Introduction
Colorectal cancer (CRC) is one of the four most
prevalent solid tumors that cause cancer-associated mortality
globally. Early detection of CRC improves the 5-year survival rate
from 12–13% in stage IV metastatic disease to 90% in stage I–II
early-stage disease (1). Fecal occult
blood test is typically used for CRC screening. Carcinoembryonic
antigen (CEA) and carbohydrate antigen 19–9 (CA19-9) have been used
for diagnosis and for disease monitoring following treatment.
Although CEA and CA19-9 have exhibited a certain level of
sensitivity, their sensitivity remains low. The pathological
testing of tumor tissue is the optimal method for histological
diagnosis, and the detection of mutations in rat sarcoma viral
oncogene homolog [RAS; Kirsten RAS (KRAS) and neuroblastoma RAS
(NRAS)] gene and B-raf serine/threonine kinase proto-oncogene
(BRAF) gene in tumor tissue are used as predictive biomarkers
aiding in the selection of targeted drug treatments (2). Colonoscopy of the primary tumor and
needle biopsy of metastatic tumors are the techniques used for
histological and genomic diagnosis; however, these methods are
invasive, uncomfortable and costly (3–5). Thus,
novel non-invasive methods for diagnosis and the detection of
mutations are required.
Exosomes are small stable vesicles of 30–100 nm in
diameter in the circulating blood, in which microRNA (miRNA/miR),
mRNA and DNA fragments are coated in numerous proteins and
bioactive lipids (6–9). Previous studies have identified that
exosomes may be directly released from cells through the outward
budding of the plasma membrane in a calcium-dependent manner, and
be shuttled from donor cells to recipient cells (10–14). The
level of exosomes released from cancer cells has been demonstrated
to be increased compared with normal cells, and exosomal RNAs and
proteins may implicate the origin of the donor cells (15,16).
Therefore, exosomes may serve as a highly sensitive and specific
diagnostic tool for the repetitive and non-invasive monitoring of
patients with cancer, aiding clinicians in the diagnosis,
classification and treatment of cancer (17).
To validate the potential of the serum exosome as a
novel biomarker for the monitoring of cancer, the current study
investigated whether established cancer-associated mutations could
be amplified from mRNA in the serum exosome of patients with
cancer. In the present study, KRAS and BRAF gene mutations were
detected in patients with CRC, and the consistency of the detection
of these mutations between primary tumor tissues and the matched
serum exosomes were compared. The results of the current study
indicated that serum exosomal mRNA had the potential to be used for
gene mutation detection in patients with CRC.
Materials and methods
Clinical samples
The current study consisted of 35 patients (age,
40–75 years; mean ± standard deviation, 60.0±9.5 years) from The
First Affiliated Hospital of The People's Liberation Army General
Hospital (Beijing, China). Patients underwent tumor resection
surgery between July 2013 and December 2013 with histologically
confirmed colorectal adenocarcinoma prior to the surgery.
Colorectal primary tumor tissue samples were obtained from the
surgical specimens and the matched blood samples were obtained from
the patients prior to the surgery. Detailed information of the
patients were presented in Table I.
The classification of tumor differentiation and stage were assessed
according to the 2000 World Health Organization (WHO)
classification system for tumors of digestive system and the
American Joint Committee on Cancer (AJCC) staging system,
respectively (18). The present study
obtained ethical approval from the Ethics Committees of The First
Affiliated Hospital of The People's Liberation Army General
Hospital (no. 2013067) and informed written consents were obtained
for all patients.
 | Table I.Clinicopathological characteristics
of 35 CRC patients. |
Table I.
Clinicopathological characteristics
of 35 CRC patients.
| Clinicopathological
characteristic | No. of patients
(%) |
|---|
| Gender |
|
|
Male | 22 (62.9) |
|
Female | 13 (37.1) |
| Age, years |
|
|
>65 | 10 (28.6) |
|
≤65 | 25 (71.4) |
| Tumor site |
|
|
Colon | 21 (60) |
|
Rectum | 14 (40) |
| Tumor
differentiation |
|
| G1 | 4 (11.4) |
| G2 | 20 (57.2) |
| G3 | 11 (31.4) |
| Tumor stage |
|
| I | 4 (11.4) |
| II | 17 (48.6) |
|
III | 11 (31.4) |
| IV | 3 (8.6) |
Exosomes were obtained from blood
serum
Exosomes were prepared using the differential
ultra-centrifugation method, as previously described (19). Blood serum (5 ml) was centrifuged at
500 × g for 10 min, at 2,000 × g for 20 min and at 10,000 × g for
10 min, all at 4°C. The supernatant was filtered through 0.22 µm
disposable filter units, and transferred to an Amicon®
Stirred Ultrafiltration Cell (Model 8050) with a 100,000 KDa
molecular weight cutoff ultrafiltration membrane (all EMD
Millipore, Billerica, MA, USA) at a nitrogen gas pressure of <75
psi (5.3 kg/cm2). The samples were stirred and the rate
of stirring was adjusted so that the vortex created was 1/3 the
depth of the liquid volume. Following the supernatant
ultrafiltration, 10 ml PBS was added and ultra-filtrated three
times. Following washing twice, 0.1 ml PBS was added to suspend the
exosomes. Subsequently, exosomes were isolated using
ultra-centrifugation at 120,000 × g for 1 h at 4°C and stored at
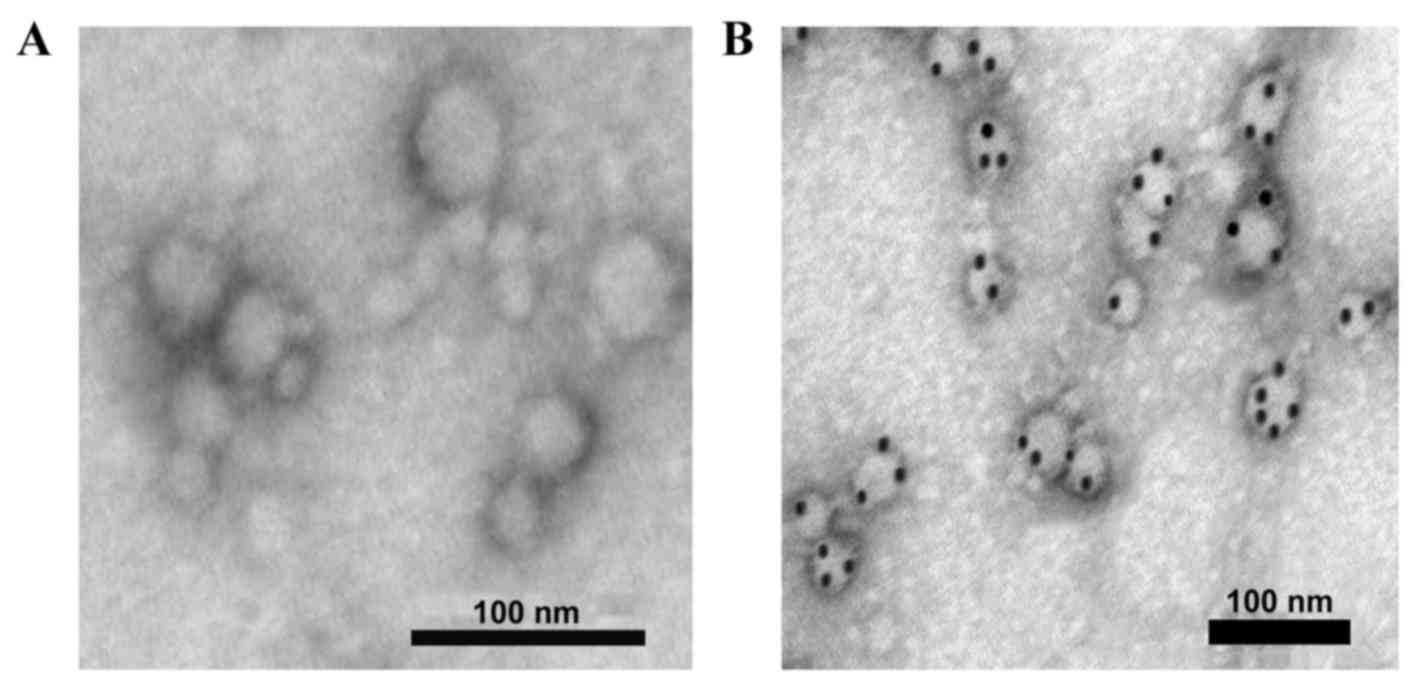
−80°C. The purified exosomes from the microcapsule membrane
structure (30–80 nm) were observed using a Hitachi H-7500
transmission electron microscope (Hitachi, Ltd., Tokyo, Japan;
Fig. 1A). For immunoelectron
microscopy, exosomes were incubated with a rabbit anti-human
cluster of differentiation (CD)63 monoclonal antibody (dilution,
1:1,000; #EXOAB-CD63A-1; System Biosciences Inc., Palo Alto, CA,
USA) for 1 h, followed by 20 µl Staphylococcal protein A Immunogold
(dilution, 1:15; Meridian Life Science, Inc., Memphis, TN, USA) for
30 min at room temperature. Samples incubated with PBS were used as
the blank control. The positively labeled exosomes were confirmed
as vesicles containing black colloidal gold particles using a
transmission electron microscope (Fig.
1B).
cDNA synthesized from exosomal
mRNA
Total RNA were extracted from serum exosomes using
the RNeasy Mini Spin kit (Qiagen GmbH, Hilden, Germany) according
to the manufacturer's protocol. A total of 1 µl Oligo dT (5′-d(TTT
TTT TTT TTT TTT TTT)-3′) and 1 µl random sequence (Takara Bio, Inc.
Otsu, Japan) were added into the 10 µl of RNA elution fluids,
incubated at 70°C for 10 min and then on ice for 2 min. For cDNA
synthesis, reagents were added to the 12 µl RNA mixture described
above, resulting in final concentrations (in 20 µl) of 200 U RNase
M-MLV (RNase H-), 40 U ribonuclease inhibitor, 10 mM of each dNTP,
4 µl 5X M-MLV buffer (all from Takara Bio, Inc.) and RNase-free
distilled H2O. This reaction was incubated at 42°C for 1
h, 70°C for 15 min and then placed on ice. The cDNA products were
stored at −20°C.
Polymerase chain reaction (PCR) of
KRAS and BRAF
Genomic DNA were extracted from 100 mg of tumor
tissue using a standard protein kinase K procedure (20) and observed through separation on a
0.5% agarose gel and visualized by ethidium bromide (DL2000 DNA
Marker, Takara Bio, Inc.). A total of 2 µl DNA isolated from tumor
tissue or 2 µl cDNA from serum exosomes were added to result in the
following final concentrations in a 20 µl reaction: 5 units Ex Taq
(Takara Bio, Inc.); 0.2 µM of forward primer; 0.2 µM of reverse
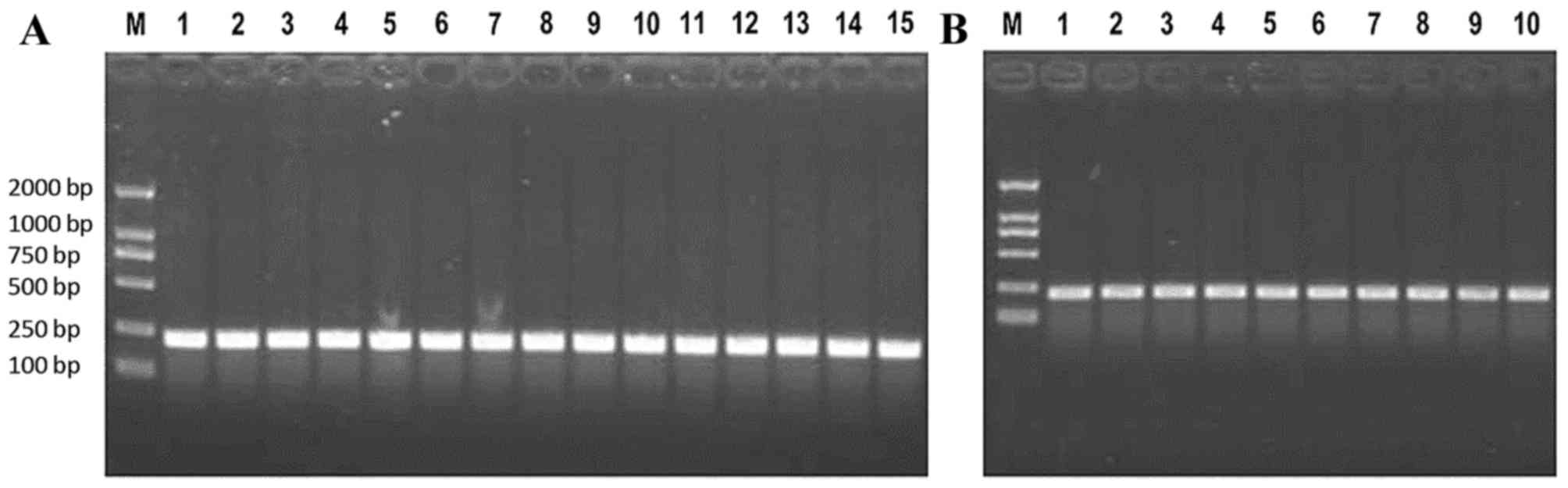
primer. The primers used in the PCR are presented in Table II. These reactions were incubated at
95°C for 40 sec, followed by 40 cycles of 95°C for 10 sec, 60°C for
20 sec and 72°C for 20 sec. PCR products were separated on a 2%
agarose gel (Fig. 2) and the target
products were purified using UNIQ-10 Column DNA Gel Extraction kit
(Sangon Biotech Co., Ltd., Shanghai, China) according to the
manufacturer's protocol. The remaining elution containing the
purified DNA products was stored at −20°C.
 | Table II.Primers used for polymerase chain
reaction amplification of the KRAS and BRAF genes. |
Table II.
Primers used for polymerase chain
reaction amplification of the KRAS and BRAF genes.
| Gene target | Primer sequence
(5′-3′) | Predicted product
size (bp) |
|---|
| KRAS-exon 2 | F:
TACTGGTGGAGTATTTGATAG | 243 |
|
| R:
TCCTGCACCAGTAATATGCATAT |
|
| KRAS-exon 3 | F:
AAGTAAAAGGTGCACTGTAATAA | 235 |
|
| R:
AACCCACCTATAATGGTGAATATCT |
|
| BRAF-exon 15 | F:
TTCATAATGCTTGCTCTGATAG | 243 |
|
| R:
AACTCAGCAGCATCTCAGGGCCAA |
|
| KRAS-CDS | F:
ATGACTGAATATAAACTTGT | 230 |
|
| R:
AGTCCTCATGTACTGGTCCCTC |
|
| BRAF-CDS | F:
TGGATTACTTACACGCCAAGTCA | 238 |
|
| R:
AATGCATATACATCTGACTGAAAGC |
|
Cloning of the PCR products and gene
sequencing
For the ligation of the PCR products and vector, 500
ng of PCR product was added to a total reaction volume of 10 µl
that contained 0.1 µg T-Vector, 2 µl T4 DNA Ligase buffer (10X),
0.5 µl T4 DNA ligase (all from Takara Bio, Inc.) and distilled
H2O. These reactions were incubated at 22°C for 2 h. DNA
ligation product (5 µl) was added to 100 µl of competent DH5α
Escherichia coli cells (Takara Bio, Inc.), mixed and incubated on
ice for 30 min. The mixture was subjected to heat shock at 42°C for
90 sec and subsequently incubated on ice for 2 min. A total of 900
µl super optimal broth with catabolite repression (SOC, Sangon
Biotech Co., Ltd.) was added and the mixture was agitated at 37°C
for 1 h at 150 rpm. Subsequently, 100 µl or 900 µl of DH5α cells
were spread onto ampicillin-containing Luria broth plates and
incubated overnight at 37°C. Plasmid extraction was performed using
a TIANprep Mini Plasmid kit (Tiangen Biotech Co., Ltd., Beijing,
China) according to the manufacturer's protocol. A total of 30 µl
of each elution sample was sent to Invitrogen (Thermo Fisher
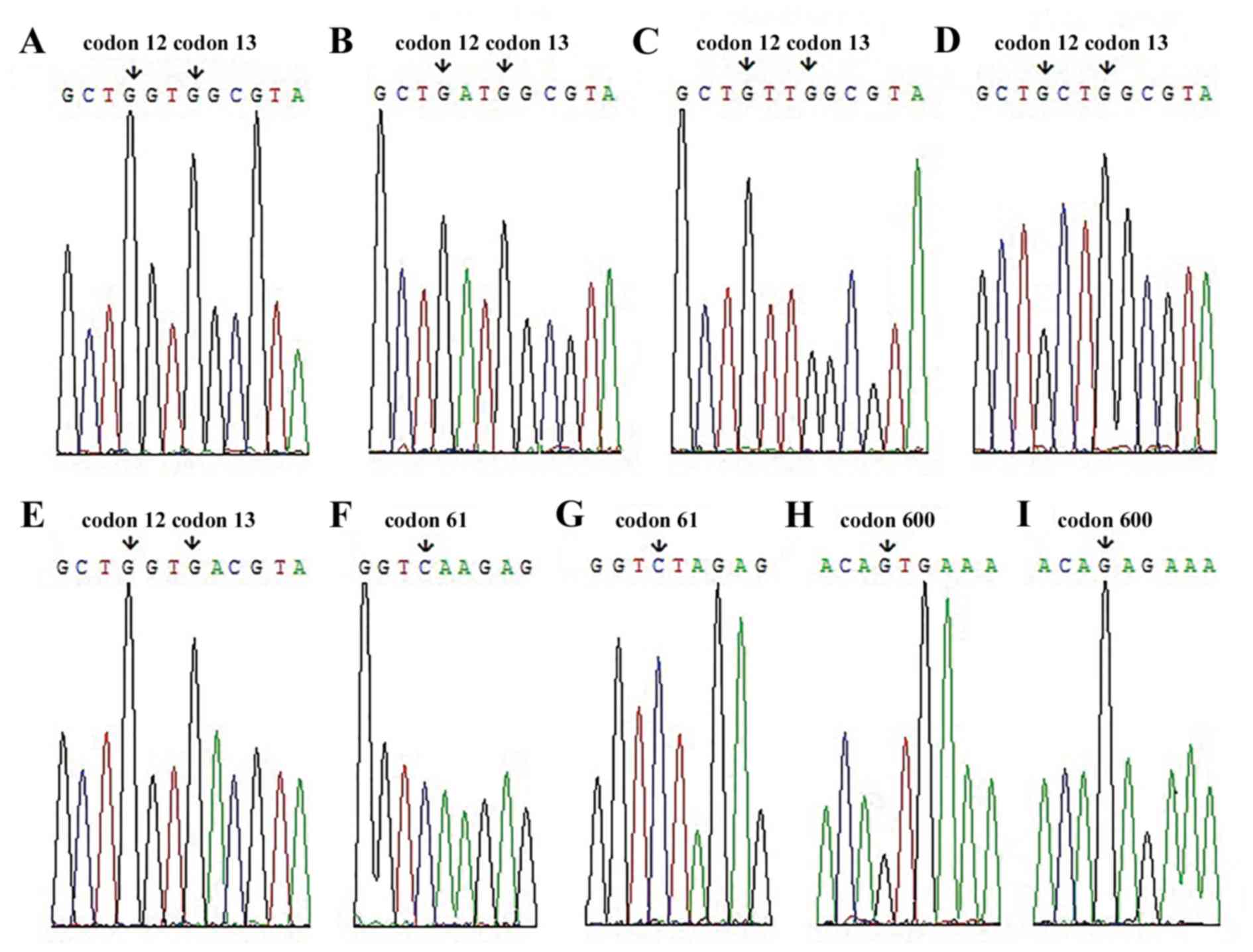
Scientific, Inc., Waltham, MA, USA) for gene sequencing. The gene
mutations identified in the samples are presented in Fig. 3.
Statistical analysis
Statistical analysis was performed using SPSS
software (version 22.0; IBM SPSS, Armonk, NY, USA). The McNemar
test was used to compare the distribution differences of KRAS and
BRAF gene mutations between primary tumor tissues and matched serum
exosomes in patients with CRC. The kappa statistic was used to
assess the reliability of mutation detection in the tumor tissue
and serum exosome samples. The consistency rate was the proportion
of the samples with same mutation in the matched serum exosome and
tumor tissue of total samples. The gene mutations distribution
according to the clinicopathological characteristics of patients
were analyzed using the χ2 test. The significance of
association between gene mutations and the age of patients were
performed using the Student's t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
KRAS mutation detection in tumor
tissues and matched serum exosomes
The mutation status of the KRAS gene was analyzed in
35 tumor tissues and the matched serum exosomes. The spectrum of
these mutations is presented in Table
III. The KRAS gene was not detected in 2 serum exosome samples.
For the other 33 tumor tissue-serum exosome matched samples, 19
(57.6%) tumor tissues had a mutation in exon 2 or exon 3, of which
12 (36.4%) were at codon 12 of exon 2, 5 (15.2%) at codon 13 of
exon 2 and 2 (6.1%) at codon 61 of exon 3. The most frequent
mutation was G12D (42.1%) followed by G13D (26.3%) of all KRAS
mutations in tumor tissues. A total of 14 (42.4%) matched serum
exosomes contained a mutation in exon 2 or exon 3, of which there
were 8 (24.2%) had a mutation at codon 12 of exon 2, 4 (12.1%) at
codon 13 of exon 2 and 2 (6.1%) at codon 61 of exon 3. The most
frequent mutation in serum exosomes was also G12D followed by G13D,
which represented 35.7 and 28.6% of all mutations, respectively.
There was no significant difference found in the distribution of
detected KRAS mutations between tumor tissues and the serum
exosomes of patients with CRC (P=0.063).
 | Table III.KRAS and BRAF gene mutations in tumor
tissues and matched serum exosomes. |
Table III.
KRAS and BRAF gene mutations in tumor
tissues and matched serum exosomes.
|
|
|
|
| Detection in
exosome |
|---|
|
|
|
|
|
|
|---|
| Gene | Tumor tissue, no.
(%) | Exosome, no.
(%) | McNemar test
P-value | Sensitivity
(%) | Total consistency
rate (%) | κ score |
|---|
| KRAS |
|
| 0.063 | 73.7 | 94.9 | 0.819 |
| Exon2 (codon
12) |
|
| 0.125 | 66.7 | 87.9 | 0.718 |
|
G12D | 8 (24.2) | 5 (15.1) |
|
|
|
|
|
G12V | 3 (9.1) | 3 (9.1) |
|
|
|
|
|
G12A | 1 (3.0) | 0 (0.0) |
|
|
|
|
|
Wild-type | 21 (63.7) | 25 (75.8) |
|
|
|
|
| Exon2 (codon
13) |
|
| 1.000 | 80.0 | 97.0 | 0.872 |
|
G13D | 5 (15.2) | 4 (12.1) |
|
|
|
|
|
Wild-type | 28 (84.8) | 29 (87.9) |
|
|
|
|
| Exon3 (codon
61) |
|
| 1.000 | 100.0 | 100.0 | 1.000 |
|
Q61L | 2 (6.1) | 2 (6.1) |
|
|
|
|
|
Wild-type | 31 (93.9) | 31 (93.9) |
|
BRAF |
| Exon15 (codon
600) |
|
| 0.500 | 75.0 | 93.9 | 0.820 |
|
V600E | 8 (24.2) | 6 (18.2) |
|
|
|
|
|
Wild-type | 25 (75.8) | 27 (81.8) |
|
|
|
|
The sensitivity, consistency rate and κ score of
KRAS gene mutation detection in the serum exosome of patients was
calculated (Table III). Comparison
of the KRAS mutation status between tumor tissue and the matched
serum exosome identified that 84.8% (28/33) of patients with CRC
had the same three KRAS mutations (codon 12, 13 and 61; data not
shown). At codon 12 of exon 2, 8 (24.2%) patients with CRC were
positive for a KRAS mutation in their tumor tissue and 5 (15.1%) in
the serum exosome, giving a total consistency rate of 87.9%. At
codon 13 of exon 2, 4 (12.1%) patients with CRC were identified to
have mutant KRAS in their tumor tissue and serum exosome, with 1
(3.0%) patient positive for a KRAS mutation in their tumor tissue
only, giving a total consistency rate of 97.0%. The KRAS mutation
status was identical in tumor tissue-serum exosome matched samples
at codon 61 of exon 3 and therefore the consistency rate was 100%.
Overall, the sensitivity of exosomal mRNA KRAS mutation detection
was 73.7%, the specificity was 100% and the total consistency rate
was 94.9% (κ=0.819).
There was no significant association between KRAS
mutations and the gender, age, tumor site, tumor differentiation
and tumor stage of the patients, analyzed using the χ2
test (P>0.05; Table IV). However,
using the Student's t-test, the patients with a KRAS gene mutation
at codon 12 of exon 2 were found to be significantly older compared
with those patients without this mutation (tumor tissue, P=0.002;
serum exosome, P=0.022).
 | Table IV.Distribution of KRAS and BRAF
mutations according to the clinicopathological characteristics of
patients with colorectal cancer. |
Table IV.
Distribution of KRAS and BRAF
mutations according to the clinicopathological characteristics of
patients with colorectal cancer.
|
| KRAS mutation | BRAF mutation |
|---|
|
|
|
|
|---|
|
| Tumor tissue | Exosome | Tumor tissue | Exosome |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | No. (%) |
P-valuea | No. (%) |
P-valuea | No. (%) |
P-valuea | No. (%) |
P-valuea |
|---|
| Gender |
|
|
|
|
|
|
|
|
|
Male | 11/22 (50.0) | 0.508 | 7/20 (35.0) | 0.284 | 5/22 (22.7) | 1.000 | 3/20 (15.0) | 0.900 |
|
Female | 8/13 (61.5) |
| 7/13 (53.8) |
| 3/13 (23.1) |
| 3/13 (23.1) |
|
| Age, years |
|
>65 | 8/10 (80.0) | 0.120 | 6/10 (60.0) | 0.335 | 3/10 (30.0) | 0.849 | 3/10 (30.0) | 0.503 |
|
≤65 | 11/25 (44.0) |
| 8/23 (34.8) |
| 5/25 (20.0) |
| 3/23 (13.0) |
|
| Tumor site |
|
|
|
|
|
|
|
|
|
Colon | 14/21 (66.7) | 0.072 | 9/20 (45.0) | 0.710 | 5/21 (23.8) | 1.000 | 4/20 (20.0) | 1.000 |
|
Rectum | 5/14 (35.7) |
| 5/13 (38.5) |
| 3/14 (21.4) |
| 2/13 (15.4) |
|
| Tumor
differentiation |
|
|
|
|
|
|
|
|
| G1 | 0/4 (0.0) | 0.067 | 0/3 (0.0) | 0.203 | 0/4 (0.0) | 0.507 | 0/3 (0.0) | 0.589 |
| G2 | 12/20 (60.0) |
| 10/19 (52.6) |
| 5/20 (25.0) |
| 3/19 (15.8) |
|
| G3 | 7/11 (63.6) |
| 4/11 (36.4) |
| 3/11 (27.3) |
| 3/11 (27.3) |
|
| Tumor stage |
|
I–II | 10/21 (47.6) | 0.332 | 9/19 (47.4) | 0.503 | 3/21 (14.3) | 0.285 | 2/19 (10.5) | 0.383 |
|
III–IV | 9/14 (64.3) |
| 5/14 (35.7) |
| 5/14 (35.7) |
| 4/14 (28.6) |
|
BRAF mutation detection in tumor
tissues and matched serum exosomes
BRAF mutations were detected in tumor tissues and
matched serum exosomes (Table III).
For the 33 tumor tissue-serum exosome matched samples, a BRAF
mutation at codon 600 in exon 15 was detected in 8 (24.2%) of the
tumor tissues and 6 (18.2%) of the serum exosomes. Gene detection
was not possible in 2 serum exosomes, potentially due to a low
level of exosomes in the serum. A total of 93.9% (31/33) of
patients with CRC had the same BRAF mutation in their tumor tissue
and serum exosome matched samples (data not shown), and therefore
there was no significant difference in BRAF mutation distribution
(P=0.500; Table III). In 6 (18.2%)
patients with CRC, a mutated BRAF gene was detected in the tumor
tissue and serum exosome, and in 2 (6.1%) patients with CRC the
BRAF mutation was detected in their tumor tissue only. The
sensitivity of BRAF gene mutation detection in the serum exosome
was 75%, the specificity was 100% and the total consistency rate
was 93.9% (κ=0.820). There was no significant association between
BRAF mutations and the clinicopathological characteristics of the
patients through the χ2 test or the Student's t-test
(all P>0.05; Table IV).
Discussion
Colorectal cancer is becoming the fourth most common
malignant carcinoma in recent years (2). Early diagnosis and effective treatment
may significantly reduce the mortality rate of this disease and
these factors rely on accurate diagnosis, precise tumor staging and
gene mutation status analysis. Aside from the traditional
chemotherapy recommended by pathological diagnosis, epidermal
growth factor receptor (EGFR) -targeted therapy may be administered
according to the KRAS and BRAF gene mutation status of patients
with CRC (5). Gene evaluation of the
tumor tissue is the optimal standard for assessing mutation status,
but it is typically performed a single time as it is invasive and
costly. However, genomic alterations may differ in primary and
metastatic tumor tissues as the disease progresses, and monitoring
this requires repetitive genotyping. Other techniques that are in
clinical use or are the focus of previous studies are not
consistently successful; therefore novel methods allowing
repetitive monitoring of these genetic events are being
investigated (21–23).
It has been established that cancer initiation and
progression are associated with numerous genetic and epigenetic
factors, which may be detected through gene alternations in the
tumor tissue. DNA, mRNA and miRNA are released into the blood and
other bodily fluids from tumor tissues, and may be used to identify
tumor-associated genetic and epigenetic alterations. These products
may be more informative, specific and accurate compared with
protein biomarkers. Sorenson et al (24) and Vasioukhin et al (25) identified that a RAS gene mutation may
be detected in blood from cell-free DNA (cfDNA). Koyanagi et
al (26) and Mori et al
(27) identified that the association
between circulating tumor cells and methylated cfDNA can aid in the
assessment of disease severity and treatment efficacy in metastatic
melanoma. Qiu et al (28)
compared the diagnostic value of cfDNA and tumor tissue pathology,
the current optimal standard, using a meta-analysis of EGFR
mutations in non-small cell lung cancer, with the results
suggesting that cfDNA is a highly specific biomarker, but has low
sensitivity. However, other sources of cfDNA besides tumors exist
and cfDNA is not stable for longer periods of time, therefore cfDNA
has low sensitivity as a cancer biomarker (29–31).
Previous studies have suggested that quantitatively assaying fecal
DNA may provide a non-invasive method with improved sensitivity and
specificity for the detection and monitoring of cancer (32,33).
Ahlquist (34) identified mutated
KRAS and tumor protein 53 (p53) genes in fecal samples from
patients with CRC and associated these with the pathogenesis of the
disease. However, fecal DNA testing is clinically challenging, as
it is costly, time consuming and the results are variable due to
DNA degradation (32,35). Despite the challenges, peripheral
blood cfDNA and fecal DNA may provide the opportunity to
repetitively monitor patients with cancer that are difficult to
biopsy, but these methods have not yet been sufficiently
successful.
In addition, RNAs are detectable in serum and other
bodily fluids, and may also be a stable representation of exosomes
(14,36–40).
Exosomes are small membrane vesicles that are derived from the
endosomal membrane compartment, following the fusion of
multi-vesicular bodies with the plasma membrane, and have been
found in a number of body fluids, including serum, malignant
pleural effusion and urine (41–44).
Previous studies have reported that exosomes released from a number
of cell types, including immune, mesenchymal and cancer cells,
contained identical proteins, mRNAs, miRNAs and DNA fragments
(6–9).
Ogata-Kawata et al (45)
demonstrated that the serum exosome levels of seven miRNAs (let-7a,
miR-1229, miR-1246, miR-150, miR-21, miR-223 and miR-23a) were
significantly increased in patients with CRC compared with healthy
controls, and significantly downregulated following surgical tumor
resection. Furthermore, these miRNAs were also identified to be
secreted at significantly higher levels in colon cancer cell lines
compared with a normal colon-derived cell line (45). Skog et al (9) reported that serum exosomes were positive
for the EGFR variant III mutation when the parental glioblastoma
cells expressed the same mutation, and that the parental cells
exhibited a lower rate of this mutation (28 vs. 47%). It has been
reported that exosomes contain fragments of double-stranded genomic
DNA of >10 kb, which spans all chromosomes, and that mutations
in KRAS and p53 have been detected in pancreatic cancer cell lines
and the serum from patients with pancreatic cancer (46). Although previous studies have reported
that exosomes contain mitochondrial DNA, single-stranded DNA and
double-stranded genomic DNA (13,46,47), no
DNA was detected in exosomes derived from MC/9, BMMC or HMC-1 cells
(14). Therefore, the presence of
exosomal DNA is not consistent or it may be at low quantities that
cannot be analyzed. In addition, compared with membrane-binding
proteins, RNA/DNA-binding proteins and lipoprotein complexes,
exosomes remain stable despite the presence of RNases, proteases
and adverse physical conditions, and may be stored at 4°C for 96 h,
at −70°C for long periods of time and endure multiple freeze-thaw
cycles (14,48–51). Due
to these characteristics, exosomal RNA has been considered as a
potential novel biomarker for predictive analysis in patients with
cancer.
According to previous studies, the KRAS and BRAF
gene mutation rate of tumor tissue differs from 20–50%, but is
considered to be 35–45% in patients with CRC (52–54). The
fraction of plasma exosomes in patients with CRC has been reported
to be statistically higher compared with healthy controls (55). However, there is a lack of previous
studies investigating the association between gene mutations in
serum exosome and tumor tissue in patients with CRC. In the present
study, KRAS and BRAF gene mutations were detected and the
consistency of detected gene mutations was compared between tumor
tissues and matched serum exosomes from patients with CRC. The
mutation status of tumor tissue served as the reference for
detectable mutations and was therefore compared with that of the
matched serum exosome. These results demonstrated that the KRAS
mutation rate was 57.6 and 42.4% and BRAF mutation rate was 24.2
and 18.2%, in the tumor tissues and the matched serum exosomes,
respectively. In serum exosomes, the sensitivity of KRAS and BRAF
mutation detection was 73.7 and 75% with the total consistency rate
of 94.9 and 93.9%, respectively, and the specificity of these two
gene mutations were 100%. Previous studies have reported a
detection rate of KRAS mutation in cfDNA of 3–50% in patients with
CRC (21–23), thus the efficacy of cfDNA screening
remains to be elucidated. The current study hypothesized that the
detection rate of mutation in exosomal RNA is higher compared with
that in cfDNA, as exosomal RNA were found to be enriched and stable
(9). However, the results of the
current study demonstrated that the KRAS mutation rate of serum
exosomal RNA was similar to that of cfDNA. The similar detection
rate may be due to the small sample size, the serum sample
preparation, the exosome collection method, the RNA or DNA
extraction method, or the sequencing method. In future, purifying
the serum exosome RNA may increase the mutation detection rate.
Aging is a consequence of the accumulation of
unrepaired naturally-occurring DNA damage. DNA damage typically
causes errors in DNA replication or repair, and these errors are
the primary source of mutations. Epigenetic alterations may also
occur as a result of environmental exposure. The present study
demonstrated that CRC patients with a KRAS mutation at codon 12 of
exon 2 in their tumor tissue and serum exosome were significantly
older compared with those without this mutation. It has also been
established that KRAS mutation is significantly higher in CRC
patients who are >50 years old in the Indian population
(56). However, no statistically
significant difference in the distribution of age was identified
according to KRAS mutation status of patients with CRC in American
(57) or Chinese (58) populations. Although the association
between KRAS mutation and age has not yet been fully elucidated, it
may be suggested that the KRAS gene mutation rate increases with
age and other factors, including chemotherapy, radiotherapy and
disease progression.
The present study identified that serum exosomal
mRNA detection may be effective for the repetitive and non-invasive
genotyping of patients with CRC, particularly in patients without
the opportunity for a biopsy prior to treatment selection. However,
the results of the current study were obtained from a small sample
size, thus further studies with a larger sample size in multicenter
settings are required to validate these results. The application of
exosomes as a cancer biomarker is the focus of current studies, but
not yet sufficiently optimized for clinical use. Whether the
diagnostic and predictive value of exosomal RNA is similar to DNA
from cancer tissues remains to be elucidated, but exosomes have the
potential to replace tissue samples in certain situations. Whether
exosomes may be used for clinical assessment, including overall
survival and progression-free survival, also remains to be
elucidated.
In conclusion, exosomal RNA has the potential to
replace existing cancer tissue and blood biomarkers to provide
information for diagnostic screens, personalized medicine and
treatment efficacy.
Acknowledgements
The present study was supported by the The First
Affiliated Hospital of The People's Liberation Army General
Hospital (Beijing, China). The authors would like to thank Dr Ning
Dong (The First Affiliated Hospital of The People's Liberation Army
General Hospital) for aiding with exosome collection and exosomal
cDNA synthesis and Dr Jayashri Ghosh (Temple University,
Philadelphia, PA, USA) for her assistance in editing the
language.
References
|
1
|
Hofsli E, Sjursen W, Prestvik WS, Johansen
J, Rye M, Tranø G, Wasmuth HH, Hatlevoll I and Thommesen L:
Identification of serum microRNA profiles in colon cancer. Br J
Cancer. 108:1712–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
National Comprehensive Cancer Network.
Guidelines for Treatment of Cancer By Site and Guidelines for
Dectection, Prevention, & Risk Reduction. https://www.nccn.org/professionals/physician_gls/f_guidelines.asp#siteAccessed.
December 24–2016.
|
|
3
|
Geiger TM and Ricciardi R: Screening
options and recommendations for colorectal cancer. Clin Colon
Rectal Surg. 22:209–217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hol L, de Jonge V, van Leerdam ME, van
Ballegooijen M, Looman CW, van Vuuren AJ, Reijerink JC, Habbema JD,
Essink-Bot ML and Kuipers EJ: Screening for colorectal cancer:
Comparison of perceived test burden of guaiac-based faecal occult
blood test, faecal immunochemical test and flexible sigmoidoscopy.
Eur J Cancer. 46:2059–2066. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
National Comprehensive Cancer Network.
Clinical Practice Guidelines in Oncology. Colon cancer Version 2.
2015 http://www.nccn.org/professionals/physician_gls/pdf/colon.pdfAccessed.
October 20–2015.
|
|
6
|
Baj-Krzyworzeka M, Szatanek R, Weglarczyk
K, Baran J, Urbanowicz B, Brański P, Ratajczak MZ and Zembala M:
Tumour-derived microvesicles carry several surface determinants and
mRNA of tumour cells and transfer some of these determinants to
monocytes. Cancer Immunol Immunother. 55:808–818. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deregibus MC, Cantaluppi V, Calogero R, Lo
Iacono M, Tetta C, Biancone L, Bruno S, Bussolati B and Camussi G:
Endothelial progenitor cell derived microvesicles activate an
angiogenic program in endothelial cells by a horizontal transfer of
mRNA. Blood. 110:2440–2448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ratajczak J, Miekus K, Kucia M, Zhang J,
Reca R, Dvorak P and Ratajczak MZ: Embryonic stem cell-derived
microvesicles reprogram hematopoietic progenitors: Evidence for
horizontal transfer of mRNA and protein delivery. Leukemia.
20:847–856. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Skog J, Würdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim CW, Lee HM, Lee TH, Kang C, Kleinman
HK and Gho YS: Extracellular membrane vesicles from tumor cells
promote angiogenesis via sphingomyelin. Cancer Res. 62:6312–6317.
2002.PubMed/NCBI
|
|
11
|
Ratajczak J, Wysoczynski M, Hayek F,
Janowska-Wieczorek A and Ratajczak MZ: Membrane-derived
microvesicles: Important and underappreciated mediators of
cell-to-cell communication. Leukemia. 20:1487–1495. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gould SJ, Booth AM and Hildreth JE: The
Trojan exosome hypothesis. Proc Natl Acad Sci USA. 100:10592–10597.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy
SL, Breakefield XO and Skog J: Tumour microvesicles contain
retrotransposon elements and amplified oncogene sequences. Nat
Commun. 2:1802011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim HK, Song KS, Park YS, Kang YH, Lee YJ,
Lee KR, Kim HK, Ryu KW, Bae JM and Kim S: Elevated levels of
circulating platelet microparticles, VEGF, IL-6 and RANTES in
patients with gastric cancer: Possible role of a metastasis
predictor. Eur J Cancer. 39:184–191. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobayashi M, Salomon C, Tapia J, Illanes
SE, Mitchell MD and Rice GE: Ovarian cancer cell invasiveness is
associated with discordant exosomal sequestration of Let-7 miRNA
and miR-200. J Transl Med. 12:42014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao W, Dong W, Zhang C, Saren G, Geng P,
Zhao H, Li Q, Zhu J, Li G, Zhang S and Ye M: Effects of the
epigenetic drug MS-275 on the release and function of
exosome-related immune molecules in hepatocellular carcinoma cells.
Eur J Med Res. 18:612013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu P, Liang H, Xue L, Yang C, Liu Y, Zhou
K and Jiang X: Potential clinical significance of plasma-based KRAS
mutation analysis using the COLD-PCR/TaqMan(®)-MGB probe
genotyping method. Exp Ther Med. 4:109–112. 2012.PubMed/NCBI
|
|
21
|
Thierry AR, Mouliere F, El Messaoudi S,
Mollevi C, Lopez-Crapez E, Rolet F, Gillet B, Gongora C, Dechelotte
P, Robert B, et al: Clinical validation of the detection of KRAS
and BRAF mutations from circulating tumor DNA. Nat Med. 20:430–435.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuo YB, Chen JS, Fan CW, Li YS and Chan
EC: Comparison of KRAS mutation analysis of primary tumors and
matched circulating cell-free DNA in plasmas of patients with
colorectal cancer. Clin Chin Acta. 433:284–289. 2014. View Article : Google Scholar
|
|
23
|
Perrone F, Lampis A, Bertan C, Verderio P,
Ciniselli CM, Pizzamiglio S, Frattini M, Nucifora M, Molinari F,
Gallino G, et al: Circulating free DNA in a screening program for
early colorectal cancer detection. Tumori. 100:115–121.
2014.PubMed/NCBI
|
|
24
|
Sorenson GD, Pribish DM, Valone FH, Memoli
VA, Bzik DJ and Yao SL: Soluble normal and mutated DNA sequences
from single-copy genes in human blood. Cancer Epidemiol Biomarkers
Prev. 3:67–71. 1994.PubMed/NCBI
|
|
25
|
Vasioukhim V, Anker P, Maurice P, Lyautery
J, Lederrey C and Stroun M: Point mutations of the N-ras gene in
the blood plasma DNA of patients with myelodysplastic syndrome or
acute myelogenous leukaemia. Br J Haematol. 86:774–779. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koyanagi K, Mori T, O'Day SJ, Martinez SR,
Wang HJ and Hoon DS: Association of circulating tumor cells with
serum tumor-related methylated DNA in peripheral blood of melanoma
patients. Cancer Res. 66:6111–6117. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mori T, O'Day SJ, Umatani N, Martinez SR,
Kitago M, Koyanagi K, Kuo C, Takeshima TL, Milford R, Wang HJ, et
al: Predictive utility of circulating methylated DNA in serum of
melanoma patients receiving biochemotherapy. J Clin Oncol.
23:9351–9358. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu M, Wang J, Xu Y, Ding X, Li M, Jiang
F, Xu L and Yin R: Circulating tumor DNA is effective for the
detection of EGFR mutation in non-small cell lung cancer: A
meta-analysis. Cancer Epidemiol Biomarkers Prev. 24:206–212. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fleischhacker M and Schmidt B: Circulating
nucleic acids (CNAs) and cancer-a survey. Biochim Biophys Acta.
1775:181–232. 2007.PubMed/NCBI
|
|
30
|
Emlen W and Mannik M: Effect of DNA size
and strandedness on the in vivo clearance and organ localization of
DNA. Clin Exp Immunol. 56:185–192. 1984.PubMed/NCBI
|
|
31
|
Wimberger P, Roth C, Pantel K,
Kasimir-Bauer S, Kimmig R and Schwarzenbach H: Impact of
platinum-based chemotherapy on circulating nucleic acid levels,
protease activities in blood and disseminated tumor cells in bone
marrow of ovarian cancer patients. Int J Cancer. 128:2572–2580.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ahlquist DA, Sargent DJ, Loprinzi CL,
Levin TR, Rex DK, Ahnen DJ, Knigge K, Lance MP, Burgart LJ,
Hamilton SR, et al: Stool DNA and occult blood testing for screen
detection of colorectal neoplasia. Ann Intern Med. 149:441–450,
W481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Potack J and Itzkowitz SH: Practical
advances in stool screening for colorectal cancer. J Natl Compr
Canc Netw. 8:81–92. 2010.PubMed/NCBI
|
|
34
|
Ahlquist DA: Next-generation stool DNA
testing: Expanding the scope. Gastroenterology. 136:2068–2073.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Davies RJ, Miller R and Coleman N:
Colorectal cancer screening: Prospects for molecular stool
analysis. Nat Rev Cancer. 5:199–209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cocucci E, Racchetti G and Meldolesi J:
Shedding microvesicles: Artefacts no more. Trends Cell Biol.
19:43–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zomer A, Vendrig T, Hopmans ES, van
Eijndhoven M, Middeldorp JM and Pegtel DM: Exosomes: Fit to deliver
small RNA. Commun Integr Biol. 3:447–450. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Russo F, Di Bella S, Nigita G, Macca V,
Laganà A, Giugno R, Pulvirenti A and Ferro A: miRandola:
Extracellular circulating microRNAs database. PLoS One.
7:e477862012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Andre F, Schartz NE, Movassagh M, Flament
C, Pautier P, Morice P, Pomel C, Lhomme C, Escudier B, Le Chevalier
T, et al: Malignant effusions and immunogenic tumour-derived
exosomes. Lancet. 360:295–305. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bard MP, Hegmans JP, Hemmes A, Luider TM,
Willemsen R, Severijnen LA, van Meerbeeck JP, Burgers SA,
Hoogsteden HC and Lambrecht BN: Proteomic analysis of exosomes
isolated from human malignant pleural effusions. Am J Respir Cell
Mol Biol. 31:114–121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pisitkun T, Shen RF and Knepper MA:
Identification and proteomic profiling of exosomes in human urine.
Proc Natl Acad Sci USA. 101:13368–13373. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ogata-Kawata H, Izumiya M, Kurioka D,
Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H,
et al: Circulating exosomal microRNAs as biomarkers of colon
cancer. PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kahlert C, Melo SA, Protopopov A, Tang J,
Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A and Kalluri R:
Identification of double-stranded genomic DNA spanning all
chromosomes with mutated KRAS and p53 DNA in the serum exosomes of
patients with pancreatic cancer. J Biol Chem. 289:3869–3875. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guescini M, Guidolin D, Vallorani L,
Casadei L, Gioacchini AM, Tibollo P, Battistelli M, Falcieri E,
Battistin L, Agnati LF and Stocchi V: C2C12 myoblasts release
micro-vesicles containing mtDNA and proteins involved in signal
transduction. Exp Cell Res. 316:1977–1984. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Onco. 110:13–21. 2008. View Article : Google Scholar
|
|
49
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vickers KC, Palmisano BT, Shoucri BM,
Shamburek RD and Remaley AT: MicroRNAs are transported in plasma
and delivered to recipient cells by high-density lipoproteins. Nat
Cell Biol. 13:423–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Khalyfa A and Gozal D: Exosomal miRNAs as
potential biomarkers of cardiovascular risk in children. J Transl
Med. 12:1622014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Benvenuti S, Sartore-Bianchi A, Di
Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S and Bardelli
A: Oncogenic activation of the RAS/RAF signaling pathway impairs
the response of metastatic colorectal cancers to antie-epidermal
growth factor receptor antibody therapies. Cancer Res.
67:2643–2648. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Neumann J, Zeindl-Eberhart E, Kirchner T
and Jung A: Frequency and type of KRAS mutations in routine
diagnostic analysis of metastatic colorectal cancer. Pathol Res
Pract. 205:858–862. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shen H, Yuan Y, Hu H, Zhong X, Ye XX, Li
MD, Fang WJ and Zheng S: Clinical significance of K-ras and BRAF
mutations in Chinese colorectal cancer patients. World J
Gastroenterol. 17:809–816. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Silva J, Garcia V, Rodriguez M, Compte M,
Cisneros E, Veguillas P, Garcia JM, Dominguez G, Campos-Martin Y,
Cuevas J, et al: Analysis of exosome release and its prognostic
value in human colorectal cancer. Gens Chromosomes Cancer.
51:409–418. 2012. View Article : Google Scholar
|
|
56
|
Bisht S, Ahmad F, Sawaimoon S, Bhatia S
and Das BR: Molecular spectrum of KRAS, BRAF and PIK3CA gene
mutation: Determination of frequency, distribution pattern in
Indian colorectal carcinoma. Med Oncol. 31:1242014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Phipps AI, Buchanan DD, Makar KW, Win AK,
Baron JA, Lindor NM, Potter JD and Newcomb PA: KRAS-mutation status
in relation to colorectal cancer survival: The joint impact of
correlated tumour markers. Br J Cancer. 108:1757–1764. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mao C, Zhou J, Yang Z, Huang Y, Wu X, Shen
H, Tang J and Chen Q: KRAS, BRAF and PIK3CA mutations and the loss
of PTEN expression in Chinese patients with colorectal cancer. PLoS
One. 7:e366532012. View Article : Google Scholar : PubMed/NCBI
|