and protein
levels. No effect of miR-1258 on HPSE mRNA levels was exhibited 48
h after transfection (Fig. 4A).
However, a marked decrease in translational level in cells
overexpressing miR-1258 was observed, which was normalized to an
endogenous reference β-actin protein (Fig. 4B). The transfection efficiency of
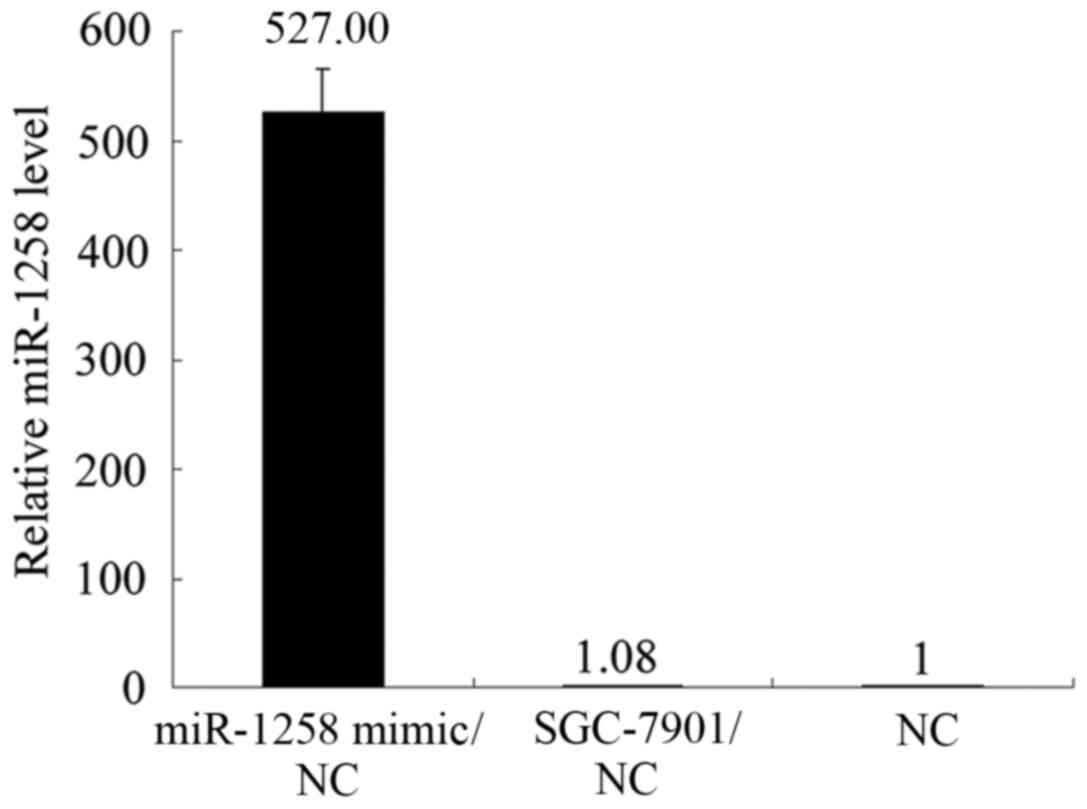
miR-1258 mimic was marked (Fig. 5).
The results indicate that miR-1258 interacts with HPSE and
inversely regulates the expression of HPSE at the translational
level.
Discussion
miRNAs have been studied extensively in numerous
types of cancer. In the last 20 years, >2,000 miRNAs have been
discovered in humans, and are reported to regulate ~1/3 of the
genes in the human genome (19).
There are numerous miRNAs that exhibit similar functions in
different types of cancer. For example, miR-203 is able to inhibit
the proliferation, migration and metastasis of triple-negative
breast and lung cancer cells (20,21). The
expression of miR-203 is associated with tumor size, advanced
Borrmann type and pT category of the GC (22). In addition, miR-148b is able to
suppress cell growth in GC and colorectal cancer by targeting
cholecystokinin B receptor (8,23),
indicating that a number of miRNAs have the same function in
different types of cancer.
It has been reported that the low expression of
miR-1258 is associated with the development, progression,
metastasis, and prognosis of cancer: For example, Zhang et
al (15) identified that
miR-1258, a candidate miRNA, directly targeted HPSE and suppressed
brain metastatic breast cancer. Furthermore, it has been
demonstrated that HPSE is upregulated in GC and facilitates
invasion and metastasis of GC cells (24–27). A
number of molecular mechanisms underlying single nucleotide
polymorphisms of HPSE have been reported (28,29).
Furthermore, Liu et al (16)
identified that miR-1258 regulates the expression level of HPSE to
influence the morbidity and metastasis of non-small cell lung
cancer. Therefore, the aim of the present study was to determine
whether these effects were associated only with breast cancer and
lung cancer, or whether they also existed in GC.
In the present study, the expression level of
miR-1258 was significantly decreased in GC tissues and cells. The
patients with GC that exhibited decreased expression of miR-1258
may exhibit a more advanced pT category and positive lymphatic
vessel invasion. The decreased expression of miR-1258 was inversely
associated with increased age of the patients. It is well-known
that miR-1258 facilitates invasion and metastasis of breast cancer
or non-small cell lung cancer cells (15,16).
Migration and invasion assays, and metastasis formation assay
identified that the upregulation of miR-1258 suppressed SGC-7901
cell invasion in vitro and inhibited SGC-7901 cell
metastasis in vivo.
RT-qPCR results indicated that miR-1258 exhibited a
limited effect on the HPSE mRNA level; however, a marked inverse
association was observed between miR-1258 and HPSE protein
expression. These results indicate that HPSE is a target of
miR-1258, and that miR-1258 negatively regulates HPSE expression at
the translational level. Furthermore, it was demonstrated
previously that, following knockdown of HPSE by siRNA, SGC-7901
cell invasion was significantly decreased (13). Therefore, the results of the present
study suggest that miR-1258 was downregulated in GC, and influenced
the invasion and metastasis of GC cells by regulating the
expression level of HPSE.
Although an inverse association between miR-1258 and
HPSE protein expression has been demonstrated in GC in the present
study, a number of previous studies have identified that the same
miRNA potentially regulates distinct targets in various cell types
or in the same cell type, or is dependent on distinct binding
regions (30–34). Similarly, a single target gene may be
regulated by a number of miRNAs (35,36).
Therefore, further investigation of the target genes of miR-1258
and the other miRNAs that regulate HPSE expression is
warranted.
The results of the present study have demonstrated
downregulation of miR-1258 in GC tissues and cell lines compared
with NATs. Furthermore, decreased miR-1258 expression was
identified to be associated with pT stage depth of invasion and
positive lymphatic vessel invasion in patients with GC. In
addition, it was identified that overexpression of miR-1258 was
able to suppress SGC-7901 cell invasion in vitro and inhibit
SGC-7901 cell metastasis in vivo. As miR-1258 downregulates
the expression of HPSE protein in GC cells and inhibits cell
invasion, miR-1258 may serve as a novel biomarker and therapeutic
target for the treatment of GC.
Acknowledgements
The present study was supported by the National
Science Foundation of China (grant nos. 81201888, 81372549 and
81172370), the Natural Science Foundation of Liaoning Province
(grant no. 2014029201) and the Program of Education of the
Department of Liaoning Province (grant no. L2014307). The authors
acknowledge the Department of Surgical Oncology of the First
Hospital of China Medical University for providing human GC
samples. The authors also thank the College of China Medical
University for technical assistance in experiments.
References
|
1
|
Chen WQ, Zheng RS, Zhang SW, Zeng HM and
Zou XN: The incidences and mortalities of major cancers in China,
2010. Chin J Cancer. 33:402–405. 2014.PubMed/NCBI
|
|
2
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Crone SG, Jacobsen A, Federspiel B,
Bardram L, Krogh A, Lund AH and Friis-Hansen L: microRNA-146a
inhibits G protein-coupled receptor-mediated activation of NF-κB by
targeting CARD10 and COPS8 in gastric cancer. Mol Cancer.
11:712012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carthew RW and Sontheimer EJ: Origins and
Mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao Y, Deng C, Lu W, Xiao J, Ma D, Guo M,
Recker RR, Gatalica Z, Wang Z and Xiao GG: let-7 microRNAs induce
tamoxifen sensitivity by downregulation of estrogen receptor α
signaling in breast cancer. Mol Med. 17:1233–1241. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song Y, Xu Y, Wang Z, Chen Y, Yue Z, Gao
P, Xing C and Xu H: MicroRNA-148b suppresses cell growth by
targeting cholecystokinin-2 receptor in colorectal cancer. Int J
Cancer. 131:1042–1051. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang H, Yu WW, Wang LL and Peng Y:
miR-130a acts as a potential diagnostic biomarker and promotes
gastric cancer migration, invasion and proliferation by targeting
RUNX3. Oncol Rep. 34:1153–1161. 2015.PubMed/NCBI
|
|
11
|
Agaoglu Yaman F, Kovancilar M, Dizdar Y,
Darendeliler E, Holdenrieder S, Dalay N and Gezer U: Investigation
of miR-21, miR-141, and miR-221 in blood circulation of patients
with prostate cancer. Tumour Biol. 32:583–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ilan N, Elkin M and Vlodavsky I:
Regulation, function and clinical significance of heparanase in
cancer metastasis and angiogenesis. Int J Biochem Cell Biol.
38:2018–2039. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yingying X, Yong Z, Zhenning W, Xue Z, Li
J, Yang L and Huimian X: Role of heparanase-1 in gastric carcinoma
invasion. Asian Pac J Cancer Prev. 10:151–154. 2009.PubMed/NCBI
|
|
14
|
Wang Z, Xu H, Jiang L, Zhou X, Lu C and
Zhang X: Positive association of heparanase expression with tumor
invasion and lymphatic metastasis in gastric carcinoma. Mod Pathol.
18:205–211. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang L, Sullivan PS, Goodman JC,
Gunaratne PH and Marchetti D: MicroRNA-1258 suppresses breast
cancer brain metastasis by targeting heparanase. Cancer Res.
71:645–654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu H, Chen X, Gao W and Jiang G: The
expression of heparanase and microRNA-1258 in human non-small cell
lung cancer. Tumour Biol. 33:1327–1334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sobin LH, Gospodarowicz MK and Wittekind
C: International Union Against Cancer (UICC)TNM Classification of
Malignant Tumours. 7th. Wiley-Blackwell; New York: pp. 117–126.
2010
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang N, Liang H, Zhou Y, Wang C, Zhang S,
Pan Y, Wang Y, Yan X, Zhang J, Zhang CY, et al: miR-203 suppresses
the proliferation and migration and promotes the apoptosis of lung
cancer cells by targeting SRC. PLoS One. 9:e1055702014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiang Y, Song Y, Wang Z, Chen Y, Yue Z,
Xu H, Xing C and Liu Z: Aberrant expression of miR-203 and its
clinical significance in gastric and colorectal cancers. J
Gastrointest Surg. 15:63–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu
HM, Zhang X, Jiang L, Xing CZ and Zhang Y: MicroRNA-148b is
frequently down-regulated in gastric cancer and acts as a tumor
suppressor by inhibiting cell proliferation. Mol Cancer. 10:12011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li HL, Gu J, Wu JJ, Ma CL, Yang YL, Wang
HP, Wang J, Wang Y, Chen C and Wu HY: Heparanase mRNA and protein
expression correlates with clinicopathologic features of gastric
cancer patients: A Meta- analysis. Asian Pac J Cancer Prev.
16:8653–8658. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X, Xu S, Tan Q and Liu L: High
expression of heparanase-2 is an independent prognostic parameter
for favorable survival in gastric cancer patients. Cancer
Epidemiol. 37:1010–1013. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma XM, Shen ZH, Liu ZY, Wang F, Hai L, Gao
LT and Wang HS: Heparanase promotes human gastric cancer cells
migration and invasion by increasing Src and p38 phosphorylation
expression. Int J Clin Exp Pathol. 7:5609–5621. 2014.PubMed/NCBI
|
|
27
|
Tang B, Xie R, Qin Y, Xiao YF, Yong X,
Zheng L, Dong H and Yang SM: Human telomerase reverse transcriptase
(hTERT) promotes gastric cancer invasion through cooperating with
c-Myc to upregulate heparanase expression. Oncotarget.
7:11364–11379. 2016.PubMed/NCBI
|
|
28
|
Yue Z, Song Y, Wang Z, Luo Y, Jiang L,
Xing L, Xu H and Zhang X: Association of heparanase gene (HPSE-1)
single nucleotide polymorphisms with gastric cancer. J Surg Oncol.
102:68–72. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li AL, Song YX, Wang ZN, Gao P, Miao Y,
Zhu JL, Yue ZY and Xu HM: Polymorphisms and a haplotype in
heparanase gene associations with the progression and prognosis of
gastric cancer in a northern Chinese population. PLoS One.
7:e302772012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dyrskjøt L, Ostenfeld MS, Bramsen JB,
Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL,
Andersen CL, Zieger K, et al: Genomic profiling of microRNAs in
bladder cancer: miR-129 is associated with poor outcome and
promotes cell death in vitro. Cancer Res. 69:4851–4860. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Strillacci A, Griffoni C, Sansone P,
Paterini P, Piazzi G, Lazzarini G, Spisni E, Pantaleo MA, Biasco G
and Tomasi V: MiR-101 downregulation is involved in
cyclooxygenase-2 overexpression in human colon cancer cells. Exp
Cell Res. 315:1439–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duursma AM, Kedde M, Schrier M, le Sage C
and Agami R: miR-148 targets human DNMT3b protein coding region.
RNA. 14:872–877. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R,
Sun Z and Zheng X: Downregulation of CCND1 and CDK6 by miR-34a
induces cell cycle arrest. FEBS Lett. 582:1564–1568. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Takahashi Y, Forrest AR, Maeno E,
Hashimoto T, Daub CO and Yasuda J: MiR-107 and MiR-185 can induce
cell cycle arrest in human non small cell lung cancer cell lines.
PLoS One. 4:e66772009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu J, Qian J, Li C, Kwok L, Cheng F, Liu
P, Perdomo C, Kotton D, Vaziri C, Anderlind C, et al: miR-129
regulates cell proliferation by downregulating Cdk6 expression.
Cell Cycle. 9:1809–1818. 2010. View Article : Google Scholar : PubMed/NCBI
|