Introduction
Pituitary adenoma (PA) accounts for between 10 and
25% of all types of intracranial neoplasm, with an estimated
prevalence in the general population of ~17% (1). PA may develop at any age, but primarily
occurs at between 30 and 40 years old, with an equal prevalence
among males and females. Pathologically, PA is classified into the
following subtypes: Non-functioning (NF), growth hormone
(GH)-secreting, prolactin (PRL)-secreting, thyroid-stimulating
hormone (TSH)-secreting and adrenocorticotrophic hormone
(ACTH)-secreting (2,3). Furthermore, NFPA is classified into null
cell adenoma, which does not secrete hormones and gonadotroph
adenoma (GA), which secretes luteinizing hormone (LH),
follicle-stimulating hormone (FSH), and their subunits (4). Currently, surgery is the first-line
treatment for patients with PA.
Postoperative residual tissues/cells and subsequent
recurrence remain problematic (5–7). However,
gene therapy presents an effective approach to solve this problem.
Previous studies have demonstrated that mitochondrial dysfunction,
oxidative stress, cell-cycle dysregulation and abnormal
mitogen-activated protein kinase signaling are significantly
associated with the occurrence of PA (8–10). These
studies provide novel insights into the underlying molecular
mechanism of human PA pathogenesis, and provide opportunities for
in-depth investigations on PA and gene therapy discovery (8–10).
The Notch signaling pathway is a short-range
communication transducer that is involved in the regulation of
numerous cellular processes, such as cell fate, maintenance of stem
cells, apoptosis during development and renewal of adult tissue
(11–13). In humans, there are four known Notch
receptors (Notch1, 2, 3 and 4), two Jagged ligands (Jagged 1 and 2)
and three δ-like canonical Notch ligands [(Dll) 1, Dll3 and Dll4]
(14). The Notch receptor is
activated by its ligands, and the intracellular domain of Notch
(Notch-IC) is separated by α- and γ-secretase prior to entry into
the nucleus. Binding of Notch-IC to a ubiquitous transcription
factor activates the transcription of Notch-targeted genes
(12,13,15).
Notch-ligand interactions participate in the pathogenesis of a
number of human diseases, including the formation and progression
of PA. A study conducted by Moreno et al (16) revealed that Notch3 was overexpressed
in human NFPA, but not in GH- and PRL-secreting adenoma. In
addition, this was coupled with the downregulation of Dll1 ligand
expression, which was identified through gene expression profiling,
reverse transcription-quantative polymerase chain reaction
(RT-qPCR) and proteomic analyses. Another previous study
demonstrated a significant upregulation of Notch3 mRNA and protein
expression in NFPA (17). In our
previous study, RT-qPCR and western blot analyses demonstrated that
the upregulation of the Notch3 receptor and Jagged1 ligand occurs
in human NFPA, but not in normal human PA or hormone-secreting
adenoma (18). However, the
expression levels and functions of the other Notch receptors, and
ligands in PA remain to be reported. In the present study, the role
of Notch1, and its ligands Dll1, Dll3 and Dll4, was investigated in
various types of PA. To the best of our knowledge, the present
study is the first to describe the associations between the
differential expression of Notch1, Dll1, Dll3 and Dll4. This may
aid in the development of gene therapies for treatment of patients
with PA.
Materials and methods
Patients and tissue samples
A total of 16 PA tissue samples were collected from
patients who underwent endoscopic transsphenoidal surgery at the
Beijing Tiantan Hospital (Beijing, China). Written informed consent
was obtained from all patients and the study protocol was approved
by the Ethics Committee of Beijing Tiantan Hospital. All samples
were rinsed in sterile saline, snap-frozen in liquid nitrogen and
subsequently stored in liquid nitrogen until required for analysis.
Clinicopathological characteristics of the patients are summarized
in Table I. Individual PA samples
were classified on the basis of the profile of adenohypophyseal
hormone content by histological and immunohistochemical analyses
prior to molecular analysis.
 | Table I.Clinicopathological characteristics
of 16 patients with pituitary adenoma. |
Table I.
Clinicopathological characteristics
of 16 patients with pituitary adenoma.
| Patient ID | Sex | Age, years | Tumor size, cm | Clinical
characteristic | Immunohistochemical
analysis |
|---|
| 1 | M | 43 | 2.8 | Headache and
hypopituitarism | NF- |
| 2 | F | 50 | 4.3 | Headache and visual
defects | NF- |
| 3 | M | 57 | 2.0 | Headache | NF- |
| 4 | M | 46 | 3.5 | Visual defects | NF- |
| 5 | F | 42 | 1.7 | Headache | NF+: FSH+ |
| 6 | M | 32 | 3.6 | Headache and visual
defects | NF+: LH+, FSH+ |
| 7 | F | 53 | 3.0 | Visual defects | NF+: FSH+ |
| 8 | M | 45 | 2.1 | Symptomless | NF+: FSH+ |
| 9 | M | 31 | 1.5 | Acromegaly | GH+ |
| 10 | F | 47 | 1.6 | Acromegaly | GH+ |
| 11 | M | 39 | 2.0 | Acromegaly | GH+ |
| 12 | F | 29 | 3.1 | Acromegaly | GH+ |
| 13 | F | 63 | 3.4 |
Hyperprolactinemia | PRL+ |
| 14 | M | 29 | 2.2 |
Hyperprolactinemia | PRL+ |
| 15 | M | 43 | 1.9 |
Hyperprolactinemia | PRL+ |
| 16 | F | 44 | 2.6 |
Hyperprolactinemia | PRL+ |
Protein preparation and western blot
analysis
The resected PA samples were thawed and homogenized
in lysis buffer (Abcam., Cambridge, MA, USA) using a handheld
microtissue homogenizer. The homogenate was subsequently
centrifuged at 12,000 × g for 15 min at 4°C and the
supernatant was denatured for 5 min at 95°C in loading buffer.
Protein concentrations were measured using the bicinchoninic acid
protein assay with bovine serum albumin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) as the standard control. Total proteins (60 µg)
were separated using SDS-PAGE on 8 or 10% gels, subsequently
transferred onto nitrocellulose membranes and incubated with 5%
non-fat milk in Tris-buffered saline containing Tween-20 (TBST) for
1 h at room temperature. Membranes were then probed overnight with
the corresponding primary antibody (Ab) at 4°C, followed by three
10-min washes with TBST. Subsequently, membranes were incubated
with horseradish peroxidase-conjugated secondary Abs at room
temperature for 1 h. The following rabbit primary Abs were used:
Monoclonal Notch1 (dilution, 1:2,000; catalog no., ab52627;
Abcam,), polyclonal DLL1 (dilution, 1:2,000; catalog no., ab84620;
Abcam), polyclonal DLL3 (dilution, 1:1,000; catalog no., ab63707;
Abcam) and polyclonal DLL4 (dilution, 1:2,000; catalog no., ab7280;
Abcam). Goat anti-rabbit IgG H&L (horseradish peroxidase)
secondary antibody was used (dilution, 1:5,000; catalog no.,
ab6721; Abcam). An enhanced chemiluminescence system (GE Healthcare
Life Sciences, Chalfont, UK) was used according to the
manufacturer's protocol in order to visualize the positive bands on
transparent medical X-ray film. The final data were subjected to
grayscale scanning and semi-quantitative analysis using Quantity
One software version 4.6.2 (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
RNA extraction and qPCR analysis
Total RNA was extracted from frozen PA samples
(40–60 mg) using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and first-strand cDNA
was synthesized from total RNA using the SuperScript™ First-Strand
Synthesis system with SuperScript II reverse transcriptase
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. qPCR was performed in an Applied
Biosystems 7500 Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using Platinum SYBR-Green/ROX qPCR
Supermix-UDG (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR was
performed using a 25-µl reaction volume containing 2X Master mix
(12.5 µl), forward/reverse primers (0.5 µl each, 10 µM; Table II), sample cDNA (1 µl), and double
distilled water (10.5 µl). The thermocycling conditions were as
follows: 50°C for 120 sec, 95°C for 120 sec, followed by 40 cycles
at 95°C for 15 sec and 60°C for 30 sec. Fluorescence of the PCR
products was measured following completion of the extension step.
mRNA expression levels of the genes of interest were determined
from the threshold cycle (Cq) and the relative expression levels of
the genes examined were normalized relative to that of GAPDH and
quantified using the 2−ΔΔCq method (7500 software
version 2.3; Thermo Fisher Scientific, Inc.) (19).
 | Table II.Quantitative polymerase chain
reaction primers. |
Table II.
Quantitative polymerase chain
reaction primers.
| Gene name | Product size,
bp | Forward sequence
(5′-3′) | Reverse sequence
(5′-3′) | Temperature,
°C |
|---|
| Notch1 | 172 |
AAGCTGCATCCAGAGGCAAAC |
TGGCATACACACTCCGAGAACAC | 64.0 |
| DLL1 | 112 |
GATGTGATGAGCAGCATGGA |
CCATGGAGACAGCCTGGATA | 60.8 |
| DLL3 | 205 |
AATCGCCCTGAAGATGTAGACC |
GCACCACCGAGCAAATACAA |
61.70 |
| DLL4 | 115 |
GGCCAACTATGCTTGTGAATGTC |
ACCTCGGTTCAGGCACTGTC | 63.0 |
Statistical analysis
All data are presented as the mean ± standard error.
Statistical analyses of protein expression between tumor types were
performed using Student's t-test or non-parametric Mann-Whitney U
test. Correlations were identified using Pearson's rank-sum test.
P<0.05 was considered to indicate a statistically significant
difference. All analyses were performed using SPSS software
(version 17.0; SPSS Inc., Chicago, IL, USA).
Results
Tumor classification
The clinicopathological characteristics of the 16
adenoma tissue samples used in the present study are listed in
Table I. There were 9 male and 7
female patients. The mean patient age was 43.3 years (range, 29–63
years) and the mean tumor diameter was 2.6 cm (range, 1.5–4.3 cm).
There were 8 NFPA, 4 GH-secreting and 4 PRL-secreting adenoma
samples. The 4 NFPA samples were identified to be anterior
pituitary hormone-negative using immunohistochemical analysis and
were designated as NF- tumors. The remaining 4 NFPA samples were
stained with LH and/or FSH, and designated immunohistochemically
positive (NF+). The 4 PRL-secreting adenoma samples manifested as
hyperprolactinemia, whereas the 4 GH-secreting adenoma samples were
characterized as acromegaly. For the 8 patients with NFPA, headache
and visual defects were the main symptoms.
Western blot analysis
Western blot analysis demonstrated that Notch1
protein expression (Fig. 1A) was
highest in GA and GH-secreting adenoma samples, as compared with
null cell and PRL-secreting adenoma samples. Dll1 protein
expression (Fig. 1B) was increased in
GH-secreting and PRL-secreting adenoma samples, as compared with
null cell adenoma and GA (P<0.05). The highest level of Dll3
protein expression (Fig. 1C) occurred
in null cell adenoma and the lowest level was identified in
PRL-secreting adenoma. Significant differences were identified
between any two groups (P<0.05). Dll4 protein expression
(Fig. 1D) was highest in GH-secreting
adenoma and PRL-secreting adenoma, and lowest in null cell adenoma.
As a group, NFPA demonstrated lower Dll4 expression levels,
compared with GH-secreting and PRL-secreting adenoma samples (n=4;
P<0.05).
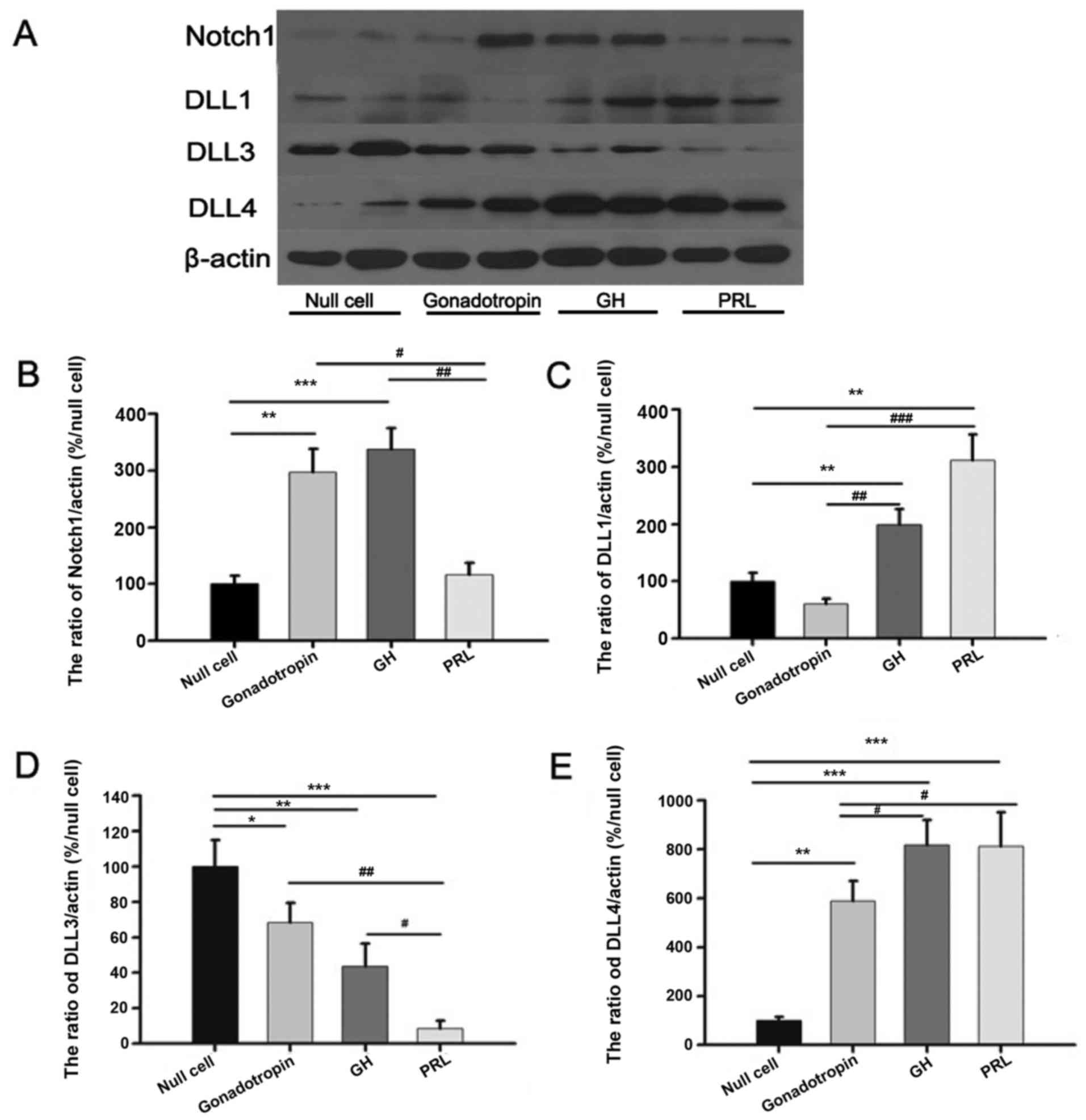 | Figure 1.Western blot analysis of tissue
samples from patients with pituitary adenoma. (A) Representative
western blot analysis of Notch1, Dll1, Dll3 and Dll4 protein
expression. Quantification of (B) Notch1, (C) Dll1, (D) Dll3 and
(E) Dll4 protein expression in null cell adenoma (n=4), GA (n=4),
GH-secreting (n=4) and PRL-secreting adenoma (n=4). β-actin was
used for normalization. Western blot analysis revealed increased
Notch1 expression in GA and GH-secreting adenoma (P=0.023).
Increased Dll1 expression was demonstrated in GH-secreting and
PRL-secreting adenoma (P=0.036). Increased Dll3 expression was
demonstrated in null cell adenoma (P=0.041). Decreased Dll3
expression was evident in PRL-secreting adenoma (P=0.037).
Increased Dll4 expression was detected in GH-secreting and
PRL-secreting adenoma (P=0.030). Decreased Dll4 expression was
evident in null cell adenoma (P=0.016). Dll, δ-like canonical Notch
ligand; GH, growth hormone; PRL, prolactin; GA, gonadotroph
adenoma. *Comparison between null cell and the other groups;
#comparison between the three groups except null
cell. |
qPCR analysis
Notch1 mRNA expression (Fig. 2A) was highest in GA and there were
significant differences compared with the other three groups
(P<0.05). Dll1 mRNA expression (Fig.
2B) was lowest in GH-secreting adenoma, whereas no significant
differences were identified between the other three groups
(P>0.05). Dll3 mRNA expression (Fig.
2C) was highest in GA and lowest in GH-secreting adenoma.
Significant differences compared with the other three groups were
identified (P<0.05). Dll4 mRNA expression (Fig. 2D) was lowest in GA and there were
significant differences compared with the other three groups
(P<0.05). No significant differences were identified between the
other three groups (P>0.05).
 | Figure 2.qPCR analysis results of tissue
samples from patients with pituitary adenoma. (A) qPCR analysis of
relative Notch1 mRNA expression in null cell adenoma (n=4), GA
(n=4), GH-secreting (n=4) and PRL-secreting adenoma (n=4).
Increased Notch1 expression was demonstrated in GA (P=0.048). (B)
qPCR analysis of relative Dll1 mRNA expression in null cell
adenoma, GA, GH-secreting and PRL-secreting adenoma samples. Dll1
expression was lowest in GH-secreting adenoma (P=0.036). (C) qPCR
analysis of relative Dll3 mRNA expression in null cell adenoma, GA,
GH-secreting and PRL-secreting adenoma. Increased Dll3 expression
was demonstrated in GA (P=0.028). Decreased Dll3 expression was
evident in GH-secreting adenoma (P=0.045). (D) qPCR analysis of
relative Dll4 mRNA expression in null cell adenoma, GA,
GH-secreting and PRL-secreting adenoma. Decreased Dll4 expression
was evident in GA (P=0.047). qPCR, quantitative polymerase chain
reaction; Dll, δ-like canonical Notch ligand; GH, growth hormone;
PRL, prolactin; GA, gonadotroph adenoma. *Comparison between null
cell and the other groups; #comparison between the three
groups except null cell. |
Associations between the expression
levels of Notch1, and Dll1, Dll3 and Dll4
Notch1 protein expression was demonstrated to be
positively associated with Dll4 protein expression (Fig. 3A), with a Pearson's correlation
coefficient of 0.825 (P=0.016). However, Notch1 protein expression
was identified to be negatively correlated with Dll3 protein
expression (Fig. 3B), with a
Pearson's correlation coefficient of −0.703 (P=0.012). No
significant association was identified between Notch1 and Dll1
protein expression.
Discussion
The Notch signaling pathway is highly conserved
among the majority of multicellular organisms, and is used to
determine cell fates and regulate pattern formation, whereas its
dysfunction results in a variety of developmental defects, and
adult pathologies (20). The Notch
signaling pathway may convey antitumor or tumor-promoting effects
in different tumor types depending on the microenvironment. Thus,
inhibition of the Notch signaling pathway may either inhibit or
promote tumor growth (21,22).
Abundant evidence indicates that Notch1 is involved
in the angiogenesis and tumorigenesis of various types of
malignancy. It has been revealed previously that Notch1 signaling
is the convergence point of numerous signaling pathways (23). The dysfunction of Notch1 may inhibit
cell differentiation, resulting in the malignant transformation of
undifferentiated cells. In addition, Notch1 has been reported to
induce cell cycle arrest and apoptosis in certain tumors, and may
induce epithelial-mesenchymal transition, which is consistent with
the cancer stem cell phenotype of pancreatic cancer cells (24). Previous research has demonstrated that
Notch1 has different functions in the various phases of tumor
development. Notch1 serves a promoting role in the majority of
carcinoma types, but serves an antitumor role in skin, lung, liver,
thyroid and breast cancer. Notch1 has a promoting effect in the
early stage of cervical cancer, while inhibiting tumor growth in
end-stage cervical cancer (25). A
possible explanation for the differences in Notch1 function may be
that it is highly conserved during biological evolution and
expressed in various tissues and cells. Furthermore, crosstalk
between Notch1 and other signaling pathways may give rise to the
diversity and complexity of Notch1 function (26).
The results of the present study provide evidence of
Notch1 expression in human PA. Notch1 expression was demonstrated
to be increased in GA and GH-secreting adenoma, as compared with
null cell and PRL-secreting adenoma, indicating that Notch1 can
stimulate and inhibit tumor growth, and hormone production of GA
and, GH-secreting adenoma, although the underlying molecular
mechanism remains unclear. However, ACTH- and TSH-secreting adenoma
were not investigated in the present study.
In a study by Gao et al (27), DLL1 overexpression was observed in
hepatic carcinoma, but not in healthy liver tissue. Increased DLL1
expression was able to increase the proliferative ability of
hepatic carcinoma, whereas decreased DLL1 expression may decrease
this proliferative ability, indicating the tumor-promoting effect
of DLL1 in hepatic carcinoma. Another study identified
overexpression of Notch1 and DLL1 mRNA, and protein levels in six
human glioma cell lines and brain glioma (28). Similarly, Somasundaram et al
(29) demonstrated DLL1
overexpression in diffuse astrocytoma, anaplastic astrocytoma and
secondary glioblastoma multiforme. Previous studies have
demonstrated that Dll1 expression was increased in GH-secreting and
PRL-secreting adenoma, compared with null cell adenoma and GA
(17,20). Coincidently, the results of the
present study demonstrated that DLL1 expression was decreased in
NFPA, as compared with GH-secreting adenoma and PRL-secreting
adenoma, as previously mentioned. It is hypothesized that Dll1
serves an essential role in the tumorigenesis of GH-secreting and
PRL-secreting adenoma.
Dll3 has also been implicated in tumorigenesis and
histogenesis. Maemura et al (30) reported that Dll3 was silenced by
methylation in human hepatocellular carcinoma and that it inhibits
the growth of hepatocellular carcinoma cells. Targeted deletion of
Dll3 in mouse causes a developmental defect in somite segmentation
and consequently severe disruption to vertebral formation,
resembling spondylocostal dysostosis in humans (31). Another study (32) demonstrated that mutations in the
Notch1 receptor and Dll3 ligand caused global disruption to axial
segmental patterning, as 30% of Dll3-Notch1 double heterozygous
animals exhibited localized segmental anomalies similar to
congenital vertebral defects in humans. In the present study, it
was demonstrated that Dll3 protein expression was highest in null
cell and lowest in PRL-secreting adenoma tissue samples.
Significant differences were identified between any two groups.
Therefore, Dll3 may be more important in NFPA, including null cell
adenoma and GA compared with hormone-secreting adenoma. However,
the function and specific underlying molecular mechanism of Dll3 in
PA remain to be elucidated.
As an important ligand of Notch, Dll4 was
demonstrated to be significantly overexpressed in tumor vessels
compared with the surrounding blood vessels when it was first
detected in a previous study (33).
Abundant evidence describes the participation of Dll4 in tumor
angiogenesis. A study conducted by Mailhos et al (34) reported that Dll4 expression was
elevated in tumor vascular endothelial cells. Tumor hypoxia induces
the upregulation of Dll4, which regulates downstream gene
expression by activating the Dll4-Notch signaling pathway to reduce
dysfunctional tumor vascular density and promote tumor growth
(35–37). Another study (38) concluded that vascular endothelial
growth factor (VEGF) upregulates angiogenesis, whereas Dll4
inhibits VEGF-induced endothelial cell function to prevent
excessive blood vessel formation. The results of the present study
demonstrated that Dll4 protein expression was highest in GH- and
PRL-secreting adenoma, and lowest in null cell adenoma tissue. As a
group, Dll4 protein expression was revealed to be lower in NFPA
compared with GH- and PRL-secreting adenoma. These data suggest
that Dll4 serves a more important role in the regulation of
angiogenesis, and growth of GH- and PRL-secreting adenoma.
To the best of our knowledge, the results of the
present study provide the first evidence to demonstrate the
differential expression of the Notch1 receptor, and its ligands
Dll1, Dll3 and Dll4 in various types of human PA at the mRNA and
protein level. It was identified that Notch1 protein expression was
positively associated with Dll4 protein expression, but negatively
associated with Dll3 protein expression (r=0.815 and −0.703,
respectively), indicating possible synergistic effects of the
Notch1 receptor and its Dll4 ligand. Furthermore, the Dll3 ligand
may act as an inhibitor of the Notch1 receptor, indicating an
antagonistic association between the two. However, the molecular
mechanism underlying the interactions between Notch1 and its
ligands remains to be elucidated. In addition, no significant
association was identified between the expression levels of the
Notch1 receptor and Dll1 ligand.
The results of the present study are consistent with
previous studies on other tumors. Previous studies have revealed
that the Notch1-DLL4 signaling pathway participates in a range of
processes, including the formation, development, invasion and
metastasis of malignant tumors (39).
Dll4 interacts with Notch1 and associated transcription factors to
promote cell proliferation or apoptosis. A previous study
identified that Dll4 is an important ligand of the Notch1 signaling
pathway and involved in the regulation of tumor angiogenesis
(35). The Notch1-DLL4 signaling
pathway inhibits excessive vascularization to decrease the number
of new blood vessels, whereas in other tumors it accelerates the
development of new vessels and maturation to improve the function
of the new blood vessels (40).
The results of the present study suggest that there
is a potential Notch1-Dll4 positive feedback loop in PA cells. Dll4
in one cell can bind to Notch1 in a neighboring cell. This
interaction results in the proteolytic release of Notch-IC, which
translocates to the nucleus, and induces the expression of target
genes, including Hes/Hey family genes, cyclin D and nuclear
factor-κB (41). These genes activate
the Notch1 signaling pathway in neighboring cells to promote the
proliferation of adjacent PA cells. In the present study, Dll4
ligand expression was demonstrated to be upregulated in PA cells,
which further upregulates the activation of the Notch1 signaling
pathway. Therefore, Notch1 and Dll4 may promote cell proliferation
in PA or induce apoptosis and regulate angiopoiesis via synergistic
effects. Therefore, the blockade of the Notch1-DLL4 signaling
pathway may serve as a novel gene therapeutic strategy for the
treatment of patients with PA. It is plausible that modulators of
the Notch signaling pathway, such as γ-secretase inhibitors
developed for Alzheimer's disease, may be useful as pharmacological
regulators of PA.
Regarding the association between Notch1 and Dll3,
Ladi et al (42) performed
multiple assays that demonstrated that Dll3 does not activate Notch
signaling, as Notch did not bind to Dll3-expressing cells. However,
in a cell-autonomous manner, Dll3 suppressed Notch signaling. Dll3
functions as a dedicated inhibitor of Notch signaling. Chapman
et al (31) reported that Dll3
interacts with Notch1 in the late endocytic compartment and the
mechanism for Dll3-mediated cis-inhibition of Notch
signaling may involve Dll3 targeting newly synthesized Notch1 for
lysosomal degradation prior to post-translational processing and
cell-surface presentation of the receptor. An inhibitory role for
Dll3 in vivo is further supported by the observation of Dll3
protein and Notch1 signaling juxtaposition in the presomitic
mesoderm (31).
Following the results of the present study, it is
hypothesized that the Dll3 ligand does not activate Notch1 in PA,
but rather functions to autonomously inhibit signaling. The Dll3
ligand does not bind to Notch1 in neighboring cells and can
cis-inhibit ligand-dependent Notch1 activation when
expressed on the surface of the same cell as the Notch1 receptor.
Dll3 is a potent antagonist of ligand-induced Notch1 signaling when
co-expressed with Notch1. It is suggested that a potential
Notch1-Dll3 negative feedback loop exists in PA. Therefore,
elucidation of the negative feedback mechanism of Notch1 and Dll3
could aid in guiding the development of novel treatments for
patients with PA. However, the underlying molecular mechanism of
the interactions between Notch1 and Dll3/Dll4 in PA remains to be
elucidated.
In conclusion, the results of the present study
provide evidence of Notch1, Dll1, Dll3 and Dll4 expression in human
PA, indicating that synergistic interactions occur between Notch1
and DLL4 in PA, and the negative correlation between Notch1 and
DLL3 expression suggests the presence of a negative feedback loop.
Further studies are warranted to clarify the precise mechanism
underlying the involvement of the Notch1 signaling pathway in the
pathogenesis of PA.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 30971005,
2013BAI09B03 and 201402008).
References
|
1
|
Ezzat S, Asa SL, Couldwell WT, Barr CE,
Dodge WE, Vance ML and McCutcheon IE: The prevalence of pituitary
adenomas. Cancer. 101:613–619. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ironside JW: Best Practice No 172:
Pituitary gland pathology. J Clin Pathol. 56:561–568. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Scanarini M and Mingrino S: Functional
classification of pituitary adenomas. Acta Neurochir (Wien).
52:195–202. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korbonits M and Carlsen E: Recent clinical
and pathophysiological advances in non-functioning pituitary
adenomas. Horm Res. 71 Suppl 2:S123–S130. 2009.
|
|
5
|
Jaffe CA: Clinically non-functioning
pituitary adenoma. Pituitary. 9:317–321. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Daly AF, Rixhon M, Adam C, Dempegioti A,
Tichomirowa MA and Beckers A: High prevalence of pituitary
adenomas: A cross-sectional study in the province of Liege,
Belgium. J Clin Endocrinol Metab. 91:4769–4775. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Greenman Y and Stern N: Non-functioning
pituitary adenomas. Best Pract Res Clin Endocrinol Metab.
23:625–638. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhan X and Desiderio DM: Signaling pathway
networks mined from human pituitary adenoma proteomics data. BMC
Med Genomics. 3:132010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Indraccolo S, Minuzzo S, Masiero M and
Amadori A: Ligand-driven activation of the notch pathway in T-ALL
and solid tumors: Why Not(ch)? Cell Cycle. 9:80–85. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Melmed S: Pathogenesis of pituitary
tumors. Nature Rev Endocrinol. 7:257–266. 2011. View Article : Google Scholar
|
|
11
|
Bianchi S, Dotti MT and Federico A:
Physiology and pathology of notch signalling system. J Cell
Physiol. 207:300–308. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai EC: Notch signaling: Control of cell
communication and cell fate. Development. 131:965–973. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greenwald I: LIN-12/Notch signaling:
Lessons from worms and flies. Genes Dev. 12:1751–1762. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Artavanis-Tsakonas S, Matsuno K and
Fortini ME: Notch signaling. Science. 268:225–232. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moreno CS, Evans CO, Zhan X, Okor M,
Desiderio DM and Oyesiku NM: Novel molecular signaling and
classification of human clinically nonfunctional pituitary adenomas
identified by gene expression profiling and proteomic analyses.
Cancer Res. 65:10214–10222. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miao Z, Miao Y, Lin Y and Lu X:
Overexpression of the Notch3 receptor in non-functioning pituitary
tumours. J Clin Neurosci. 19:107–110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu R, Gao H, Wang H, Cao L, Bai J and
Zhang Y: Overexpression of the Notch3 receptor and its ligand
Jagged1 in human clinically non-functioning pituitary adenomas.
Oncol Lett. 5:845–851. 2013.PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Z, Li Y, Kong D and Sarkar FH: The
role of Notch signaling pathway in epithelial-mesenchymal
transition (EMT) during development and tumor aggressiveness. Curr
Drug Targets. 11:745–751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu XB, Feng F, Wang YC, Wang L, He F, Dou
GR, Liang L, Zhang HW, Liang YM and Han H: Blockade of Notch
signaling in tumor-bearing mice may lead to tumor regression,
progression, or metastasis, depending on tumor cell types.
Neoplasia. 11:32–38. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Roy M, Pear WS and Aster JC: The
multifaceted role of Notch in cancer. Curr Opin Genet Dev.
17:52–59. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weijzen S, Rizzo P, Braid M, Vaishnav R,
Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC,
et al: Activation of Notch-1 signaling maintains the neoplastic
phenotype in human Ras-transformed cells. Nat Med. 8:979–986. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bao B, Wang Z, Ali S, Kong D, Li Y, Ahmad
A, Banerjee S, Azmi AS, Miele L and Sarkar FH: Notch-1 induces
epithelial-mesenchymal transition consistent with cancer stem cell
phenotype in pancreatic cancer cells. Cancer Lett. 307:26–36. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, de Marco MA, Graziani I, Gazdar
AF, Strack PR, Miele L and Bocchetta M: Oxygen concentration
determines the biological effects of Notch-1 signaling in
adenocarcinoma of the lung. Cancer Res. 67:7954–7959. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koch U and Radtke F: Notch and cancer: A
double-edged sword. Cell Mol Life Sci. 64:2746–2762. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao J, Chen C, Hong L, Wang J, Du Y, Song
J, Shao X, Zhang J, Han H, Liu J and Fan D: Expression of Jagged1
and its association with hepatitis B virus X protein in
hepatocellular carcinoma. Biochem Biophys Res Commun. 356:341–347.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Purow BW, Haque RM, Noel MW, Su Q, Burdick
MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, et al:
Expression of Notch-1 and its ligands, Delta-Like-1 and Jagged-1,
is critical for glioma cell survival and proliferation. Cancer Res.
65:2353–2363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Somasundaram K, Reddy SP, Vinnakota K,
Britto R, Subbarayan M, Nambiar S, Hebbar A, Samuel C, Shetty M,
Sreepathi HK, et al: Upregulation of ASCL1 and inhibition of Notch
signaling pathway characterize progressive astrocytoma. Oncogene.
24:7073–7083. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maemura K, Yoshikawa H, Yokoyama K, Ueno
T, Kurose H, Uchiyama K and Otsuki Y: Delta-like 3 is silenced by
methylation and induces apoptosis in human hepatocellular
carcinoma. Int J Oncol. 42:817–822. 2013.PubMed/NCBI
|
|
31
|
Chapman G, Sparrow DB, Kremmer E and
Dunwoodie SL: Notch inhibition by the ligand DELTA-LIKE 3 defines
the mechanism of abnormal vertebral segmentation in spondylocostal
dysostosis. Hum Mol Genet. 20:905–916. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Loomes KM, Stevens SA, O'Brien ML,
Gonzalez DM, Ryan MJ, Segalov M, Dormans NJ, Mimoto MS, Gibson JD,
Sewell W, et al: Dll3 and Notch1 genetic interactions model axial
segmental and craniofacial malformations of human birth defects.
Dev Dyn. 236:2943–2951. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Patel NS, Li JL, Generali D, Poulsom R,
Cranston DW and Harris AL: Up-regulation of delta-like 4 ligand in
human tumor vasculature and the role of basal expression in
endothelial cell function. Cancer Res. 65:8690–8697. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mailhos C, Modlich U, Lewis J, Harris A,
Bicknell R and Ish-Horowicz D: Delta4, an endothelial specific
notch ligand expressed at sites of physiological and tumor
angiogenesis. Differentiation. 69:135–144. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li JL, Sainson RC, Shi W, Leek R,
Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E,
Hainfellner JA and Harris AL: Delta-like 4 Notch ligand regulates
tumor angiogenesis, improves tumor vascular function, and promotes
tumor growth in vivo. Cancer Res. 67:11244–11253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Djokovic D, Trindade A, Gigante J, Badenes
M, Silva L, Liu R, Li X, Gong M, Krasnoperov V, Gill PS and Duarte
A: Combination of Dll4/Notch and Ephrin-B2/EphB4 targeted therapy
is highly effective in disrupting tumor angiogenesis. BMC Cancer.
10:6412010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Al Haj, Zen A, Oikawa A, Bazan-Peregrino
M, Meloni M, Emanueli C and Madeddu P: Inhibition of
delta-like-4-mediated signaling impairs reparative angiogenesis
after ischemia. Circ Res. 107:283–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Williams CK, Li JL, Murga M, Harris AL and
Tosato G: Up-regulation of the Notch ligand Delta-like 4 inhibits
VEGF-induced endothelial cell function. Blood. 107:931–939. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Weng AP and Aster JC: Multiple niches for
Notch in cancer: Context is everything. Curr Opin Genet Dev.
14:48–54. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hellström M, Phng LK, Hofmann JJ, Wallgard
E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N,
et al: Dll4 signalling through Notch1 regulates formation of tip
cells during angiogenesis. Nature. 445:776–780. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bray SJ: Notch signalling: A simple
pathway becomes complex. Nat Rev Mol Cell Biol. 7:678–689. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ladi E, Nichols JT, Ge W, Miyamoto A, Yao
C, Yang LT, Boulter J, Sun YE, Kintner C and Weinmaster G: The
divergent DSL ligand Dll3 does not activate Notch signaling but
cell autonomously attenuates signaling induced by other DSL
ligands. J Cell Biol. 170:983–992. 2005. View Article : Google Scholar : PubMed/NCBI
|

















