Introduction
Cervical cancer (CC) is the second most common type
of female-specific cancer, ranking only behind breast cancer, and
the fourth leading cause of cancer-associated mortality in females
worldwide (1). The majority of cases
of CC, ~80%, occur in developing countries (2). As detection of CC precursors by
cytological examination and their eradication by laser vaporization
or cone biopsy are the most effective methods of preventing CC, the
carcinoma is treatable, and, in many cases, curable. However, once
cancer advances and spreads to the distant lymph nodes, the
prognosis is poor (3). Therefore, it
is essential to identify metastasis-associated molecules and to
improve our understanding of the underlying molecular mechanisms of
the metastasis of CC.
Ultraconserved regions (UCRs) are non-coding gene
sequences that are strictly conserved across mice, rats and humans.
UCRs regulate the expression and translation of mRNAs. It has been
demonstrated that UCRs encode non-coding RNAs (ncRNAs) that serve
as modulators of gene expression (4,5).
Previous genome expression profiling studies
demonstrated that transcribed ultraconserved RNAs (TUC-RNAs)
exhibit distinct profiles in human carcinomas (6,7).
Furthermore, certain TUC-RNAs are able to serve as oncogenes or
tumor suppressor genes in tumor development. For example,
transcribed ultraconserved element 73 (TUC73) has been demonstrated
to regulate cell proliferation and apoptosis in colorectal cancer
cell lines (6). However, the
regulation of the majority of TUC-RNAs and their underlying
molecular mechanisms remain unknown in CC.
TUC338 is a TUC-RNA that serves as a tumor oncogene.
Braconi et al (8) demonstrated
that TUC338 is markedly overexpressed in hepatocellular cancer
(HCC) cell lines and tissue, and it promotes tumor cellular
proliferation and modulates progression through regulation of the
cell cycle. However, the expression level and biological role of
TUC338 in CC remains unclear.
In the present study, the differential expression of
TUC338 in human CC samples was identified using reverse
transcription-quantitative polymerase chain reaction (RT-qPCR), and
the function of TUC338 in the migration and invasion of CC cells
was investigated. In addition, to understand the underlying
molecular mechanism of CC metastasis, the target gene of TUC338 was
further investigated. To the best of our knowledge, the present
study is the first to investigate the expression and mechanism of
TUC338 in CC migration and invasion. The results of the present
study identify TUC338 as a novel target for further therapeutic
studies of CC.
Materials and methods
CC tissue collection
CC tissue specimens (n=40) and normal cervical
tissues (n=40) were obtained from female patients with CC (age
range, 40–68 years) at the Department of Obstetrics and Gynecology
of the Jiangsu Subei People's Hospital, affiliated to Yangzhou
University (Yangzhou, China) from January 2015 to January 2016. At
the time of sample collection, the patients were untreated, and the
samples were taken during initial diagnosis as part of routine
examination.
The absence of tumor cells in the matched normal
tissues was confirmed by a pathologist. All tissues were obtained
during surgery and immediately stored in liquid nitrogen prior to
use. Approval for the present study was granted by the
Institutional Medical Research Ethics Committee of the Medical
College of Yangzhou University (Yangzhou, China). All patients
provided written informed consent.
Cell line culture
Human CC cell lines (HeLa and C33A) were purchased
from the Chinese Peking Union Medical College Cell Bank (Beijing,
China). All cell lines were maintained in RPMI-1640 ‘medium
(HyClone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented
with 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life
Sciences), 100 IU/ml penicillin and 100 mg/ml streptomycin
(Beyotime Institute of Biotechnology, Haimen, China) at 37°C in a
humidified atmosphere containing 5% CO2.
RT-qPCR detection and
quantification
Total RNA was isolated from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
Purification of RNA (1 µg) was performed using TRIzol®
Plus RNA Purification kit (Thermo Fisher Scientific, Inc.). RNA was
reverse-transcribed into cDNA using the PrimeScript First Strand
cDNA Synthesis kit (Takara Bio, Inc., Otsu, Japan), according to
the manufacturer's protocol. qPCR was performed using the UltraSYBR
Mixture kit (CWBIO; Kang Wei Biological Technology Co. Ltd.,
Beijing, China) on an Applied Biosystems 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions for qPCR were; pre-denaturation at 95°C
for 10 min, denaturing at 95°C for 15 sec and annealing/extending
at 60°C for 1 min for 40 cycles. U6 small nuclear ribonucleoprotein
RNA and β-actin mRNA were used as internal controls. All reactions
were run in triplicate using the following primers: TUC338 forward,
5′-GCAGCGACAGTGCGAGCT-3′ and reverse, 5′-TCCGAGTGAGTTAGGAAG-3′, U6
forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse,
5′-TTCACGAATTTGCGTGTCAT-3′, tissue inhibitor of metalloproteinase 1
(TIMP1) forward, 5′-CTTCTGGCATCCTGTTGT-3′ and reverse,
5′-ACTGCAGGTAGTGATGTG-3′, matrix metalloproteinase-9 (MMP9)
forward, 5′-GTGATTGACGACGCCTTT-3′ and reverse,
5′-CAACTCGTCATCGTCG-3′ and β-actin forward,
5′-GTCACCAACTGGGACGACAT-3′ and reverse, 5′-GAGGCGTACAGGGATAGCAC-3′.
The 2−ΔΔCq method was used for quantification
(9).
Transfection of CC cells
Full-length murine TIMP1 mRNA was cloned into the
MigRI-green fluorescent protein (GFP) retroviral vector at the
Noncoding RNA Center, Yangzhou University (Yangzhou, China) and the
sequence was confirmed by Sangon Biotech Co., Ltd. (Shanghai,
China). The vector was obtained from Dr Xiaoyun Dong (Temple
university, PA, USA). Small interfering RNAs (siRNAs) against
TUC338 were purchased from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA). The siRNA sequences (5′-3′) were as follows: siRNA-1,
UGACAGCCCUGGAGACUGA; siRNA-2, CCACAGGACAGGUACAGCA (8). HeLa and C33A cells were transfected with
60 nM TUC338 siRNA (8) or 0 nM siRNA
as the negative control (NC) by Lipofectamine® 2000
(Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.) for
48 h prior to further experiments, according to the manufacturer's
instructions. The transfected cells were incubated at 37°C with 5%
CO2. The TUC338 and TIMP1 RNA levels in the transfected
CC cells were quantified using RT-qPCR.
Nucleotide blast
In order to identify potential targets of TUC338,
the nucleotide Basic Local Alignment Search Tool (blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYTYPE=BlastSearch&LINK_LOC=blasthome)
was used. This provided complementary sequences of two genes TUC338
and TIMP1.
Dual-luciferase reporter assay
The full-length 3′-untranslated region (UTR) of
TIMP1 was amplified by PCR from genomic DNA and cloned into the
EcoRI and XhaI sites of a pGL3-BS vector (Promega Corporation,
Madison, WI, USA). The primers for TIMP1 3′-UTR were as follows:
forward, 5′-GTGAATTCATCCTGCCCGGAGTGGAA-3′ and reverse,
5′-GTTCTAGATTTCTGCTGGGTGGTAAC-3′. The mutant construct of TIMP1
3′-UTR was generated using a QuikChange™ mutagenesis kit
(Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA).
Co-transfection of reporter vectors and TUC338 siRNA or NC was
performed using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
After 48 h, dual-luciferase activity was determined using a
Dual-Luciferase Reporter Assay System (Promega Corporation)
according to the manufacturer's protocol.
Migration and invasion assay
In vitro cell migration and invasion assays were
performed using Transwell chambers as described previously
(10). For the migration assays,
5×104 cells (HeLa and C33A, transfected with TUC338
siRNA or NC) were added into the upper Transwell chamber (pore
size, 8 µm; BD Biosciences, Franklin Lakes, NJ, USA). For the
invasion assays, 1×105 cells (HeLa and C33A, transfected
with TUC338 siRNA or NC) were added into the upper Transwell
chamber (pore size, 8 µm) precoated with Matrigel (BD Biosciences).
In both assays, cells were plated in medium without serum and
medium containing 10% FBS in the lower chamber served as the
chemoattractant. Following incubation (14 h) at 37°C in a 5%
CO2 humidified atmosphere, the cells that had not
migrated or invaded through the pores were removed. The filters
were fixed in 90% alcohol and stained with crystal violet (0.1%, 20
min). A total of 5 random fields/chamber were counted using an
inverted microscope (Olympus, Japan), and each test was performed
in triplicate.
Western blot analysis
Proteins were extracted using cell lysis buffer for
western and immunoprecipitation (Beyotime Institute of
Biotechnology) according to the manufacturer's instructions.
Protein concentration was quantified using an enhanced
bicinchoninic acid assay kit (Beyotime Institute of Biotechnology),
according to the manufacturer's protocol. For western blot
analysis, equal amounts (5–10 µg) of total protein were heated to
100°C, and separated by 8.5% SDS-PAGE. Following electrophoresis,
proteins were blotted onto a polyvinylidene difluoride membrane and
blocked using bovine serum albumin for 2 h at room temperature.
Membranes were incubated with anti-TIMP1 antibody (1:1,000; catalog
no. 8946S; Cell Signaling Technology, Inc., Danvers, MA, USA),
anti-MMP9 antibody (1:1,000; catalog no. 13667; Cell Signaling
Technology, Inc.) or β-actin mouse monoclonal antibody (1:1,000;
catalog no. cat-AF0003; Beyotime Institute of Biotechnology)
overnight at 4°C. TIMP1 protein level was detected using a
horseradish peroxidase-conjugated anti-rabbit secondary
immunoglobulin G antibody (1:1,000; catalog no. A0208; Beyotime
Institute of Biotechnology) for 2 h at room temperature. Protein
bands were detected using a FluorChem FC2 Imaging system
(ProteinSimple, San Jose, CA, USA).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 16.0; SPSS, Inc., Chicago, IL, USA). All plots
were created using Microsoft Office Excel 2010 software (Microsoft
Corporation, Redmond, WA, USA) and GraphPad Prism software (version
5; GraphPad Software, Inc., La Jolla, CA, USA). Results are
expressed as the mean ± standard deviation from three independent
experiments. Differences were assessed using a two-tailed Student's
t-test. The association between TIMP1 and TUC338 expression was
tested using a two-tailed Pearson's correlation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of TUC338 is upregulated in
CC tissues and cell lines
As TUC338 has been reported to be upregulated in
HCC, TUC338 expression levels were investigated in human CC tissues
in the present study. The endogenous expression of TUC338 in human
CC tissues and adjacent normal cervical tissues was determined
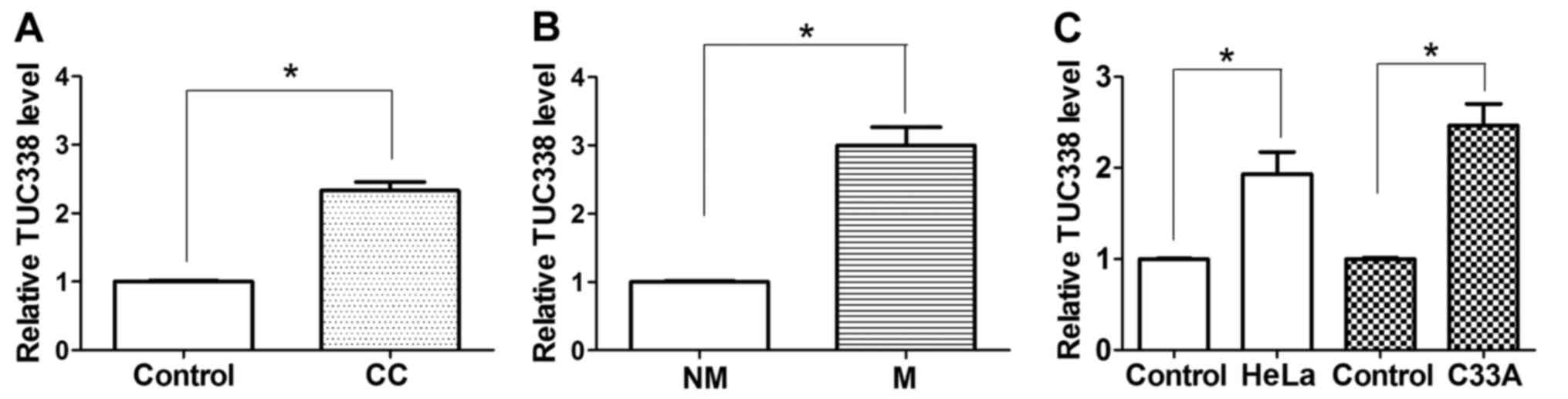
using RT-qPCR. As presented in Fig.
1A, expression of TUC338 was upregulated in 92.5% (37/40) of CC
tissues compared with the corresponding adjacent normal cervical
tissues. It was also identified that TUC338 expression was further
upregulated in 92.59% (25/27) of patients with CC with lymph node
metastasis compared with patients with CC without lymph node
metastasis (P<0.05) (Fig. 1B).
Similarly, the expression of TUC338 was observed to be
significantly increased in two CC cell lines compared with normal
human cervical tissue cells (Fig.
1C). These results suggest that increased TUC338 expression
serves an important role in CC development and metastasis.
Downregulated expression of TUC338 via
siRNA inhibits CC cell migration and invasion
On the basis of the aforementioned results, it was
investigated whether TUC338 served a role in regulating the
migration and invasion of CC cells. HeLa and C33A cells were
transfected with TUC338 siRNA or NC, and the transfection
efficiency was evaluated using cell invasion and migration assays.
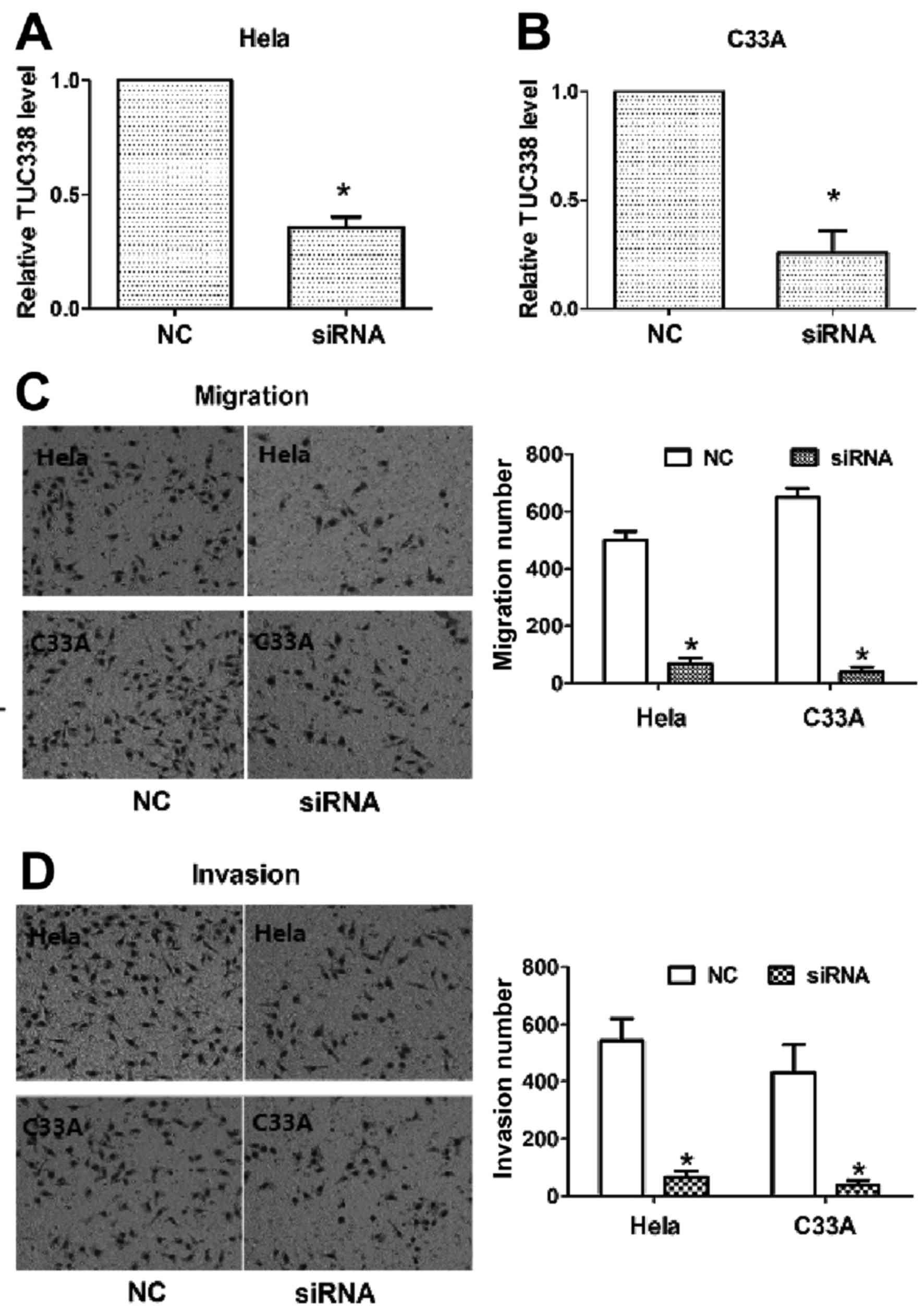
As expected, transfection with TUC338 siRNAs significantly
decreased TUC338 expression compared with the NC in HeLa and C33A
cells (Fig. 2A and B). Furthermore,
the cell migration and invasion assay demonstrated that
downregulated expression of TUC338 resulted in a significantly
decreased migration rate and invasion rate in HeLa and C33A cells
compared with the NC (Fig. 2C and D).
These results indicate that TUC338 acts as a tumor oncogene ncRNA
and contributed to the promotion of migration and invasion of CC
cells.
TIMP1 is a direct target of
TUC338
ncRNAs are known to modulate hundreds of mRNA
targets, resulting in global changes in the cell phenotype. In
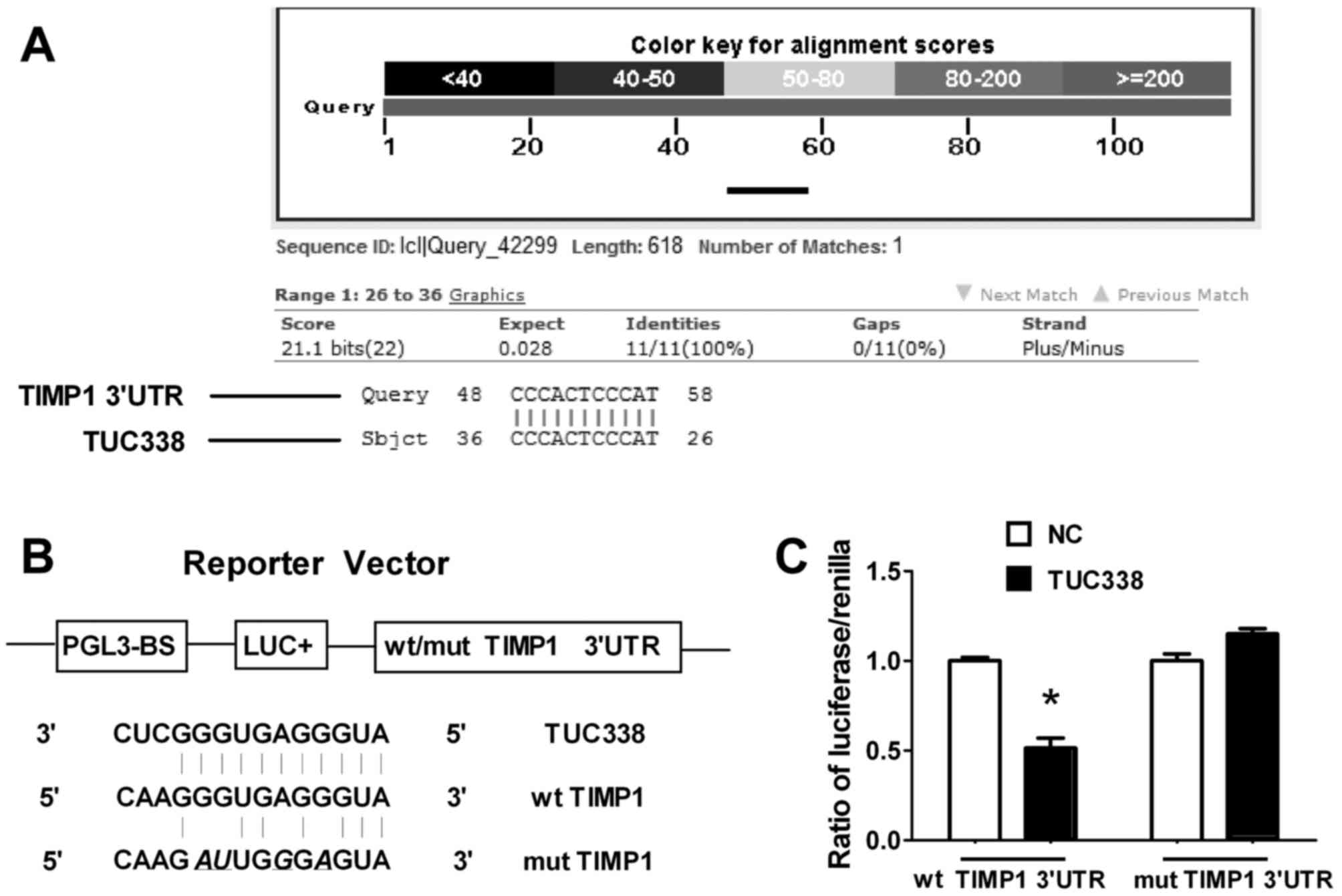
order to identify potential targets of TUC338, the nucleotide Basic
Local Alignment Search Tool (blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome)
was used. TIMP1 was identified as a putative target gene for TUC338
that mediates cell migration and invasion. To verify whether TUC338
targeted TIMP1 directly, a Dual-Luciferase reporter assay was
performed. As presented in Fig. 3, in
HeLa cells co-transfected with TIMP1-3′-UTR/pGL3-BS, transfection
with TUC338 siRNA led to a significant decrease in luciferase
activity compared with the group transfected with the NC
(P<0.05). This repressive effect was eliminated by point
mutations in the binding site of the TIMP1 3′-UTR. These results
indicated that TUC338 exerts inhibitory effects on TIMP1 expression
via interaction with the 3′-UTR of TIMP1.
TUC338 negatively regulates TIMP1
expression and upregulates matrix metalloproteinase 9 (MMP9)
expression
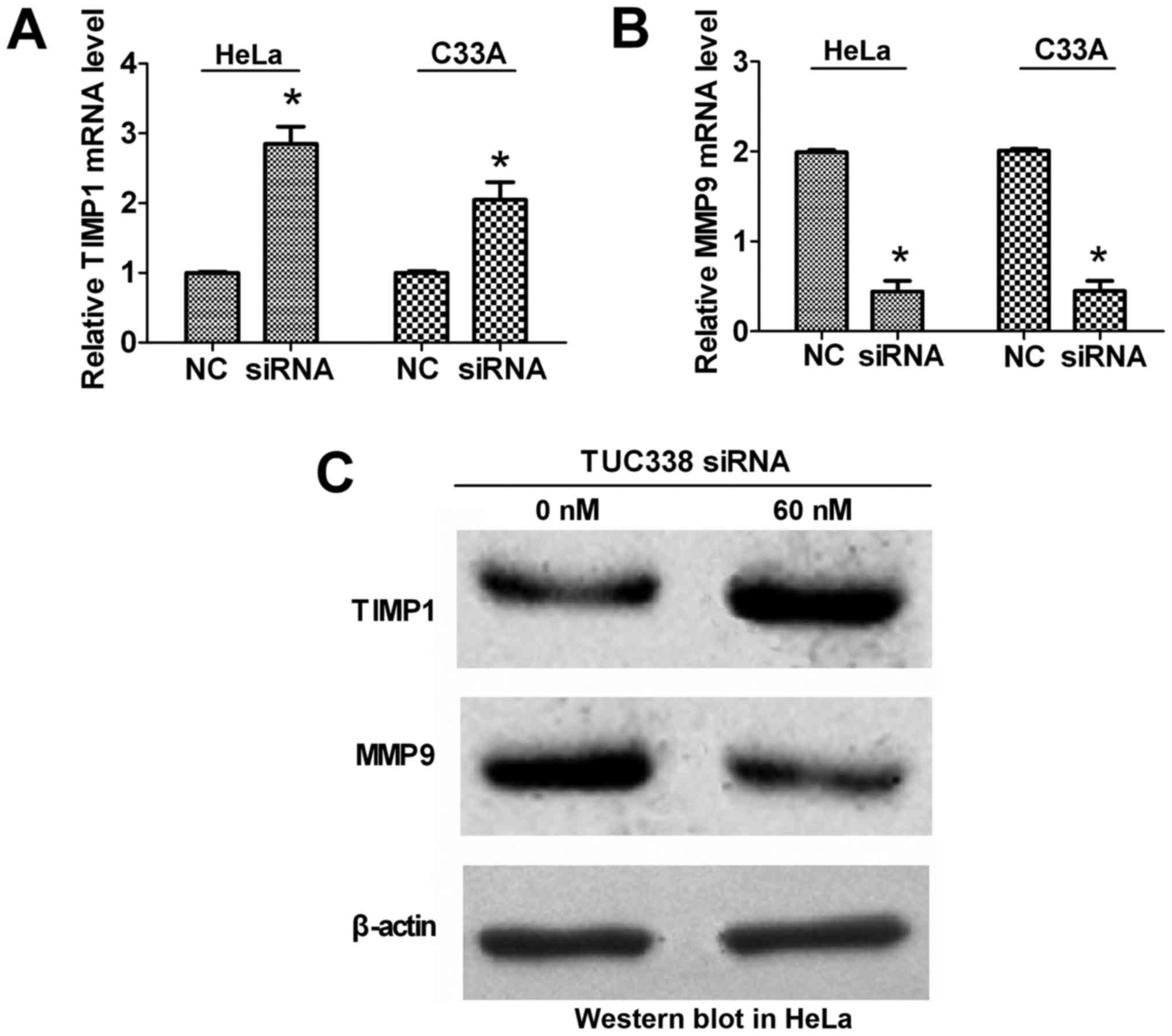
To further confirm that TIMP1 is a target gene of
TUC338 and that TIMP1 is a natural inhibitor of MMP9 (11), RT-qPCR and western blot analysis were
performed to detect the expression of TIMP1 and MMP9 in association
with TUC338 expression in HeLa and C33A cells. The results
indicated that expression of TIMP1 was significantly upregulated
and MMP9 was significantly downregulated at the mRNA level
(Fig. 4A and B) following siRNA
knockdown of TUC338 compared with the NC. At the protein level
(Fig. 4C), TIMP1 was markedly
upregulated and MMP9 was markedly downregulated following siRNA
knockdown of TUC338 compared with NC. These results suggest that
TUC338 negatively regulates the expression of its target gene TIMP1
at the post-transcriptional level, and that decreased expression of
TIMP1 results in increased expression of MMP9.
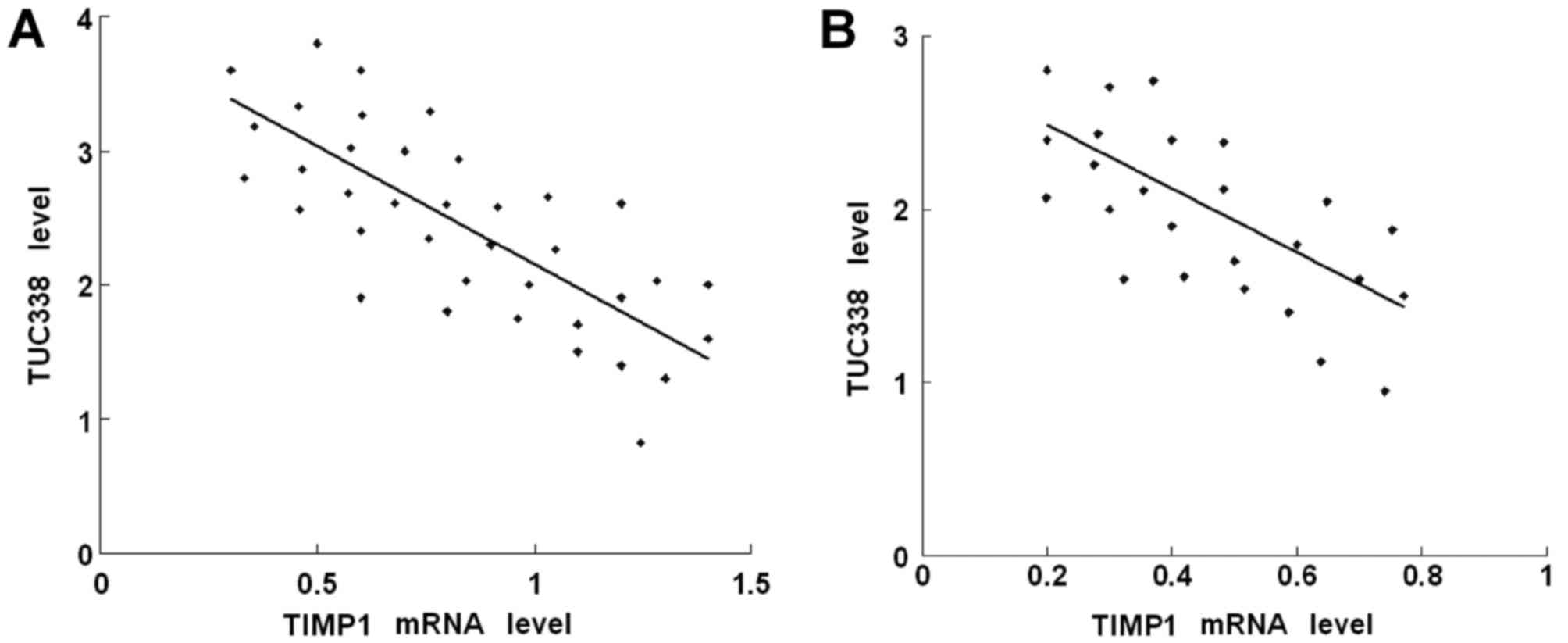
Expression levels of TUC338 and TIMP1
are inversely correlated at the RNA level
To assess the association between TUC338 and TIMP1
in CC, the mRNA levels of TUC338 and TIMP1 were determined in CC
and adjacent normal cervical tissues using RT-qPCR. As presented in
Fig. 5A, when the TIMP1 mRNA
expression levels were plotted against TUC338 mRNA expression
levels in up-regulated CC tissues, a significant inverse
association was identified (two-tailed Pearson's correlation
analysis, n=37, r=−0.57; P<0.05). Similarly, a significant
inverse association was also identified in CC specimens with lymph
node metastasis (Fig. 5B; two-tailed
Pearson's correlation analysis, n=25, r=−0.61; P<0.05).
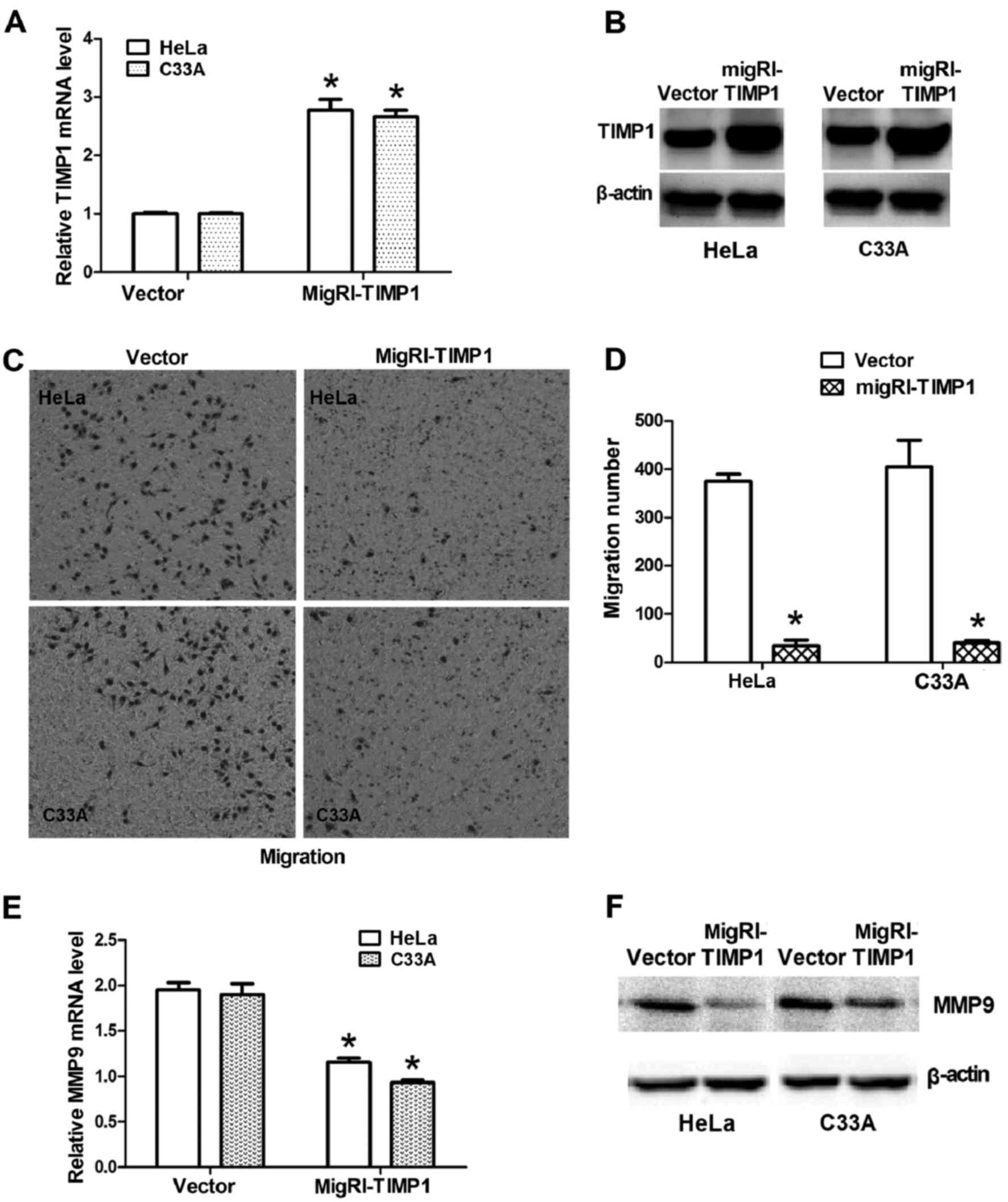
Overexpression of TIMP1 decreases the
migration potential of CC cells and upregulates MMP9
Expression of TIMP1 in CC cells was forced using
transfection with MigRI-TIMP1-GFP plasmids. TIMP1 mRNA levels were
significantly upregulated in HeLa and C33A cells following MigRI-
TIMP1-GFP transfection (Fig. 6A);
furthermore, TIMP1 protein levels were markedly upregulated in HeLa
and C33A cells following MigRI-TIMP1-GFP transfection (Fig. 6B). Consistently, overexpression of
TIMP1 markedly inhibited migration in CC cells (Fig. 6C and D), which resembled the
inhibitory effects of TUC338 knockdown. It was also confirmed that
the expression of MMP9 was significantly downregulated at the mRNA
level (Fig. 6E) and at the protein
level (Fig. 6F) following
overexpression of TIMP1. These results suggested that TIMP1
modulates the migration of CC cells and acts as an inhibitor of
MMP9 by negatively regulating its gene expression.
Discussion
UCRs are non-coding gene sequences that are strictly
conserved across mice, rats and humans. UCRs regulate the
expression and translation of mRNAs. By regulating protein
production post-transcriptionally, a number of UCRs act as
oncogenes or tumor suppressor genes (6–8,12,13).
However, the regulation of TUC338 and its underlying molecular
mechanisms of action in CC are unknown.
The results of the present study indicated that
TUC338 exhibited increased expression in CC tissues compared with
in matched normal cervical tissues. The results also indicated that
TUC338 siRNAs inhibited the invasion and migration of two CC cell
lines. These results suggest that TUC338, as a novel tumor
oncogene, serves a role in the metastasis and infiltration of CC.
In addition, TIMP1 was identified as a potential target of TUC338
using theoretical prediction software. TIMP1 is a member of the
TIMP family, which are natural inhibitors of the MMPs, a group of
peptidases involved in degradation of the extracellular matrix.
TIMP1 participates in the majority of the physiological and
pathological processes involved in cell proliferation, migration,
matrix remodeling, cell survival and matrix degradation (14–18). It
has also been reported that TIMP1 is overexpressed in colorectal
and lung tumors (19,20), and is associated with metastasis and
invasion in breast, upper urinary tract and oral squamous cell
tumors (19–22).
In the present study, an inverse association between
TUC338 and TIMP1 expression was identified in CC tissues. The
results also demonstrated that TIMP1 was negatively regulated by
TUC338 at the post-transcriptional level, via a specific target
site within the 3′-UTR, and that TUC338 inhibited CC cell migration
and invasion through the TIMP1-MMP9 signaling pathway. These
findings suggest that a decrease in TIMP1 expression, induced by
the suppression of TUC338, allows the progression of CC.
Furthermore, this implicates dysregulation of the TIMP1 signaling
pathway by UCRs as an important mechanism underlying cancer
metastasis, particularly in cancer cell migration and cell
invasion.
Metastasis is the movement of cancer cells from one
organ or tissue to another. Its sequential events include
detachment, migration, local invasion, formation of tumor embolus,
extravasation and establishment in different organs (23,24).
Certain ncRNAs are able to regulate tumor metastasis signaling
pathways (25). The identification of
TUC338 as an important regulator of tumor cell migration and
invasion in vitro emphasizes an essential role for this UCR
in mediating CC oncogenesis and tumor behavior.
In summary, the results of the present study
demonstrate that TUC338 is upregulated in CC, and that its
upregulation is significantly associated with lymph node
metastasis. Furthermore, siRNAs against TUC338 can inhibit the
invasion and migration of CC cells. TUC338 can directly inhibit
TIMP1 expression by targeting its 3′-UTR; TIMP1 was identified to
be downregulated and inversely associated with TUC338 levels in CC.
The results of the present study suggest that TUC338 is an
important oncogene in CC, and this knowledge may lead to novel
treatments to prevent CC metastasis.
Acknowledgements
The present study was supported by Yangzhou Key
Research Project-Social Development Plan (grant no. YZ2016065),
2016 High-level Talents Science Start-up Foundation in Yangzhou
University (grant no. 2016 WCH) and the China National Science
Foundation (grant no. 81273214).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kent A: HPV vaccination and testing. Rev
Obstet Gynecol. 3:33–34. 2010.PubMed/NCBI
|
|
3
|
Kyung MS, Kim HB, Seoung JY, Choi IY, Joo
YS, Lee MY, Kang JB and Park YH: Tumor size and lymph node status
determined by imaging are reliable factors for predicting advanced
cervical cancer prognosis. Oncol Lett. 9:2218–2224. 2015.PubMed/NCBI
|
|
4
|
Bejerano G, Lowe CB, Ahituv N, King B,
Siepel A, Salama SR, Rubin EM, Kent WJ and Haussler D: A distal
enhancer and an ultraconserved exon are derived from a novel
retroposon. Nature. 441:87–90. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katzman S, Kern AD, Bejerano G, Fewell G,
Fulton L, Wilson RK, Salama SR and Haussler D: Human genome
ultraconserved elements are ultraselected. Science. 317:9152007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lujambio A, Portela A, Liz J, Melo SA,
Rossi S, Spizzo R, Croce CM, Calin GA and Esteller M: CpG island
hypermethylation-associated silencing of non-coding RNAs
transcribed from ultraconserved regions in human cancer. Oncogene.
29:6390–6401. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Braconi C, Valeri N, Kogure T, Gasparini
P, Huang N, Nuovo GJ, Terracciano L, Croce CM and Patel T:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang C, Yan G, Zhang Y, Jia X and Bu P:
Long non-coding RNA MEG3 suppresses migration and invasion of
thyroid carcinoma by targeting of Rac1. Neoplasma. 62:541–549.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pietruszewska W, Bojanowska-Poźniak K and
Kobos J: Matrix metalloproteinases MMP1, MMP2, MMP9 and their
tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An
immunohistochemical study. Otolaryngol Pol. 70:32–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mestdagh P, Fredlund E, Pattyn F, Rihani
A, van Maerken T, Vermeulen J, Kumps C, Menten B, de Preter K,
Schramm A, et al: An integrative genomics screen uncovers ncRNA
T-UCR functions in neuroblastoma tumours. Oncogene. 29:3583–3592.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Catucci I, Verderio P, Pizzamiglio S,
Manoukian S, Peissel B, Barile M, Tizzoni L, Bernard L, Ravagnani
F, Galastri L, et al: SNPs in ultraconserved elements and familial
breast cancer risk. Carcinogenesis. 30:544–546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma J, Wang J, Fan W, Pu X, Zhang D, Fan C,
Xiong L, Zhu H, Xu N, Chen R and Liu S: Upregulated TIMP-1
correlates with poor prognosis of laryngeal squamous cell
carcinoma. Int J Clin Exp Pathol. 7:246–254. 2014.PubMed/NCBI
|
|
15
|
Lorente L, Martín MM, López P, Ramos L,
Blanquer J, Cáceres JJ, Solé-Violán J, Solera J, Cabrera J, Argueso
M, et al: Association between serum tissue inhibitor of matrix
metalloproteinase-1 levels and mortality in patients with severe
brain trauma injury. PLoS One. 9:e943702014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ramer R, Fischer S, Haustein M, Manda K
and Hinz B: Cannabinoids inhibit angiogenic capacities of
endothelial cells via release of tissue inhibitor of matrix
metalloproteinases-1 from lung cancer cells. Biochem Pharmacol.
91:202–216. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Niewiarowska K, Pryczynicz A,
Dymicka-Piekarska V, Gryko M, Cepowicz D, Famulski W, Kemona A and
Guzińska-Ustymowicz K: Diagnostic significance of TIMP-1 level in
serum and its immunohistochemical expression in colorectal cancer
patients. Pol J Pathol. 65:296–304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rojiani MV, Ghoshal-Gupta S, Kutiyanawalla
A, Mathur S and Rojiani AM: TIMP-1 overexpression in lung carcinoma
enhances tumor kinetics and angiogenesis in brain metastasis. J
Neuropathol Exp Neurol. 74:293–304. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ramos-DeSimone N, Hahn-Dantona E, Sipley
J, Nagase H, French DL and Quigley JP: Activation of matrix
metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1
cascade enhances tumor cell invasion. J Biol Chem. 274:13066–13076.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Morini M, Mottolese M, Ferrari N, Ghiorzo
F, Buglioni S, Mortarini R, Noonan DM, Natali PG and Albini A: The
alpha 3 beta 1 integrin is associated with mammary carcinoma cell
metastasis, invasion, and gelatinase B (MMP-9) activity. Int J
Cancer. 87:336–342. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Farina AR and Mackay AR: Gelatinase
B/MMP-9 in tumour pathogenesis and progression. Cancers (Basel).
6:240–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Groblewska M, Siewko M, Mroczko B and
Szmitkowski M: The role of matrix metalloproteinases (MMPs) and
their inhibitors (TIMPs) in the development of esophageal cancer.
Folia Histochem Cytobiol. 50:12–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klein CA: Cancer. The metastasis cascade.
Science. 321:1785–1787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sreekumar R, Sayan BS, Mirnezami AH and
Sayan AE: MicroRNA control of invasion and metastasis pathways.
Front Genet. 2:582011. View Article : Google Scholar : PubMed/NCBI
|