Introduction
There are numerous strategies with inherent
advantages and disadvantages that may be used for the evaluation of
DNA damage and repair. DNA is the primary target following exposure
to stimuli such as ultraviolet (UV) radiation, DNA alkylators,
certain environmental carcinogens, oxidative stress and
chemotherapeutic drugs (1). All these
damaging factors produce lesions on DNA and a base alteration
promoting a break in the DNA helix (2). Double-strand breaks (DSBs) are lethal to
cells, as they affect both strands of DNA and promote the loss of
genetic information (3). DNA damage,
which frequently occurs in eukaryotic cells, may promote genomic
instability and aid the development of disease, including cancer
(4). Following DNA damage, cellular
responses are induced and allow the cell to repair the damage or
process the damage via a variety of mechanisms (5). Therefore, DNA repair proteins are
important biomarkers for predicting the response of tumors to
genotoxic stress and the prognosis of patients with more accuracy.
This highlights the importance of detecting and quantifying DNA
damage. There are a number of strategies that allow the
investigation of these underlying mechanisms and the current review
discusses these strategies and highlights their importance. These
techniques may be separated into two perspectives: Techniques for
detecting DNA damage and techniques for evaluating the underlying
repair mechanisms.
Molecular strategies
Polymerase chain reaction (PCR) and
agarose gel electrophoresis
Breaks in DNA reduce the molecular weight of a
single DNA strand, and this may be caused by physical, chemical or
enzymatic reagents (6). DNA breaks
and lesions may be detected by PCR or using agarose gel
electrophoresis (7).
PCR is one of the most frequently used techniques
for detecting DNA damage (7). DNA
amplification is stopped at the sites of damage via the blocking of
the progression of Taq polymerase, which results in a
decrease in the quantity of PCR product and a reduced number of DNA
templates, which do not contain the Taq-blocked lesions as
they are not amplified (8). This is
considered to be a simple and reliable method in which particular
segments of DNA are specifically replicated and visualized using
agarose gels that resolve a range of DNA fragments (50–50,000 bp)
dependent on the agarose percentage (8).
Quantitative PCR (qPCR) has been performed to
quantify the amount of DNA damage on both strands, as well as the
kinetics of DNA damage removal in the mitochondrial DNA (mtDNA) of
human and other organisms (7,9). The technique has been used to measure
the formation and repair of UV-induced photoproducts in a 1.2-kb
fragment of the LacI gene from Escherichia coli
(8) and to measure the damage to
mtDNA in Schizosaccharomyces pombe cells treated with
hydrogen peroxide (10). The
frequency of cisplatin-induced lesions has been investigated in a
series of fragments ranging from 150 to 2,000 bp from the hamster
aprt gene (11). Taken
together, these previous studies have demonstrated the ability to
detect and analyze gene-specific DNA damage and repair with PCR
(12). The qPCR method is dependent
on high-molecular weight DNA, DNA quantification, qPCR conditions,
quantification of amplification products and the calculation of
lesion frequencies (8), and has the
advantage of quantitative detection of DNA damage in a specific
gene that is expressed mathematically in terms of lesions per kb
and the requirement of only 1–2 ng of total genomic DNA (9).
Ligation-mediated PCR (LMPCR) analyzes the
distribution of the two types of UV-induced DNA photoproducts,
namely cyclobutane pyrimidine dimers and 6–4 photoproducts. The
technique has the capability to detect an individual DNA
photoproduct at low UV doses (10–20 J/m2) and is also
highly sensitive for studying the interactions of proteins and DNA
in vivo (13), and for
measuring the repair of cyclobutane pyrimidine dimers (14). By contrast, terminal
transferase-dependent PCR (TDPCR) is a technique that adds a
terminal transferase prior to ligation to an oligonucleotide, and
as with LMPCR, this method is able to map pyrimidine 6–4 pyrimidone
photoproducts and obtain information on the in vivo
chromatin structure (15).
Immuno-coupled PCR (ICPCR) combines nucleic acid
amplification with an antibody-based assay in which the detection
enzyme in the ELISA is replaced with a biotinylated reporter DNA
bound to an antigen-antibody complex (16). This methodology allows for the
quantification of thymine dimer formations in genes and these have
been established to be directly proportional to the global levels
identified in UV radiation-exposed human genomic DNA (17). PCR-based short interspersed DNA
element (SINE)-mediated is also a highly sensitive assay that
detects DNA adducts produced by drug treatment, including cisplatin
(18) or UV-B induced damage, and
detects repair in the mammalian genome (19). This assay relies on the abundance,
dispersion and conservation of the SINEs in mammalian genomes
(19). Compared with conventional PCR
and qPCR, this method differs in that it involves the amplification
of long segments of DNA in the transcribed regions of the genome in
a faster and more cost-effective manner (18).
DNA repair proteins that are used as
molecular markers
Ku protein
Ku is a heterodimer consisting of two subunits (70
and 80 kDa) that bind to a 470-kDa catalytic subunit termed the
DNA-dependent protein kinase, which is involved in repairing DNA
DSBs (20). The DSB repair pathway is
dependent on Ku protein and is the primary DNA DSB repair mechanism
in mammalian cells (21). The ability
of Ku to function affects numerous nuclear processes besides DNA
repair, including telomere maintenance and apoptosis (22). Ku protein has also been implicated in
cell survival, which suggests that the detection of Ku protein
expression may be used as a strategy for evaluating DNA damage and
repair (22). The majority of
previous studies have focused on the function of Ku in DNA DSB
repair via the non-homologous end joining pathway, and cells or
animals deficient in this protein are defective in DSB rejoining
and are hypersensitive to ionizing radiation (23). For the expression and purification of
full-length Ku heterodimer, it is necessary to have co-expression
of Ku70 and Ku80, and subsequently, the protein must be separated
and purified via chromatographic techniques (24).
Phosphorylated histone 2AX (γH2AX) protein
H2AX is a member of the histone H2A family and it
has been established that elevated phosphorylation levels of H2AX
on genomic DNA damage occur within 1–3 min of DNA damage (25). The detection of γH2AX protein
phosphorylated at Serine-139 allows an approach for detecting and
quantifying DNA DSBs, as the number of Serine-139-γH2AX molecules
is associated with the quantity of DNA damage (26), therefore it may be used as a marker of
DSBs. The primary method for detecting γH2AX is based on
immunofluorescence using a specific antibody for Serine-139-γH2AX
to demonstrate its localization in chromatin foci at the sites of
DNA damage (25). Indirect
identification has been used via flow cytometry (FCM) using
secondary antibodies tagged with fluorescein isothiocyanate (FITC),
while DNA has been counterstained with propidium iodide (PI) to
analyze an association between the presence of DSBs and cell cycle
phase (27).
X-ray repair cross complementing 1 (XRCC1)
protein
The XRCC1 protein serves an important role in
promoting efficient repair of DNA single-strand breaks (SSBs) in
mammalian cells (28). XRCC1 is able
to interact with multiple enzymatic components that are involved in
the repair process, including DNA ligase IIIa, DNA polymerase β,
apurinic/apyrimidinic endonuclease 1, polynucleotide
kinase/phosphatase, poly(ADP-ribose) polymerase 1 and 2, and
8-oxoguanine DNA glycosylase (29,30).
Previous studies have established that certain polymorphisms in the
XRCC1 gene are associated with cancer risk (31). The regulation of XRCC1 protein levels
in human cell lines has been investigated using RNA interference
and demonstrated that the reduction of XRCC1 affects the repair
pathways of SSBs, as well as having an important role in DNA base
excision repair (BER) (30,32). These events may be evaluated using the
comet assay or using fluorescent or analytical techniques that are
described in this review. For example, DNA repair assays to
evaluate the possible role of XRCC1 in the rejoining of chromosomal
SSBs are performed using alkaline elution, alkaline unwinding, or
comet assay, meanwhile, for evaluating the role of XRCC1 in the
rejoining of DSBs, neutral pH elution from a DNA filter has been
employed (33).
Fluorescence strategies
Comet assay
The comet assay, also known as single-cell gel
electrophoresis, is simple and is considered to be one of the gold
standard methods for measuring DNA strand breaks (single or double)
in eukaryotic cells (34,35). In addition to being a method for
detecting DNA breaks, it is also possible to detect UV-induced
pyrimidine dimers, oxidized bases and alkylation damage following
the introduction of lesion-specific endonucleases (36).
This technique identifies the head of the comet as a
spherical mass of undamaged DNA, and the damaged DNA (DNA loops
around strand breaks) streams out from the head as a tail (37,38). The
comet structure was first described in a study by Ostling and
Johanson (39), which explained the
tail in terms of DNA with relaxed supercoiling. In the most
frequently performed type of comet assay, cells are embedded in
agarose to immobilize the DNA and a lysis process is performed
using a detergent and high salt. The comet assay has a limited
resolution of 10–800 kb using standard conditions (40). Other variants of the comet assay are
also used to assess DNA damage and its detection.
Alkaline single-cell gel
electrophoresis
This version of the comet assay uses alkaline
denaturation surrounding a DNA break to reveal the break (single or
double) (41). This method enhances
comet tails and extends the range of DNA damage that is detected,
but sensitivity has not been increased compared to the use of
lesion-specific enzymes (34).
Neutral single-cell gel
electrophoresis
This is a variant of the comet assay that uses an
alkaline treatment, after which the conditions are restored to
neutral, followed by gel electrophoresis in neutral or mild
alkaline conditions (42). This
method is less sensitive but remains able to detect SSBs (43).
Use of lesion-specific enzymes
The use of lesion-specific enzymes may aid in the
detection of other types of DNA damage, other than SSBs or DSBs,
including oxidized bases or pyrimidine dimers (44). The enzymes create an
apurinic/apyrimidic site by removing the damaged base;
endonucleases specifically detect oxidized pyrimidines, and
formamidopyrimidine DNA glycosylases detect
8-oxo-7,8-dihydroguanine and ring opened-purines (35).
Bromodeoxyuridine-labelled DNA-comet
fluorescence in situ hybridization (FISH)
This technique combines a comet assay and FISH, and
is effective in detecting damage and repair site-specific breaks in
DNA regions in individual cells (40). This assay may be used to measure and
discriminate between SSBs or DSBs or modifications from DNA
repair.
Halo assay
This technique is based on the intercalation of PI
into the DNA helix, which causes the DNA to become a supercoiled
structure (45). Following lysis, the
nucleoids of individual cells appear as ‘halos’ that correspond to
DNA loops, which may be measured to determine the chromatin
fragility. The ‘halo’ diameter is proportional with PI
concentration and is expressed as relaxed or rewound supercoils at
low PI and high PI, respectively (45). This method may aid the study of the
effects of induced DNA damage, although it only detects alterations
in the organization of DNA if the damage has not been repaired,
which occurs at radiation doses of 2 Gy. This assay has limitations
on its sensitivity, but the advantages are that it is able to
measure the DNA damage of a single cell and no labeling of DNA with
radioactive precursors is required (46).
Terminal deoxynucleotidyl transferase
(TdT) dUTP nick-end labeling (TUNEL) assay
The TUNEL assay detects SSBs or DSBs, as well as
levels of apoptosis via the visualization of DNA fragmentation
(45). This assay primarily uses the
ability of the enzyme TdT to incorporate nucleotide analogues
conjugated with a fluorochrome onto the free 3′-OH of a DNA strand,
therefore allowing the visualization of the nuclei that contain
fragmented DNA (47). Additionally,
fluorescence may be detected using a fluorescent dye conjugated
antibody that recognizes biotin- or digoxigenin-tagged nucleotides
(48). As the assay is able to detect
the DNA fragments with fluorescence or radioactivity, microscopy
techniques, FCM, photo-multipliers and charge coupled device arrays
may be used to detect and quantify DNA damage caused by apoptosis
(49). Typically, the visualization
of DNA damage is possible as the morphological alterations occur in
the nucleus, including alterations in structural organization and
the collapse of chromatin (49).
During the degradation of DNA, a specific pattern of fragments is
generated by the activity of endonucleases enzymes, and
fragmentation of genomic DNA occurs into lower molecular weight
fragments from DNA (47).
Although this method was designed for detecting DNA
damage following apoptosis, DNA fragments with 3′-OH ends may occur
in a number of other situations where apoptosis does not take
place, including necrosis (49). The
TUNEL assay is limited in its sensitivity and specificity, but it
may also be used to stain cells undergoing DNA repair (50). TUNEL is not considered sufficient to
establish the type of cell death and must be accompanied by another
method that allows for the distinction of the origin of the DNA
fragmentation in cells undergoing apoptosis or non-apoptotic DNA
damage (51). One of the assays that
is considered to specifically detect DNA DSBs and used in
combination with TUNEL assay is the in situ ligation assay
(52), which is based on ligation of
double-stranded oligonucleotide probes by T4 DNA ligase to the ends
of the DNA breaks directly in tissue sections (53).
DNA breakage detection (DBD)-FISH
FISH is a technique for the visualization of nucleic
acids that improves resolution, speed and safety compared with
older methods that use isotopic detection (54,55). This
technology also allowed for the development of simultaneous
detection of multiple targets, quantitative analyses and live-cell
imaging (54). FISH is typically used
to locate and examine chromosomal, genetic and genomic aberrations
that are associated with the development and progression of disease
(56). Therefore, it has clinically
important applications in cytogenetic and oncology, including in
identifying gene alterations in patients with cancer (56). A modification of this technique,
DBD-FISH, has been used to investigate cervical cancer progression
by detecting and quantifying DNA breaks in genomic regions that are
sensitive to destabilization (57).
This technique allows detection and quantification of SSBs and DSBs
in the genome or in a specific DNA sequence from a single cell
(58). There are certain
disadvantages in fluorescence assays, including the reproducibility
and irregularity of the signals, and background autofluorescence
(54).
FCM-Annexin V labeling
When DNA breakage occurs, it is important to
differentiate between necrosis, autolysis and apoptosis (59). FCM was developed to detect apoptosis
(60); this method allows for the
measure of a large number of cells, and is also used to detect DNA
strand fragmentation, chromosomal aberrations and chemical adducts
in DNA (61,62).
Annexin V protein is used to quantify the number of
dead or apoptotic cells (63). The
lipid bilayer in healthy cells does not allow for Annexin V
binding, however, in cells undergoing apoptosis, Annexin V binds to
the outer surface of the cell membrane following translocation of
phosphatidylserine in the presence of Ca2+ (64). The number of apoptotic cells may be
quantified using FCM (65). With the
use of a secondary antibody tagged with FITC or PI, this method may
detect important proteins involved in DNA repair complexes
(27). FCM is able to rapidly and
sensitively measure DNA damage compared with the frequently used
comet assay method.
Radioimmunoassay (RIA)
The RIA binding assay is used to measure the
concentration of antigens using specific antibodies. The target
antigen is synthesized with a radiolabel and without a label, and
is subsequently bound to specific antibodies (66). Following the introduction of a sample,
a competitive reaction develops between the radiolabeled antigens
and the unlabeled antigens from the sample, and this releases an
amount of radiolabeled antigen. Standard curves may be obtained
from this process by mixing equal amounts of antibody and
radiolabeled antigen, with increasing concentrations of non-labeled
antigen in a constant volume; unknown antigen is similarly mixed
with antibody and radiolabeled antigen, and the concentration may
be subsequently determined (67).
This assay may be used to estimate the quantity of 6–4
photoproducts and cyclobutane dimers in DNA (45).
Chemiluminescence strategies
Enzyme-linked immunosorbent assay
(ELISA)
This is one of the most commonly used immunological
methods for the quantification of DNA damage (67) and consists of affixing an unknown
quantity of antigen to a surface and applying an unknown quantity
of antibody to the surface so that the antibody binds to the
antigen. The antibody is linked to an enzyme that may be quantified
via the addition of an appropriate substrate (colored, fluorescent
or radioactive) (45,67).
Immunohistochemical assay
This assay utilizes fixed cells that have previously
been treated with proteases and RNase. This process removes
proteins and RNA, and this ensures that cross-reaction with DNA
does not occur (67). A solution of
PI is used to counterstain the cells. The resulting
immunofluorescence allows for visualization of the nuclei in
adduct-negative cells (45).
Immunohistochemical assays, in addition to FISH, have served as a
more effective screening and diagnostic tool to detect alterations
in certain metabolites, including the case of ALK gene in non-small
cell lung cancer (68).
Immunological assay
This technique measures the presence of oxidative
DNA via the immunoslot-blot system, and uses chemiluminescent
detection and secondary antibodies that are conjugated to alkaline
phosphatase enzymes and radioactive iodine (69). This assay is effective, but is limited
by the cross-reactivity of the antibodies with normal DNA
bases.
Analytical strategies
High performance liquid chromatography
(HPLC)-electrospray tandem mass spectrometry (MS)
Oxidative stress and absorption of UV light by
nucleic acids has been established to be one of the causes of
oxidative DNA damage, which may promote cancer development
(70,71). The improvement of HPLC coupled to
tandem MS with an electrospray ionization mode, may be a sensitive
and accurate method to detect modified bases of the
oxidative-damaged DNA and UV-induced dimeric pyrimidine
photoproducts (72). Notably, during
the initial steps of the BER, the simultaneous detection and
quantification of altered and released nucleobases from genomic DNA
may be conducted using HPLC-MS (73).
Therefore, this technique may be useful for detecting SSBs, as
these lesions and base alterations are involved with proteins of
the BER pathway (74).
This assay has been used to quantify oxidized
nucleosides, including 8-oxo-7,8-dihydro-2′-deoxyguanosine,
8-oxo-7,8-dihydro-2′-deoxyadenosine, 5-formyl-2′-deoxyuridine,
5-hydroxymethyl-2′-deoxyuridine, 5-hydroxy-2′-deoxyuridine and the
four diastereomers of 5,6-dihydroxy-5,6-dihydrothymidine within
isolated and cellular DNA following exposure to γ-rays (75). It is also possible to detect tandem
DNA lesions as dinucleoside monophosphates, and in addition to
detecting the type of DNA damage, HPLC-MS may also provide
information on the location and quantity of DNA damage (75,76).
Despite the advantage of accuracy, this assay has the limitations
of a high cost and the large amount of experience that is required
to accurately use the technique to monitor the formation of low
levels of oxidized bases within cellular DNA (75). However, it remains the method of
choice for measuring modified DNA bases.
Gas chromatography-mass spectrometry
(GC-MS)
To understand diverse cellular processes, including
DNA damage, repair and its biological consequences, it is important
to characterize and quantify DNA lesions.
MS provides structural evidence for a biological or
chemical analysis, and in combination with gas chromatography, it
enables measurements of more complex samples (77). GC-MS is a technique capable of
measuring numerous products of DNA damage, including those of the
sugar moiety and heterocyclic bases, as in HPLC-MS (78). The MS analysis provides sensitive
detection of a single DNA lesion in DNA with multiple lesions or
nucleobases following chemical or enzyme degradation of the nucleic
acids (79). Additionally, this
technique measures the kinetics of a number of DNA repair enzymes
and is able to identify and quantify the expression levels of DNA
repair proteins in human tissues (80,81).
Typically, these measurements include the hydrolysis of DNA, the
derivatization of hydrolysates and the separation via gas
chromatography of hydrolysates that are identified and quantified
using MS (78). GC-MS has also been
used to identify DNA-protein crosslinks, including Thy-Gly, Thy-Ala
and Cyt-Tyr, in mammalian chromatin in vitro (82–84).
Electrochemical methods (EM)
It has been established that DNA may be damaged by
reactive oxygen species and the alterations in DNA that are formed
are detected using electrochemical methods based on the inherent
sensitivity of DNA-mediated charge transport (CT). These methods
are also capable of detecting base pair mismatches and the majority
of base damage products (85). This
methodology may detect DNA-mediated CT as a damage detection
mechanism for DNA repair enzymes (86). There have been hypotheses regarding
the development of a sensor for the detection of single base
mutations and DNA base lesions in duplex DNA to utilize the
sensitivity of this charge to transport DNA films (87). The electrochemical method,
electrocatalysis, has provided the basis for novel assays to detect
low levels of lesions and possible for use as an early diagnostic
tool. Although this is a method that provides sensitive, selective
and low cost detection of DNA damage, it has the limitation of not
being able to recognize thymidine dimer lesions until they are
connected with the distortion of DNA double helix (45).
Conclusions
Fig. 1 presents a
summary of the distinct types of DNA lesions, the repair pathways
that are involved and the experimental strategies used to evaluate
each type. The importance of the study of DNA damages and how
damage may be restored requires further study, as it has clinical
implications in multifactorial diseases, including cancer and
diabetes. There are a number of methods available for the
detection, analysis and quantification of DNA lesions and it is
important to identify the advantages and disadvantages of each
approach. The combination of these methodologies may provide an
overview of DNA lesion analysis and complementary information. In
contrast to the methodologies described in the present review,
these molecular strategies may be considered to be accurate and
sensitive, as they examine the type of DNA damage as well as the
repair mechanism involved. Notably, the accumulated research in the
current review may promote further studies to demonstrate potential
phenotypic alterations that occur from DNA lesions.
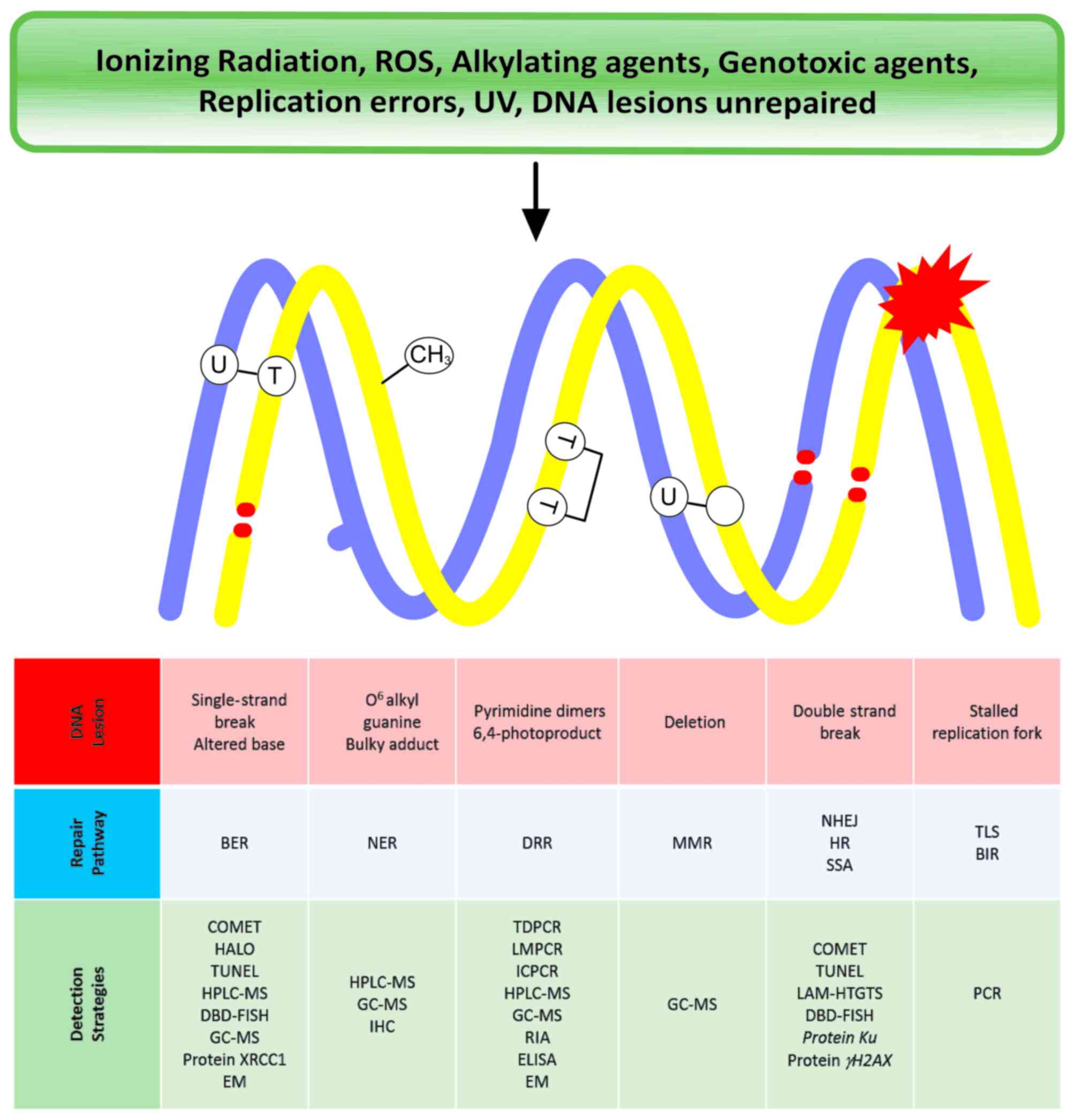 | Figure 1.Summary of distinct types of DNA
lesions, the repair pathways involved in their repair and the
experimental strategies that are used to evaluate each type. ROS,
reactive oxygen species; UV, ultraviolet; BER, base excision
repair; NER, nucleotide excision repair; DDR, DNA damage repair;
MMR, mismatch repair; NHEJ, non-homologous end joining; HR,
homologous recombination; SSR, single strand repair; TLS,
translesion synthesis; BIR, base incision repair; COMET,
single-cell gel electrophoresis; TUNEL, terminal deoxynucleotidyl
transferase dUTP nick-end labeling; HPLC-MS, high performance
liquid chromatography-mass spectrometry; DBD-FISH, DNA breakage
detection-fluorescence in situ hybridization; GC-MS, gas
chromatography-mass spectrometry; XRCC1, X-ray repair cross
complementing 1; EM, electrochemical methods; IHC,
immunohistochemistry; TDPCR, terminal transferase-dependent
polymerase chain reaction; LMPCR, ligation-mediated polymerase
chain reaction; ICPCR, immune-coupled polymerase chain reaction;
RIA, radioimmunoassay; ELISA, enzyme-linked immunosorbent assay;
LAM-HTGTS, linear amplification-mediated high-throughout
genome-wide translocation sequencing; PCR, polymerase chain
reaction. |
Acknowledgements
This review was supported by CONACyT research funds
(grant no. PN-2014-249020) and the National Autonomous University
of México (grant no. PAPIIT-IN207216).
References
|
1
|
Cadet J and Wagner JR: DNA base damage by
reactive oxygen species, oxidizing agents, and UV radiation. Cold
Spring Harb Perspect Biol. 5:a0125592013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Z, Pearlman AH and Hsieh P: DNA
mismatch repair and the DNA damage response. DNA Repair (Amst).
38:94–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Altaf M, Saksouk N and Côté J: Histone
modifications in response to DNA damage. Mutat Res. 618:81–90.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lord CJ and Ashworth A: The DNA damage
response and cancer therapy. Nature. 481:287–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cao LL, Shen C and Zhu WG: Histone
modifications in DNA damage response. Sci China Life Sci.
59:257–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schipler A and Iliakis G: DNA
double-strand-break complexity levels and their possible
contributions to the probability for error-prone processing and
repair pathway choice. Nucleic Acids Res. 41:7589–7605. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grimaldi KA, McGurk CJ, McHugh PJ and
Hartley JA: PCR-based methods for detecting DNA damage and its
repair at the sub-gene and single nucleotide levels in cells. Mol
Biotechnol. 20:181–196. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Osborne DJ: Technologies for detection of
DNA damage and mutations. Endeavour. 21:178–179. 1997. View Article : Google Scholar
|
|
9
|
Furda A, Santos JH, Meyer JN and van
Houten B: Quantitative PCR-based measurement of nuclear and
mitochondrial DNA damage and repair in mammalian cellsMolecular
Toxicology Protocols. Keohavong P and Grant GS: Humana Press;
Totowa, NJ: pp. 419–437. 2014, View Article : Google Scholar
|
|
10
|
Senoo T, Yamanaka M, Nakamura A, Terashita
T, Kawano S and Ikeda S: Quantitative PCR for detection of DNA
damage in mitochondrial DNA of the fission yeast
Schizosaccharomyces pombe. J Microbiol Methods. 127:77–81. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
De Boer JG and Glickman BW: Mutations
recovered in the Chinese hamster aprt gene after exposure to
carboplatin: A comparison with cisplatin. Carcinogenesis. 13:15–17.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Van Houten B, Cheng S and Chen Y:
Measuring gene-specific nucleotide excision repair in human cells
using quantitative amplification of long targets from nanogram
quantities of DNA. Mutat Res. 460:81–94. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Strauss EC and Orkin SH: Guanine-adenine
ligation-mediated PCR in vivo footprinting. Methods. 11:164–170.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pfeifer GP and Tornaletti S: Footprinting
with UV irradiation and LMPCR. Methods. 11:189–196. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pfeifer GP, Chen HH, Komura J and Riggs
AD: Chromatin structure analysis by ligation-mediated and terminal
transferase-mediated polymerase chain reaction. Methods Enzymol.
304:548–571. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chang L, Li J and Wang L: Immuno-PCR: An
ultrasensitive immunoassay for biomolecular detection. Anal Chim
Acta. 910:12–24. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karakoula A, Evans MD, Podmore ID,
Hutchinson PE, Lunec J and Cooke MS: Quantification of UVR-induced
DNA damage: Global-versus gene-specific levels of thymine dimers. J
Immunol Methods. 277:27–37. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang G, Hallberg LM and Englander EW:
Rapid SINE-mediated detection of cisplatin: DNA adduct formation in
vitro and in vivo in blood. Mutat Res. 434:67–74. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang G, Hallberg LM, Saphier E and
Englander EW: Short interspersed DNA element-mediated detection of
UVB-induced DNA damage and repair in the mouse genome, in vitro,
and in vivo in skin. Mutat Res. 433:147–157. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Featherstone C and Jackson SP: Ku, a DNA
repair protein with multiple cellular functions? Mutat Res.
434:3–15. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones JM, Gellert M and Yang W: A Ku
bridge over broken DNA. Structure. 9:881–884. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gullo C, Au M, Feng G and Teoh G: The
biology of Ku and its potential oncogenic role in cancer. Biochim
Biophys Acta. 1765:223–234. 2006.PubMed/NCBI
|
|
23
|
Doherty AJ and Jackson SP: DNA repair: How
Ku makes ends meet. Curr Biol. 11:R920–R924. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Walker JR, Corpina RA and Goldberg J:
Structure of the Ku heterodimer bound to DNA and its implications
for double-strand break repair. Nature. 412:607–614. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Paull TT, Rogakou EP, Yamazaki V,
Kirchgessner CU, Gellert M and Bonner WM: A critical role for
histone H2AX in recruitment of repair factors to nuclear foci after
DNA damage. Curr Biol. 10:886–895. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rogakou EP, Boon C, Redon C and Bonner WM:
Megabase chromatin domains involved in DNA double-strand breaks in
vivo. J Cell Biol. 146:905–916. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Henderson DS: DNA repair protocolsMethods
in Molecular Biology. 2nd. Humana Press; New Jersey, NJ: pp.
4982006
|
|
28
|
Levy N, Martz A, Bresson A, Spenlehauer C,
De Murcia G and Ménissier-De Murcia J: Xrcc1 is phosphorylated by
DNA-dependent protein kinase in response to DNA damage. Nucleic
Acids Res. 34:32–41. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Caldecott KW: Protein-protein interactions
during mammalian DNA single-strand break repair. Biochem Soc Trans.
31:247–251. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brem R and Hall J: XRCC1 is required for
DNA single-strand break repair in human cells. Nucleic Acids Res.
33:2512–2520. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Goode EL, Ulrich CM and Potter JD:
Polymorphisms in DNA repair genes and associations with cancer
risk. Cancer Epidemiol Biomarkers Prev. 11:1513–1530.
2002.PubMed/NCBI
|
|
32
|
Strom CE, Mortusewicz O, Finch D, Parsons
JL, Lagerqvist A, Johansson F, Schultz N, Erixon K, Dianov GL and
Helleday T: CK2 phosphorylation of XRCC1 facilitates dissociation
from DNA and single-strand break formation during base excision
repair. DNA Repair (Amst). 10:961–969. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Caldecott KW: XRCC1 and DNA strand break
repair. DNA Repair (Amst). 2:955–969. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Collins AR: The comet assay for DNA damage
and repair: Principles, applications, and limitations. Mol
Biotechnol. 26:249–261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Collins AR: Measuring oxidative damage to
DNA and its repair with the comet assay. Biochim Biophys Acta.
1840:794–800. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Collins AR and Azqueta A: DNA repair as a
biomarker in human biomonitoring studies; further applications of
the comet assay. Mutat Res. 736:122–129. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kent CR, Eady JJ, Ross GM and Steel GG:
The comet moment as a measure of DNA damage in the comet assay. Int
J Radiat Biol. 67:655–660. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jyoti S, Khan S, Naz F, Ali Rahul F and
Siddique YH: Assessment of DNA damage by panmasala, gutkha chewing
and smoking in buccal epithelial cells using alkaline single cell
gel electrophoresis (SCGE). Egyptian J Med Human Gen. 14:391–394.
2013. View Article : Google Scholar
|
|
39
|
Ostling O and Johanson KJ:
Microelectrophoretic study of radiation-induced DNA damages in
individual mammalian cells. Biochem Biophys Res Commun.
123:291–298. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Glei M, Hovhannisyan G and Pool-Zobel BL:
Use of comet-FISH in the study of DNA damage and repair: Review.
Mutat Res. 681:33–43. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Collins AR, Dobson VL, Dušinská M, Kennedy
G and Štětina R: The comet assay: What can it really tell us? Mutat
Res. 375:183–193. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rojas E, Lopez MC and Valverde M: Single
cell gel electrophoresis assay: Methodology and applications. J
Chromatogr B Biomed Sci Appl. 722:225–254. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Angelis KJ, Dusinská M and Collins AR:
Single cell gel electrophoresis: Detection of DNA damage at
different levels of sensitivity. Electrophoresis. 20:2133–2138.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Collins AR and Azqueta A: Chapter
4-single-cell gel electrophoresis combined with lesion-specific
enzymes to measure oxidative damage to DNAMethods in Cell Biology.
Conn PM: Academic Press; Burlington, MA: pp. 69–92. 2012,
View Article : Google Scholar
|
|
45
|
Kumari S, Rastogi R, Singh K, Singh S and
Sinha R: DNA damage: Detection strategies. EXCLI J. 7:44–62.
2008.
|
|
46
|
Roti Roti JL and Wright WD: Visualization
of DNA loops in nucleoids from HeLa cells: Assays for DNA damage
and repair. Cytometry. 8:461–467. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gavrieli Y, Sherman Y and Ben-Sasson SA:
Identification of programmed cell death in situ via specific
labeling of nuclear DNA fragmentation. J Cell Biol. 119:493–501.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Walker PR, Carson C, Leblanc J and
Sikorska M: Labeling DNA damage with terminal transferase, in in
situ detection of DNA damageMethods and Protocols. Didenko VV:
Humana Press; Totowa, NJ: pp. 3–19. 2002
|
|
49
|
Loo DT: TUNEL assay. An overview of
techniques. Methods Mol Biol. 203:21–30. 2002.PubMed/NCBI
|
|
50
|
Kanoh M, Takemura G, Misao J, Hayakawa Y,
Aoyama T, Nishigaki K, Noda T, Fujiwara T, Fukuda K, Minatoguchi S
and Fujiwara H: Significance of myocytes with positive DNA in situ
nick end-labeling (TUNEL) in hearts with dilated cardiomyopathy:
Not apoptosis but DNA repair. Circulation. 99:2757–2764. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Otsuki Y and Ito Y: Quantitative
differentiation of both free 3′ oh and 5′ oh DNA ends using
terminal transferase-based labeling combined with transmission
electron microscopy, in in situ detection of DNA damageMethods and
Protocols. Didenko VV: Humana Press; Totowa, NJ: pp. 41–54.
2002
|
|
52
|
Didenko VV and Hornsby PJ: Presence of
double-strand breaks with single-base 3′ overhangs in cells
undergoing apoptosis but not necrosis. J Cell Biol. 135:1369–1376.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Didenko VV: Detection of specific
double-strand DNA breaks and apoptosis in situ using T4 DNA ligase.
Methods Mol Biol. 203:143–151. 2002.PubMed/NCBI
|
|
54
|
Levsky JM and Singer RH: Fluorescence in
situ hybridization: Past, present and future. J Cell Sci.
116:2833–2838. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gall JG and Pardue ML: Formation and
detection of RNA-DNA hybrid molecules in cytological preparations.
Proc Natl Acad Sci USA. 63:378–383. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Halling KC and Kipp BR: Fluorescence in
situ hybridization in diagnostic cytology. Hum Pathol.
38:1137–1144. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cortés-Gutiérrez EI, Fernández JL,
Dávila-Rodríguez MI, López-Fernández C and Gosálvez J: Use of
DBD-FISH for the study of cervical cancer progression, in cervical
cancerMethods and Protocols. Keppler D and Lin WA: Springer; New
York, NY: pp. 291–301. 2015
|
|
58
|
Fernández JL, Vázquez-Gundín F, Rivero MT,
Genescá A, Gosálvez J and Goyanes V: DBD-fish on neutral comets:
Simultaneous analysis of DNA single- and double-strand breaks in
individual cells. Exp Cell Res. 270:102–109. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fink SL and Cookson BT: Apoptosis,
pyroptosis, and necrosis: Mechanistic description of dead and dying
eukaryotic cells. Infect Immun. 73:1907–1916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Basiji D and O'Gorman MR: Imaging flow
cytometry. J Immunol Methods. 423:1–2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Muehlbauer PA and Schuler MJ: Detection of
numerical chromosomal aberrations by flow cytometry: A novel
process for identifying aneugenic agents. Mutat Res. 585:156–169.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Henry CM, Hollville E and Martin SJ:
Measuring apoptosis by microscopy and flow cytometry. Methods.
61:90–97. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huerta S, Goulet EJ, Huerta-Yepez S and
Livingston EH: Screening and detection of apoptosis. J Surg Res.
139:143–156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vermes I, Haanen C, Steffens-Nakken H and
Reutelingsperger C: A novel assay for apoptosis. Flow cytometric
detection of phosphatidylserine expression on early apoptotic cells
using fluorescein labelled Annexin V. J Immunol Methods. 184:39–51.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Pietkiewicz S, Schmidt JH and Lavrik IN:
Quantification of apoptosis and necroptosis at the single cell
level by a combination of imaging flow cytometry with classical
Annexin V/propidium iodide staining. J Immunol Methods. 423:99–103.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Berton TR and Mitchell DL: Quantification
of DNA photoproducts in mammalian cell DNA using radioimmunoassay.
Methods Mol Biol. 920:177–187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Santella RM: Immunological methods for
detection of carcinogen-DNA damage in humans. Cancer Epidemiol
Biomarkers Prev. 8:733–739. 1999.PubMed/NCBI
|
|
68
|
Yatabe Y: ALK FISH and IHC: You cannot
have one without the other. J Thorac Oncol. 10:548–550. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kriste AG, Martincigh BS and Salter LF: A
sensitive immunoassay technique for thymine dimer quantitation in
UV-irradiated DNA. J Photochem Photobiol A: Chemistry. 93:185–192.
1996. View Article : Google Scholar
|
|
70
|
El-Yazbi AF and Loppnow GR: Detecting
UV-induced nucleic-acid damage. TrAC Trends Analytical Chemistry.
61:83–91. 2014. View Article : Google Scholar
|
|
71
|
Toyokuni S: Oxidative stress as an iceberg
in carcinogenesis and cancer biology. Arch Biochem Biophys.
595:46–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rindgen D, Turesky RJ and Vouros P:
Determination of in vitro formed DNA adducts of
2-amino-1-methyl-6-phenylimidazo [4,5-b]pyridine using capillary
liquid chromatography/electrospray ionization/tandem mass
spectrometry. Chem Res Toxicol. 8:1005–1013. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mullins EA, Rubinson EH, Pereira KN,
Calcutt MW, Christov PP and Eichman BF: An HPLC-tandem mass
spectrometry method for simultaneous detection of alkylated base
excision repair products. Methods. 64:59–66. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Caldecott KW: DNA single-strand break
repair. Exp Cell Res. 329:2–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Cadet J, Douki T, Frelon S, Sauvaigo S,
Pouget JP and Ravanat JL: Assessment of oxidative base damage to
isolated and cellular DNA by HPLC-MS/MS measurement. Free Radic
Biol Med. 33:441–449. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Pouget JP, Douki T, Richard MJ and Cadet
J: DNA damage induced in cells by gamma and UVA radiation as
measured by HPLC/GC-MS and HPLC-EC and comet assay. Chem Res
Toxicol. 13:541–549. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gowda GA and Djukovic D: Overview of mass
spectrometry-based metabolomics: Opportunities and challenges.
Methods Mol Biol. 1198:3–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Dizdaroglu M, Coskun E and Jaruga P:
Measurement of oxidatively induced DNA damage and its repair, by
mass spectrometric techniques. Free Radic Res. 49:525–548. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Sato K and Greenberg MM: Selective
detection of 2-deoxyribonolactone in DNA. J Am Chem Soc.
127:2806–2807. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Dizdaroglu M: Substrate specificities and
excision kinetics of DNA glycosylases involved in base-excision
repair of oxidative DNA damage. Mutat Res. 531:109–126. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Reddy PT, Jaruga P, Nelson BC, Lowenthal M
and Dizdaroglu M: Stable isotope-labeling of DNA repair proteins
and their purification, and characterization. Protein Expr Purif.
78:94–101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Gajewski E and Dizdaroglu M: Hydroxyl
radical induced cross-linking of cytosine and tyrosine in
nucleohistone. Biochemistry. 29:977–980. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Koivisto P and Peltonen K: Analytical
methods in DNA and protein adduct analysis. Anal Bioanal Chem.
398:2563–2572. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Dizdaroglu M and Gajewski E: Structure and
mechanism of hydroxyl radical-induced formation of a DNA-protein
cross-link involving thymine and lysine in nucleohistone. Cancer
Res. 49:3463–3467. 1989.PubMed/NCBI
|
|
85
|
Fojta M, Daňhel A, Havran L and Vyskočil
V: Recent progress in electrochemical sensors and assays for DNA
damage and repair. TrAC Trends Analytical Chemistry. 79:160–167.
2016. View Article : Google Scholar
|
|
86
|
Boal AK and Barton JK: Electrochemical
detection of lesions in DNA. Bioconjug Chem. 16:312–321. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Boon EM, Ceres DM, Drummond TG, Hill MG
and Barton JK: Mutation detection by electrocatalysis at
DNA-modified electrodes. Nat Biotechnol. 18:1096–1100. 2000.
View Article : Google Scholar : PubMed/NCBI
|















