Introduction
Esophageal cancer is the eighth most common type of
cancer in the world, making it a serious threat to human health
(1). There are ~240,000 new
esophageal cancer cases in China every year (2). Esophageal squamous cell carcinoma (ESCC)
is the major histological type of esophageal cancer. ESCC is a
highly aggressive malignancy due to late diagnosis, rapid
progression and poor prognosis of survival. Therefore, the
mortality rate of esophageal cancer is similar to the incidence of
esophageal cancer (3,4). There have been numerous improvements in
surgery, radiotherapy and chemotherapy; however, the 5-year overall
survival rate remains poor due to diagnosis of the disease at an
advanced stage (5). It is important
for ideal markers to be identified as these may help in early
diagnosis of the disease.
The B-cell specific Moloney leukemia virus insert
site 1 (Bmi-1) gene, a member of the polycomb group of proteins,
was identified in 1999 and originally isolated as an oncogene in
the generation of mouse pre-B-cell lymphomas (6). As a transcriptional repressor through
chromatin modification, Bmi-1 is involved in axial patterning, cell
cycle regulation, hematopoiesis and senescence (7,8).
Overexpression of Bmi-1 has been observed in a variety of human
cancers, including gastric (9),
breast (10), colorectal (11) and head and neck cancers (12), and it was also observed that
expression of Bmi-1 is associated with the development of tumors
(13,14). It was reported that overexpression of
Bmi-1 in primary human cancer cells may cause downregulation of the
INK4a-ARF locus and affect the pathway of p14ARF-Mdm2-p53 (15). It has been hypothesized that Bmi-1 is
an important inhibitor of the p53 pathway (13,15).
Peptidyl arginine deiminase IV (PADI4 or PAD4) is a
post-translational modification enzyme that converts arginine
residues at histone tails to citrulline, in the presence of
Ca2+ (16). The expression
of PADI4 has been detected in human CD34+
stem/progenitor cells (17), and it
is involved in the regulation of hematopoietic progenitor
proliferation (18). Our previous
study detected the overexpression of PADI4 in various malignancies,
including lung adenocarcinoma, hepatocellular cancer, breast cancer
and metastatic cancer (19). Tanikawa
et al reported that citrullination of H4R3 by PADI4 is
associated with the p53 pathway (20).
In the present study, the expression levels and
clinical significance of Bmi-1 and PADI4 were investigated in
esophageal cancer tissues and pericarcinous tissues in order to
observe differences in the expression of Bmi-1 and PADI4.
Materials and methods
ESCC patient sample preparation
The samples of esophageal cancer tissues and
adjacent noncancerous mucosal tissues were obtained from 26
patients who had received surgical treatment between January 2014
and December 2015 in the Department of Chest Surgery, Qianfoshan
Hospital of Shandong University (Shandong, China). Normal tissues
located 5 cm away from the tumor edge were collected during
surgery. Tissue microarrays containing 120 esophageal tissue
sections were commercially obtained from Chaoying Bioscience
(Shanxi, China). The slides contained ESCC tissues (n=60) and
adjacent normal tissues. Section thickness was ~5 µm. Firstly, the
sections were stained with hematoxylin and eosin (Boster, Co.,
Ltd., Beijing, China) within each tissue core. All patients had not
received radiotherapy or chemotherapy prior to surgery.
Postoperative pathologic results of the esophageal cancer biopsies
showed they were all squamous cell carcinomas. Patient information,
including gender, age and clinicopathological characteristics, was
obtained from the medical records or the manufacturer. The enrolled
patients included 61 males and 25 females, and the median age was
58 years (range, 36–76). According to the World Health Organization
standard pathology classification, the tumors of 9 patients (10.5%)
were diagnosed as well-differentiated tumors, the tumors of 47
patients (54.7%) were diagnosed as moderately differentiated
tumors, and the tumors of 30 patients (34.9%) were diagnosed as
poorly differentiated tumors. In these cases, 70 patients had no
lymph node metastasis and 16 patients had lymph node metastasis.
All patients provided informed consent prior to specimen
acquisition and the present study was approved by the Research
Ethics Committee of the Qianfoshan Hospital Affiliated to Shandong
University.
Immunohistochemical staining
Tissue samples were soaked in 10% neutral buffered
formalin, embedded in conventional paraffin, and the section
thickness was ~3 µm. Following deparaffinization, the specimens
were hydrated and incubated with an epitope retrieval solution
(Boster, Co., Ltd.; pH 6.0) in a microwave (temperature controlled
at 95–100°C) for 20 min. The slices were then cooled to room
temperature and incubated with 0.3% H2O2 for
10 min at room temperature to inactivate endogenous peroxidase, and
then rinsed with PBS. The specimens were then incubated with rabbit
polyclonal antibody for Bmi-1 (cat. no., ab85688; dilution, 1:500;
Abcam, Cambridge, UK) and mouse monoclonal antibody for PADI4 (cat.
no., ab128086; dilution, 1:500; Abcam) at 4°C overnight. Specimens
were then incubated with ready-to-use secondary antibody EliVision™
plus kit (cat. no., KIT-9901; Maixin-Bio; Lab Vision, Kalamazoo,
USA), according to the manufacturer's protocol. The specimens were
then washed using PBS, and diaminobenzidine (Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA) chromogenic reagent was
added. Termination of the chromogenic reaction was achieved with
water. Following counterstaining with hematoxylin, specimens were
dehydrated, mounted and observed under a Nikon 50i fluorescence
microscope, magnification, ×200 (Nikon Corporation, Tokyo,
Japan).
Interpretation of the results
The immunohistochemical specimens were evaluated by
German semi-quantitative statistical methods. Briefly, slices were
observed under an optical microscope (magnification, ×200) and
evaluated using positive staining intensity and percentage of
positive staining by two pathologists. Positive staining intensity
was rated as follows: 0, No staining; 1, light yellow; 2, yellow;
3, dark yellow. The extent of stained cells was ranked as follows:
0, 0–5%; 1, 5–25%; 2, 25–50%; 3, 50–75%; 4, 75–100%. The final
score was determined by multiplying the staining intensity scores
with the extent of positivity scores of cells: 0–2, negative (−);
3–5, weak (+); 6–8, moderate (++); and 9–12, strong (+++) (21).
Western blot analysis
Human esophageal cancer tissues were homogenized and
centrifuged at 12,000 × g for 30 min at 4°C. The protein
concentrations were determined using the bicinchoninic acid protein
assay kit (Boster Biological Technology, Pleasanton, CA, USA).
Total protein (30 µg) was separated by 10% SDS-PAGE, transferred to
a polyvinylidene fluoride membrane and blocked in 5% milk. The
membranes were incubated with rabbit polyclonal antibody for Bmi-1
(cat. no., ab85688, dilution, 1:8,000; Abcam) and mouse monoclonal
antibody for PADI4 (cat. no., ab128086, dilution; 1:2,000; Abcam)
or mouse antibody for GAPDH (cat. no., AF0006; dilution, 1:6,000;
Beyotime Institute of Biotechnology, Haimen, China) at 4°C
overnight. The Goat anti-Mouse secondary antibody (cat. no., A0258;
dilution, 1:8,000; Beyotime Institute of Biotechnology) and Goat
anti-Rabbit secondary antibody (cat. no., A0239; dilution, 1:8,000;
Beyotime Institute of Biotechnology) was incubated with the
membrane for 1 h at room temperature in TBS with Tween-20. Finally,
the immunoreactive protein bands were visualized with Immobilon™
Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica,
MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen specimens using
the E.Z.N.A.® total RNA kit II (Omega Bio-Tek, Inc.,
Norcross, GA, USA), according to the manufacturer's protocol. Total
RNA (1 µg) was then used to perform reverse transcription for
first-strand cDNA using the revert aid first strand cDNA synthesis
kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Subsequently, qPCR was performed in triplicate on the ABI VIIA7
real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with SuperReal PreMix Plus (SYBR-Green; Tiangen Biotech Co.,
Ltd., Beijing, China) with primers that amplified a specific single
product by melt curve. Primer sequences for Bmi-1, PADI4 and
β-actin were as follows: Bmi-1 forward, 5′-CCACCTGATGTGTGTGCTTTG-3′
and reverse, 5′-TTCAGTAGTGGTCTGGTCTTGT-3′; PADI4 forward,
5′-GGGGTGGTCGTGGATATTGC-3′ and reverse,
5′-CCCGGTGAGGTAGAGTAGAGC-3′; and β-actin forward,
5′-GACCACACCTTCTACAATGAG-3′ and reverse,
5′-GCATACCCCTCGTAGATGGG-3′.
All reactions were done in a 20 µl
reaction volume
Subsequent to pre-denaturing, PCR was performed at
95°C for 15 min, followed by 40 cycles at 95°C for 10 sec and 62°C
for 32 sec. Gene expression was analyzed with the comparative
threshold cycle (Cq) method following normalization to the
reference gene β-actin. ∆∆Cq was used to calculate the relative
amount of the transcripts in the esophageal cancer samples and the
control group, which were normalized to the endogenous control
(22). ∆∆Cq=∆Cq (esophageal
cancer)-∆Cq (control) for RNA samples. The fold change for each
esophageal cancer sample relative to the control
sample=2−∆∆Cq. When the expression showed a 2-fold
decrease or increase compared with normal counterpart tissue, it
was considered as an altered expression.
Statistical analysis
The correlation between Bmi-1 and PADI4 mRNA
expression levels was analyzed by Pearson's coefficient test. The
experimental data of protein expression was statistically analyzed
with χ2 test and Fisher's exact test. All statistical
analyses were performed using the SPSS 21.0 software package (IBM
SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Immunodetection of Bmi-1 and PADI4 in
ESCC tissues
The expression rates of Bmi-1 in ESCC tissue and
normal mucosa were 73.3 and 30.2%, respectively. The expression
rates of PADI4 in ESCC tissue and normal mucosa were 68.6 and
37.2%, respectively. The differences in the expression of Bmi-1 and
PADI4 were statistically significant between esophageal cancer
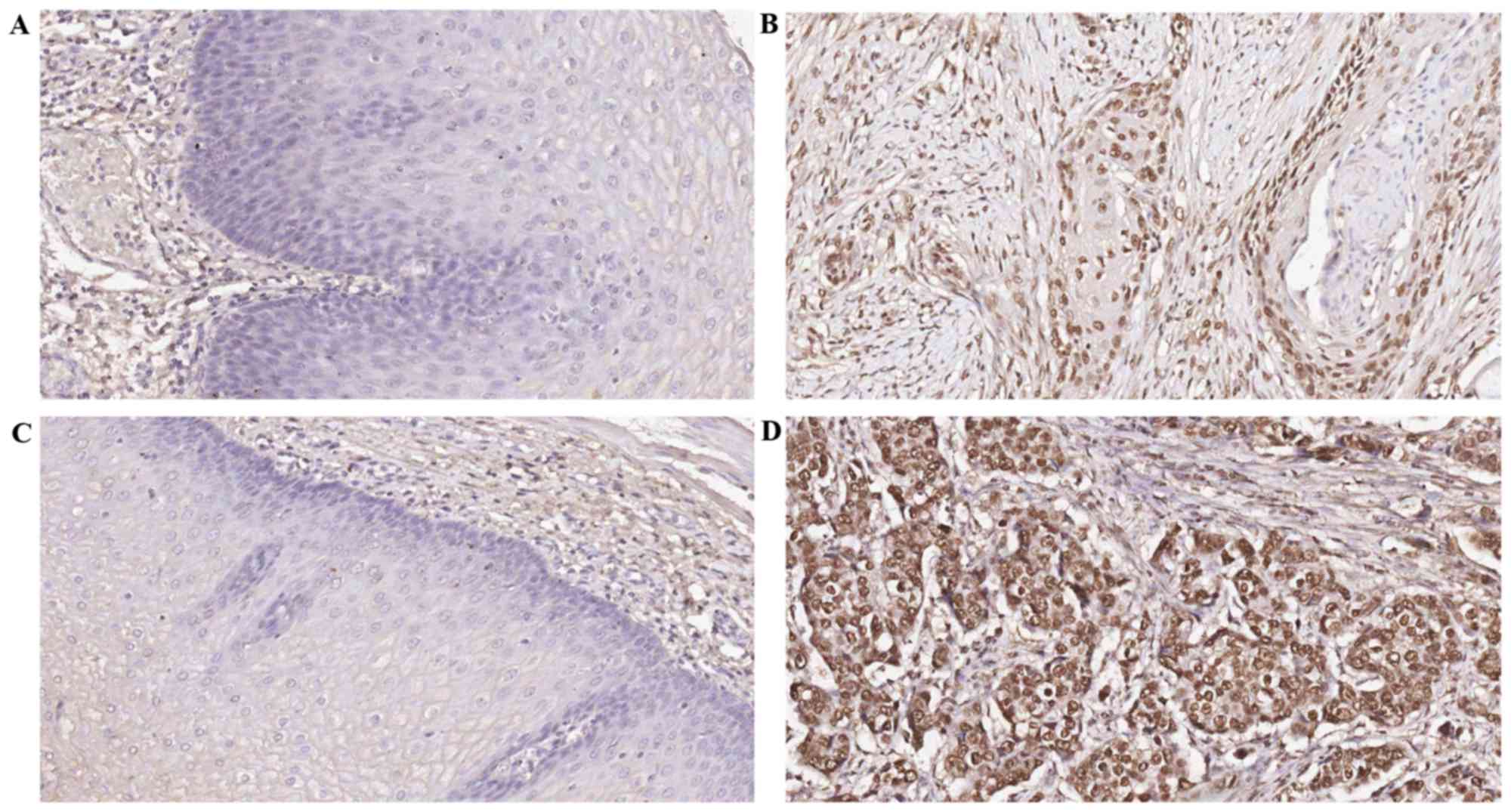
tissue and normal esophageal mucosa (P<0.05; Tables I and II). Bmi-1 and PADI4 were expressed in the
nucleus of tumor cells based on immunohistochemical staining of
tumors. The staining of Bmi-1 and PADI4 are shown at the original
magnification of ×200 (Fig. 1).
 | Table I.Bmi-1 protein expression in
esophageal cancer and normal esophageal tissues. |
Table I.
Bmi-1 protein expression in
esophageal cancer and normal esophageal tissues.
|
|
| Bmi-1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Pathological
characteristic | Total cases, n | Positive, n | Negative, n | Positive rate,
% | χ2 | P-value |
|---|
| Cancerous
tissue | 86 | 63 | 23 | 73.3 | 31.876 | P<0.05 |
| Normal tissue | 86 | 26 | 60 | 30.2 |
|
|
 | Table II.PADI4 protein expression in
esophageal cancer and normal esophageal tissues. |
Table II.
PADI4 protein expression in
esophageal cancer and normal esophageal tissues.
|
|
| PADI4
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Pathological
characteristic | Total cases, n | Positive, n | Negative, n | Positive rate,
% | χ2 | P-value |
|---|
| Cancerous
tissue | 86 | 59 | 26 | 68.6 | 17.011 | P<0.05 |
| Normal tissue | 86 | 32 | 54 | 37.2 |
|
|
Association between the expression of
Bmi-1 and PADI4 with clinical pathological parameters
The importance of Bmi-1 and PADI4 in ESCC was
evaluated by correlating its expression level with
clinicopathological features. Several of the analyzed
clinicopathological features exhibited significant associations
with the expression levels (Table
III). The results revealed that the expression of Bmi-1 was
associated with differentiation degree, clinical stage and lymph
node metastasis (P<0.05), but not with patient gender, age and
depth of invasion (P>0.05). The expression of PADI4 was
associated with clinical stage, depth of invasion and lymph node
metastasis (P<0.05), but not with patient gender, age and
differentiation degree (P>0.05).
 | Table III.Association between the expression
level of Bmi-1 or PADI4 and clinicopathological variables. |
Table III.
Association between the expression
level of Bmi-1 or PADI4 and clinicopathological variables.
|
| Bmi-1 | PADI4 |
|---|
|
|
|
|
|---|
| Variable | Total cases, n | Positive cases,
n | Positive rate,
% | χ2 | P-value | Total cases, n | Positive cases,
n | Positive rate,
% | χ2 | P-value |
|---|
| Gender | 0.028 | 0.866 |
|
|
| 2.600 | 0.107 |
|
Male | 61 | 45 | 73.8 |
|
| 61 | 45 | 73.8 |
|
|
|
Female | 25 | 18 | 72.0 |
|
| 25 | 14 | 56.0 |
|
|
| Age, years |
|
|
| 0.742 | 0.389 |
|
|
| 0.711 | 0.399 |
|
<60 | 44 | 34 | 77.3 |
|
| 44 | 32 | 72.7 |
|
|
|
≥60 | 42 | 29 | 69.0 |
|
| 42 | 27 | 64.3 |
|
|
| Differentiation
degree |
|
|
| 13.787 | 0.001 |
|
|
| 4.773 | 0.092 |
|
High | 9 | 2 | 22.2 |
|
| 9 | 5 | 55.6 |
|
|
|
Moderate | 47 | 36 | 76.6 |
|
| 47 | 29 | 61.7 |
|
|
|
Poor | 30 | 25 | 83.3 |
|
| 30 | 25 | 83.3 |
|
|
| Clinical stage |
|
|
|
| 0.005 |
|
|
| 5.771 | 0.016 |
|
I/II | 70 | 47 | 67.1 |
|
| 70 | 44 | 62.9 |
|
|
|
III/IV | 16 | 16 | 100.0 |
|
| 16 | 15 | 93.8 |
|
|
| T
classification |
|
|
| 0.618 | 0.432 |
|
|
| 6.672 | 0.010 |
|
T1/2 | 28 | 19 | 67.9 |
|
| 28 | 14 | 50.0 |
|
|
|
T3/4 | 58 | 44 | 75.9 |
|
| 58 | 45 | 77.6 |
|
|
| Lymph node
metastases |
|
|
|
| 0.005 |
|
|
| 5.771 | 0.016 |
|
Negative | 70 | 47 | 67.1 |
|
| 70 | 44 | 62.9 |
|
|
|
Positive | 16 | 16 | 100.0 |
|
| 16 | 15 | 93.8 |
|
|
Quantifying Bmi-1 and PADI4 expression
levels by western blot analysis and RT-qPCR
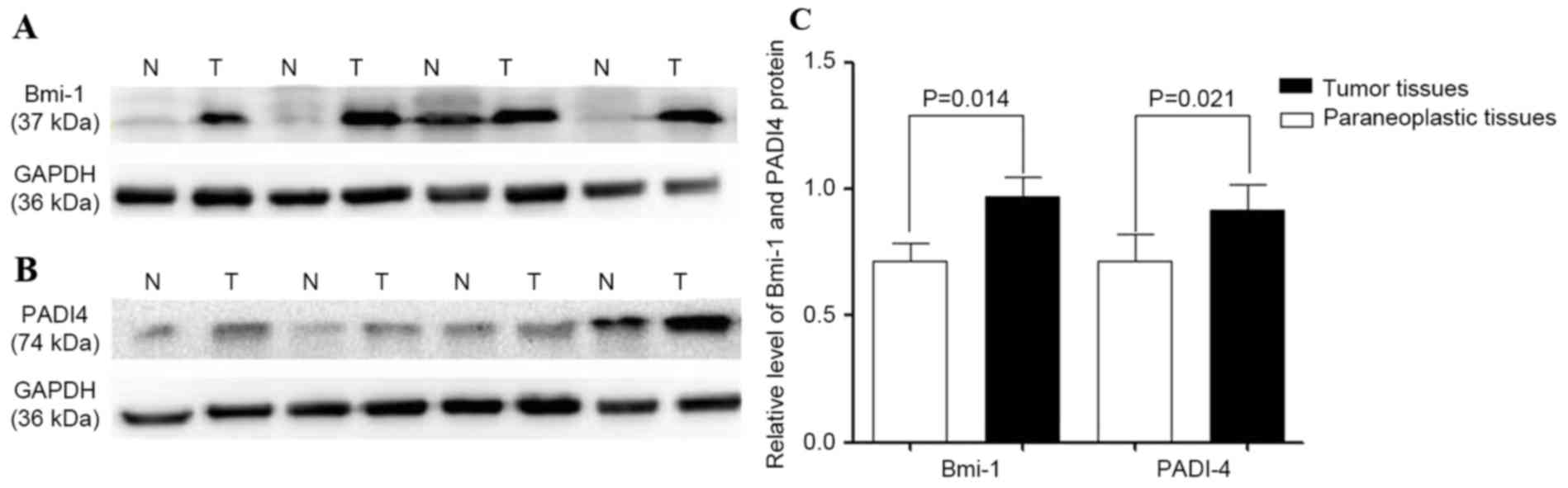
Bmi-1 and PADI4 protein levels were quantified by
western blot analysis. The protein was detected in ESCC and
corresponding para-carcinoma tissues. Compared with para-carcinoma
tissues, the expression levels of Bmi-1 and PADI4 were
significantly increased in ESCC, with statistically significant
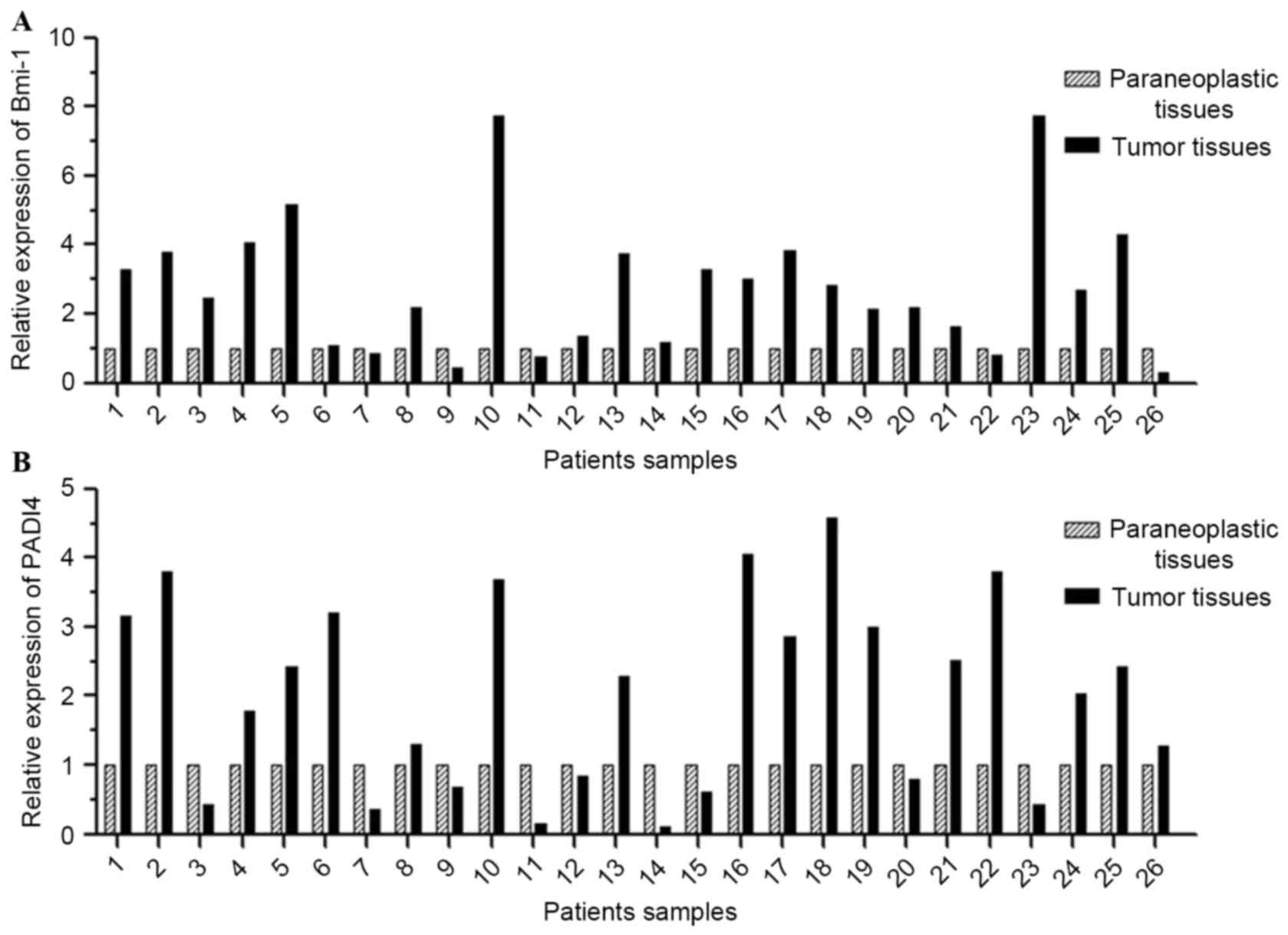
differences (P=0.014 and 0.021, respectively; Fig. 2). Transcription of Bmi-1 and PADI4 was
quantified by RT-qPCR. The results revealed that Bmi-1 mRNA was
overexpressed in 17 of 26 (65.38%) esophageal tumor tissues
(>2-fold), and PADI4 mRNA was overexpressed in 14 of 26 (53.85%;
>2-fold). As shown in Fig. 3 and
Table IV, the ESCC tissues exhibited
an increased level of Bmi-1 and PADI4 mRNA compared with the
para-carcinoma tissues.
 | Table IV.Frequencies of altered expression of
Bmi-1 and PADI4 in the 26 esophageal cancer tissues. |
Table IV.
Frequencies of altered expression of
Bmi-1 and PADI4 in the 26 esophageal cancer tissues.
|
| Decreased
expression | Normal
expression | Overexpression |
|---|
|
|
|
|
|
|---|
| Gene | Frequency, n | Percentage, % | Frequency, n | Percentage, % | Frequency, n | Percentage, % |
|---|
| Bmi-1 | 2 |
7.69 | 7 | 26.92 | 17 | 65.38 |
| PADI4 | 4 | 15.38 | 8 | 30.77 | 14 | 53.8 |
Correlation between Bmi-1 expression
and PADI4 expression in ESCC tissues
Correlation analysis was performed to test the
correlation between the expression of Bmi-1 mRNA and PADI4 mRNA.
Bmi-1 overexpression was positively associated with PADI4
expression (r=0.534; P=0.005). In addition, according to the
results of immunohistochemical studies, the correlation
coefficients indicated that there were significant positive
correlations between Bmi-1 and PADI4 genes (r=0.214; P=0.047;
Table V).
 | Table V.Association between the expression of
Bmi-1 and PADI4 in esophageal cancer. |
Table V.
Association between the expression of
Bmi-1 and PADI4 in esophageal cancer.
| Protein expression
status | Cases, n |
|---|
|
Bmi-1+/PAD14+ | 47 |
|
BMI-1+/PAD14− | 16 |
|
BMI-1−/PAD14+ | 12 |
|
BMI-1−/PAD14− | 11 |
Discussion
It is well known that ESCC, a common malignant
disease, is prone to invade adjacent regions and metastasize to
lymph nodes or distant organs. At the point of diagnosis,
metastasis has already occurred in >50% of patients with ESCC,
with no chance of resection (23),
which largely explains the poor prognosis of ESCC (5). Therefore, diagnostic markers of early
ESCC may be useful to improve prognosis, and for selecting
treatments properly. The purpose of the present study was to
investigate the expression and clinicopathological roles of Bmi-1
and PADI4 in ESCC.
Mammalian polycomb group (PcG) protein complexes are
generally classified as polycomb repressive complexes 1 or 2 (PRC1
or PRC2). Alterations in PcG expression have been observed in human
tumors (24,25). Bmi-1, a PRC1 that regulates
proliferation and senescence in mammalian cells, plays an important
role in the self-renewal of stem cells. Overexpression of Bmi-1 has
been observed in several human cancers and its overexpression is
often associated with poor prognosis in gastric cancer, bladder
cancer and esophageal squamous cell carcinoma (2,24–26). It was reported that Bmi-1, as a
proto-oncogene, plays an important role in the invasion and
metastasis of neoplasms. Overexpression of Bmi-1 in esophageal
cancer has been reported (25). The
depth of invasion, clinical stage, lymph node metastasis status and
lower survival rate have all been identified as associated with
Bmi-1 overexpression (2,25). In the present study, it was revealed
that Bmi-1 plays an important role in ESCC progression. It was
demonstrated that Bmi-1 expression is significantly upregulated
(P<0.05) in ESCC tissues compared with adjacent noncancerous
tissues. In addition, it was observed that positive expression of
Bmi-1 is associated with clinical stage, differentiation degree and
lymph node metastasis (N classification), and the difference was
statistically significant (P<0.05). This indicated that Bmi-1
protein may perform a crucial role in the carcinogenesis,
progression and metastasis of ESCC.
Genome-wide analysis has demonstrated that PADI4
functions as an activator of gene expression by citrullination of
transcription factors (27). Previous
studies demonstrated that PADI4 is part of a transcriptional
network that regulates pluripotency (28–31). It
has been suggested that PADI4 may target different histone
arginines (32,33). Kolodziej et al previously
identified PADI4 as a novel interaction partner of T-cell acute
lymphocytic leukemia protein 1 (Tal1), which is a critical
regulator of hematopoietic gene expression and may act as an
oncogene if aberrantly expressed, and identified a large number of
genes that are co-regulated by PADI4 and Tal1 (31). Overexpression of PADI4 is often
observed in growth of tumors, and the inhibitor Cl-amidine reduces
growth of a subset of cell lines (34). Chang et al confirmed that the
expression level of PADI4 was positively associated with the
pathological classification of ESCC. The previous study also
identified that apoptosis corresponded to the expression of PADI4
in cultured EC cells that were treated with dichloroacetate
(35). In the present study, the
expression of PADI4 was significantly increased in ESCC tissues
compared with the corresponding noncancerous mucosal tissues. In
addition, it was also observed that positive expression of PADI4
was correlated with the depth of cancer invasion (T
classification), clinical stage and lymph node metastasis (N
classification), with statistically significant differences
(P<0.05). This indicated that PADI4 protein is associated with
esophageal carcinogenesis, progression and metastasis, and that
PADI4 may play a crucial role in ESCC.
The present study demonstrated that Bmi-1 and PADI4
are expressed abnormally in ESCC. The expression levels of Bmi-1
and PADI4 are associated with the carcinogenesis and progression of
human tumors. However, the expression of Bmi-1 is not associated
with depth of invasion (P>0.05). No association was observed
between PADI4 expression and differentiation degree (P>0.05).
These results may be due to the small size of patient cohort, and
additional studies using larger samples are required to extend the
present findings. The present study also identified a positive
correlation between Bmi-1 and PADI4 expression at mRNA (r=0.534;
P=0.005) and protein levels (r=0.214; P=0.047). However, it remains
unknown how Bmi-1 interaction with PADI4 affects carcinogenesis,
progression and metastasis in ESCC.
The p14ARF-MDM2-p53 pathway, commonly referred to as
the p53 pathway, is an important pathway in the development and
progression of numerous human malignancies (36). The p53 pathway is usually inactivated
by TP53 mutation, amplification of MDM2 or p14ARF deletion in a
number of human cancers (37,38). Previous studies have demonstrated that
Bmi-1 is a potent repressor of the p14ARF-MDM2-p53 pathway
(13,39,40).
Inactivation of the p53 pathway by Bmi-1 has been identified in
lymphomagenesis and oncogenesis in human non-small cell lung cancer
(15,37,41). Yao
et al reported that there is an inverse correlation between
the expression of Bmi-1 and p14ARF, and thereby dysfunction of the
p53 growth regulatory pathway during the development of gastric
cardia adenocarcinoma (42). In
addition, a number of studies proposed that PADI4 plays an
important role in various cellular processes, including
proliferation, the cell cycle and apoptosis (27,30–33). PADI4
overexpression has also been considered to reduce the expression of
p53-targeted genes, resulting in the disruption of cellular
apoptosis and of the normal cell cycle (35,43,44).
Previous studies have confirmed that PADI4 disrupts the apoptotic
process via citrullination of histone H3, which acts on the
promoter of p53 target genes (44,45). Cui
et al hypothesized that PADI4 may regulate migration,
invasion and apoptosis in A2780 cells with wild-type p53 and in
p53-null SKOV3 cells, and that PADI4 may be associated with the p53
gene (29). Apoptosis PCR array
analysis demonstrated that PADI4 overexpression induced the
decreased expression of the Fas ligand gene, which is one of the
important genes downstream of p53. Additional studies proposed that
TNF receptor superfamily member (TNFRSF) 9, Bcl-2 like protein 2,
TNFRSF11B and Bcl-2 antagonist/killer 1 may perform important roles
in the mechanism of action of PADI4, which is associated with
ovarian tumorigenesis (29). The
mechanism may be associated with the p53 gene (46,47).
According to the present results, it was speculated
that Bmi-1 and PADI4 may be involved in the associated signaling
molecules of the p53 pathway, which mediate the regulation of
esophageal cancer. It was hypothesized that overexpression of Bmi-1
may inhibit the activation of p14ARF and the function of the p53
pathway. It leads to tumor formation by regulating the expression
of PADI4. Bmi-1, which is associated with PADI4 by the
p14ARF-MDM2-p53 pathway, positively regulated the carcinogenesis
and progression of ESCC.
To the best of our knowledge, this is the first
study to investigate the association between Bmi-1 and PADI4
expression in ESCC. The expression of Bmi-1 and PADI4 was
associated with esophageal cancer progression. The advanced stages
of ESCC are more likely to express increased levels of Bmi-1 and
PADI4. The present study detected that the associated gene
expression and the mechanism may be associated with p53; however,
the detailed mechanisms and how they are regulated in esophageal
cancers were not investigated.
In conclusion, the present study demonstrated that
the expression of Bmi-1 and PADI4 was associated with the
carcinogenesis and progression of ESCC. Therefore, Bmi-1 and PADI4
may be used as prognostic markers in ESCC. In addition, a positive
association was observed between Bmi-1 and PADI4, and this
mechanism may be associated with the p53 pathway.
Acknowledgements
The authors thank the Medical Research Center of
Qianfoshan Hospital of Shandong University for their excellent
technical assistance. The present study was supported by the
Ministry of Science and Technology Research Foundation (grant no.
2013GSF11836).
References
|
1
|
Zhang J, Zhu Z, Liu Y, Jin X, Xu Z, Yu Q
and Li K: Diagnostic value of multiple tumor markers for patients
with esophageal carcinoma. PLoS One. 10:e01169512015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y, Zhang YL, Chen HM, Pu HW, Ma WJ,
Li XM, Ma H and Chen X: Expression of Bmi-1 and PAI-1 in esophageal
squamous cell carcinoma. World J Gastroenterol. 20:5533–5539. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crew KD and Neugut AI: Epidemiology of
upper gastrointestinal malignancies. Semin Oncol. 31:450–464. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vaupel P and Mayer A: Hypoxia in cancer:
Significance and impact on clinical outcome. Cancer Metastasis Rev.
26:225–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sudo T, Iwaya T, Nishida N, Sawada G,
Takahashi Y, Ishibashi M, Shibata K, Fujita H, Shirouzu K, Mori M
and Mimori K: Expression of mesenchymal markers vimentin and
fibronectin: The clinical significance in esophageal squamous cell
carcinoma. Ann Surg Oncol. 20 Suppl 3:S324–S335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song LB, Zeng MS, Liao WT, Zhang L, Mo HY,
Liu WL, Shao JY, Wu QL, Li MZ, Xia YF, et al: Bmi-1 is a novel
molecular marker of nasopharyngeal carcinoma progression and
immortalizes primary human nasopharyngeal epithelial cells. Cancer
Res. 66:6225–6232. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Lugt NM, Domen J, Linders K, van
Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J,
Sofroniew M, van Lohuizen M, et al: Posterior transformation,
neurological abnormalities, and severe hematopoietic defects in
mice with a targeted deletion of the bmi-1 proto-oncogene. Genes
Dev. 8:757–769. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jacobs JJ, Kieboom K, Marino S, DePinho RA
and van Lohuizen M: The oncogene and Polycomb-group gene bmi-1
regulates cell proliferation and senescence through the ink4a
locus. Nature. 397:164–168. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu YW, Li J and Guo WJ: Expression and
clinicopathological significance of Mel-18 and Bmi-1 mRNA in
gastric carcinoma. J Exp Clin Cancer Res. 29:1432010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo BH, Feng Y, Zhang R, Xu LH, Li MZ,
Kung HF, Song LB and Zeng MS: Bmi-1 promotes invasion and
metastasis, and its elevated expression is correlated with an
advanced stage of breast cancer. Mol Cancer. 10:102011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tao J, Liu YL, Zhang G, Ma YY, Cui BB and
Yang YM: Expression and clinicopathological significance of Mel-18
mRNA in colorectal cancer. Tumour Biol. 35:9619–9625. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dalley AJ, Pitty LP, Major AG, Abdulmajeed
AA and Farah CS: Expression of ABCG2 and Bmi-1 in oral potentially
malignant lesions and oral squamous cell carcinoma. Cancer Med.
3:273–283. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li DW, Tang HM, Fan JW, Yan DW, Zhou CZ,
Li SX, Wang XL and Peng ZH: Expression level of Bmi-1 oncoprotein
is associated with progression and prognosis in colon cancer. J
Cancer Res Clin Oncol. 136:997–1006. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo P, Gao A, Zhang G, Han H and Zhou Q:
Decoding the knots of initiation of oncogenic
epithelial-mesenchymal transition in tumor progression. Curr Cancer
Drug Targets. 13:996–1011. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vonlanthen S, Heighway J, Altermatt HJ,
Gugger M, Kappeler A, Borner MM, van Lohuizen M and Betticher DC:
The bmi-1 oncoprotein is differentially expressed in non-small cell
lung cancer and correlates with INK4A-ARF locus expression. Br J
Cancer. 84:1372–1376. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hagiwara T, Nakashima K, Hirano H, Senshu
T and Yamada M: Deimination of arginine residues in
nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem
Biophys Res Commun. 290:979–983. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chang X and Han J: Expression of
peptidylarginine deiminase type 4 (PAD4) in various tumors. Mol
Carcinog. 45:183–196. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nakashima K, Arai S, Suzuki A, Nariai Y,
Urano T, Nakayama M, Ohara O, Yamamura K, Yamamoto K and Miyazaki
T: PAD4 regulates proliferation of multipotent haematopoietic cells
by controlling c-myc expression. Nat Commun. 4:18362013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang X, Han J, Pang L, Zhao Y, Yang Y and
Shen Z: Increased PADI4 expression in blood and tissues of patients
with malignant tumors. BMC Cancer. 9:402009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanikawa C, Espinosa M, Suzuki A, Masuda
K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y and Matsuda
K: Regulation of histone modification and chromatin structure by
the p53-PADI4 pathway. Nat Commun. 3:6762012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Remmele W and Stegner HE: Recommendation
for uniform definition of an immunoreactive score (IRS) for
immunohistochemical estrogen receptor detection (ERICA) in breast
cancer tissue. Pathologe. 8:138–140. 1987.(In German). PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang XW, Sheng YP, Li Q, Qin W, Lu YW,
Cheng YF, Liu BY, Zhang FC, Li J, Dimri GP and Guo WJ: BMI1 and
Mel-18 oppositely regulate carcinogenesis and progression of
gastric cancer. Mol Cancer. 9:402010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu WL, Guo XZ, Zhang LJ, Wang JY, Zhang
G, Guan S, Chen YM, Kong QL, Xu LH, Li MZ, et al: Prognostic
relevance of Bmi-1 expression and autoantibodies in esophageal
squamous cell carcinoma. BMC Cancer. 10:4672010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qin ZK, Yang JA, Ye YL, Zhang X, Xu LH,
Zhou FJ, Han H, Liu ZW, Song LB and Zeng MS: Expression of Bmi-1 is
a prognostic marker in bladder cancer. BMC Cancer. 9:612009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Gamble MJ, Stadler S, Cherrington
BD, Causey CP, Thompson PR, Roberson MS, Kraus WL and Coonrod SA:
Genome-wide analysis reveals PADI4 cooperates with Elk-1 to
activate c-Fos expression in breast cancer cells. PLoS Genet.
7:e10021122011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Christophorou MA, Castelo-Branco G,
Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, Mowen KA,
Bertone P, Silva JC, Zernicka-Goetz M, et al: Citrullination
regulates pluripotency and histone H1 binding to chromatin. Nature.
507:104–108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui YY, Yan L, Zhou J, Zhao S, Zheng YB,
Sun BH, Lv HT, Rong FN and Chang XT: The role of peptidylarginine
deiminase 4 in ovarian cancer cell tumorigenesis and invasion.
Tumour Biol. 37:5375–5383. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanikawa C, Espinosa M, Suzuki A, Masuda
K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y and Matsuda
K: Regulation of histone modification and chromatin structure by
the p53-PADI4 pathway. Nat Commun. 3:6762012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kolodziej S, Kuvardina ON, Oellerich T,
Herglotz J, Backert I, Kohrs N, Buscató El, Wittmann SK,
Salinas-Riester G, Bonig H, et al: PADI4 acts as a coactivator of
Tal1 by counteracting repressive histone arginine methylation. Nat
Commun. 5:39952014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cuthbert GL, Daujat S, Snowden AW,
Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD,
Tempst P, Bannister AJ and Kouzarides T: Histone deimination
antagonizes arginine methylation. Cell. 118:545–553. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Wysocka J, Sayegh J, Lee YH,
Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y,
et al: Human PAD4 regulates histone arginine methylation levels via
demethylimination. Science. 306:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Slack JL, Causey CP and Thompson PR:
Protein arginine deiminase 4: A target for an epigenetic cancer
therapy. Cell Mol Life Sci. 68:709–720. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chang X, Hou X, Pan J, Fang K, Wang L and
Han J: Investigating the pathogenic role of PADI4 in oesophageal
cancer. Int J Biol Sci. 7:769–781. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hashiguchi Y, Tsuda H, Yamamoto K, Inoue
T, Ishiko O and Ogita S: Combined analysis of p53 and RB pathways
in epithelial ovarian cancer. HUM Pathol. 32:988–996. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lindström MS, Klangby U and Wiman KG:
p14ARF homozygous deletion or MDM2 overexpression in Burkitt
lymphoma lines carrying wild type p53. Oncogene. 20:2171–2177.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ichimura K, Bolin MB, Goike HM, Schmidt
EE, Moshref A and Collins VP: Deregulation of the p14ARF/Mdm2/p53
pathways is a prerequisite for human astrocytic gliomas with G1-S
transition control gene abnormalities. Cancer Res. 60:417–424.
2000.PubMed/NCBI
|
|
39
|
Shafaroudi AM, Mowla SJ, Ziaee SA, Bahrami
AR, Atlasi Y and Malakootian M: Overexpression of BMI1, a polycomb
group repressor protein, in bladder tumors: A preliminary report.
Urol J. 5:99–105. 2008.PubMed/NCBI
|
|
40
|
Taran K, Wysocka A, Sitkiewicz A, Kobos J
and Andrzejewska E: Evaluation of potential prognostic value of
Bmi-1 gene product and selected markers of proliferation (Ki-67)
and apoptosis (p53) in the neuroblastoma group of tumors. Postepy
Hig Med Dosw (Online). 70:110–116. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Beà S, Tort F, Pinyol M, Puig X, Hernández
L, Hernández S, Fernandez PL, van Lohuizen M, Colomer D and Campo
E: BMI-1 gene amplification and overexpression in hematological
malignancies occur mainly in mantle cell lymphomas. Cancer Res.
61:2409–2412. 2001.PubMed/NCBI
|
|
42
|
Yao D, Wang Y, Xue L, Wang H, Zhang J and
Zhang X: Different expression pattern and significance of
p14ARF-Mdm2-p53 pathway and Bmi-1 exist between gastric cardia and
distal gastric adenocarcinoma. Hum Pathol. 44:844–851. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tanikawa C, Ueda K, Nakagawa H, Yoshida N,
Nakamura Y and Matsuda K: Regulation of protein Citrullination
through p53/PADI4 network in DNA damage response. Cancer Res.
69:8761–8769. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li P, Yao H, Zhang Z, Li M, Luo Y,
Thompson PR, Gilmour DS and Wang Y: Regulation of p53 target gene
expression by peptidylarginine deiminase 4. Mol Cell Biol.
28:4745–4758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yao H, Li P, Venters BJ, Zheng S, Thompson
PR, Pugh BF and Wang Y: Histone Arg modifications and p53 regulate
the expression of OKL38, a mediator of apoptosis. J Biol Chem.
283:20060–20068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lda C Braga, Silva LM, Piedade JB, Traiman
P and da Silva Filho AL: Epigenetic and expression analysis of
TRAIL-R2 and BCL2: On the TRAIL to knowledge of apoptosis in
ovarian tumors. Arch Gynecol Obstet. 289:1061–1069. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pavelic S Kraljevic, Cacev T and Kralj M:
A dual role of p21waf1/cip1 gene in apoptosis of HEp-2 treated with
cisplatin or methotrexate. Cancer Gene Ther. 15:576–590. 2008.
View Article : Google Scholar : PubMed/NCBI
|