Introduction
Breast cancer (BC) is the most commonly diagnosed
cancer and the leading cause of cancer-related death in women
worldwide (1). Approximately 1.7
million new cases of BC and 521,900 deaths related to BC are
estimated to have occurred worldwide in 2012 (2). Recurrent BC after resection is
frequently intractable and recognized as a major challenge in
clinical practice. Currently, adjuvant therapies such as
chemotherapy, endocrine therapy and anti-human epidermal growth
factor 2 (HER2) agent are administered to BC patients with the aim
of prevention of disease recurrences (3). However, many patients have suffered from
adverse events and other costs caused by such treatment. Risk
stratification is necessary for assessing indication for adjuvant
therapy; development of biomarkers for this purpose is therefore
needed.
Several commercial multigene expression assays
(Oncotype Dx®, MammaPrint®, PAM-50 ROR
score®, EndoPredict® and Breast Cancer
Index®) are currently available to predict patients'
prognoses and to evaluate whether adjuvant chemotherapy is
appropriate (3). However, such assays
are limited to estrogen receptor (ER)-positive, HER2-negative BC.
It's still difficult to predict prognosis of all BC subtypes.
The regulatory factor X1 (RFX1) gene is a
member of the regulatory factor X gene family which codes
transcription factors (4). Expression
and regulatory mechanisms of RFX1 transcription have been
reported in connection with several malignancies, including brain
tumors (5,6) and hepatocellular carcinoma (7). RFX1 is considered to work as a
tumor suppressor by downregulating the proto-oncogene c-myc
(8). However, the relationship
between RFX1 mRNA expression and patients'
clinicopathological factors in BC has not been studied. Here, we
focused on RFX1 and investigated its expression in BC cell
lines and patients' tumor samples to determine whether it could be
a prognostic marker for BC.
Materials and methods
Sample collection
We obtained 13 BC cell lines (BT-20, BT-474, BT-549,
HCC1419, HCC1954, Hs578T, MCF7, MDA-MB-231, MDA-MB-361, MDA-MB-415,
MDA-MB-468, SK-BR-3 and ZR-75-1) and two non-cancerous mammary
gland epithelial cell lines (MCF-10A and MCF-12A). BT-549, HCC1419,
HCC1954 and Hs578T were purchased from Japanese Collection of
Research Bioresources Cell Bank (Osaka, Japan), BT-474, MCF-7 and
MCF-12A were provided to our research team by Johns Hopkins
University and Dr. David Sidransky, the director of the Department
of Otolaryngology-Head and Neck Surgery of Johns Hopkins University
(Baltimore, MD, USA), and others were from the American Type
Culture Collection (Manassas, VA, USA). Cells were stored at −80°C
using cell preservative solution (Cell Banker; Mitsubishi Chemical
Medience Corporation, Tokyo, Japan) and cultured in RPMI-1640
(Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal
bovine serum and in an atmosphere containing 5% CO2 at
37°C (9,10). We acquired 167 primary BC tissues from
patients who underwent breast surgery at Nagoya University Hospital
between March 2002 and November 2009. All tissue samples were
diagnosed histologically as BC, frozen immediately after resection,
and stored at −80°C. Adjacent non-cancerous specimens were resected
>3 cm away from the edge of the tumor. BC specimens were
classified histologically using the 7th edition of the Union for
International Cancer Control (UICC) staging system for BC. Adjuvant
chemotherapy, endocrine therapy and anti-HER2 therapy were
administered to selected patients according to the patient's
condition, pathological factors, subtype and the physician's
discretion.
This study conforms to the ethical guidelines of the
World Medical Association Declaration of Helsinki: Ethical
Principles for Medical Research Involving Human Subjects. Written
informed consent for use of clinical samples and data was required
by the Institutional Review Board at Nagoya University (Nagoya,
Japan) and was obtained from all patients.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Levels of RFX1 mRNA were determined using
RT-qPCR. We extracted RNA from cell lines (8.0×106 cells
per each cell line), 167 primary BCs, and adjacent normal tissues
using the RNeasy Mini kit (Qiagen, Hilden, Germany) according to
the manufacture's protocol. cDNA was synthesized from total RNAs (1
µg) by M-MLV Reverse Transcriptase (Invitrogen Life Technologies,
Frederick, MD, USA) and Primer ‘random’ (Sigma-Aldrich).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels
were quantified to normalize expression levels. RT-qPCR was
performed using the SYBR-Green PCR Core Reagents kit (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) as
follows: 1 cycle at 95°C for 10 min, 40 cycles at 95°C for 5 sec,
and 60°C for 60 sec. All samples were tested in triplicate, and
samples without templates were included in each PCR plate as
negative controls. Real-time detection of SYBR Green fluorescence
was conducted using an ABI StepOnePlus Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
expression level of each sample was taken as the value of the
RFX1 amplicon divided by that of GAPDH (11,12).
Primers specific for RFX1 and GAPDH are listed in
Table I.
 | Table I.Primers. |
Table I.
Primers.
| Gene | Experiment | Type | Sequence
(5′-3′) | Product size | Annealing
temperature |
|---|
| RFX1 | RT-qPCR | Forward |
GCAGACAGAAGTGGGGAGAA | 241 bp | 60°C |
|
|
| Reverse |
CAGTATACGCCTGTGTTGCC |
|
|
|
| Bisulfite
sequencing in promoter region | Forward |
TGTTTTTGAGGGTTTAGTTTTTTTT | 207 bp | 56°C |
|
|
| Reverse |
ACAACTATTACTACCCACCCTAATTAC |
|
|
|
| Bisulfite
sequencing in intron 7 | Forward |
GGTGGAGGTTTGGAGTTT | 273 bp | 60°C |
|
|
| Reverse |
ACAAAAACAAATATAAAAACAACA |
|
|
|
| MSP | Forward |
TTTTTCGTTTTTATTTAATTTCGAC | 167 bp | 56°C |
|
|
| Reverse |
ATTTCTAACTTCTTACGCTAACGTC |
|
|
|
| U-MSP | Forward |
GGAGGTGTTATAGTTATGGTAGTTGT | 164 bp | 56°C |
|
|
| Reverse |
CTTTTCAAAAATCTCAAAATCAAT |
|
|
| GAPDH | RT-qPCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 bp | 60°C |
|
|
| Probe |
CAAGCTTCCCGTTCTCAGCC |
|
|
|
|
| Reverse |
GAAGATGGTGATGGGATTTC |
|
|
Surveillance of CpG island around the
RFX1 gene
Nucleotide sequence analysis was conducted to
determine the presence of CpG island around the promoter region of
RFX1. CpG island was defined using the following criteria:
At least 500-bp region of DNA with a high GC content (>50%) and
an observed CpG/expected CpG ratio of ≥0.6 (13). We used the MethPrime (http://www.urogene.org/methprimer/) to determine
the location of CpG island (14).
Bisulfite sequence analysis
Genomic DNA samples from 15 cell lines were
subjected to bisulfite treatment using the BisulFlash DNA
Modification Kit (EPIGENTEK, Farmingdale, NY, USA); PCR was
conducted to detect hypermethylation of the RFX1 promoter
region and intron 7 (5), using
specific primers (Table I). The PCR
amplification protocols were, for the promoter region: 50 cycles at
94°C for 20 sec, 56°C for 20 sec, and 72°C for 25 sec; and for
intron 7: 50 cycles at 94°C for 20 sec, 60°C for 20 sec, and 72°C
for 25 sec; both amplifications used an initial denaturation step
at 94°C for 2 min. PCR products were purified directly using the
MinElute PCR Purification Kit (Qiagen). Sequence analysis was
carried out by Eurofins Genomics Co. (Tokyo, Japan) (15).
Methylation-specific polymerase chain
reaction (MSP)
DNA samples from BC cell lines, clinical BC
specimens, and corresponding non-cancerous specimens were subjected
to bisulfite treatment. MSP was conducted using specific primers to
evaluate the presence of hypermethylation of RFX1 promoter
region (Table I). The PCR
amplification protocols for MSP and un-MSP (U-MSP) were as follows:
50 cycles at 94°C for 15 sec, 56°C for 15 sec, and 72°C for 30 sec;
both amplifications used an initial denaturation step at 94°C for 2
min. Each PCR product was loaded directly on to 2% agarose gels,
stained with ethidium bromide and visualized under UV
illumination.
Statistical analysis
Differences in levels of RFX1 mRNA between BC
and adjacent normal tissues were analyzed using the Mann-Whitney U
test. The χ2 test was used to analyze the significance
of the association between RFX1 expression and
clinicopathological parameters. Overall survival (OS) and
disease-free survival (DFS) rates were calculated using the
Kaplan-Meier method; difference in survival curves was analyzed
using the log-rank test. We performed multivariate regression
analyses using the Cox proportional hazards model to identify
prognostic factors, and variables for which P<0.05 were entered
into the final model. Statistical analyses were performed using JMP
11 software (SAS Institute, Inc., Cary, NC, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
RFX1 mRNA expression and DNA
methylation status of BC cell lines
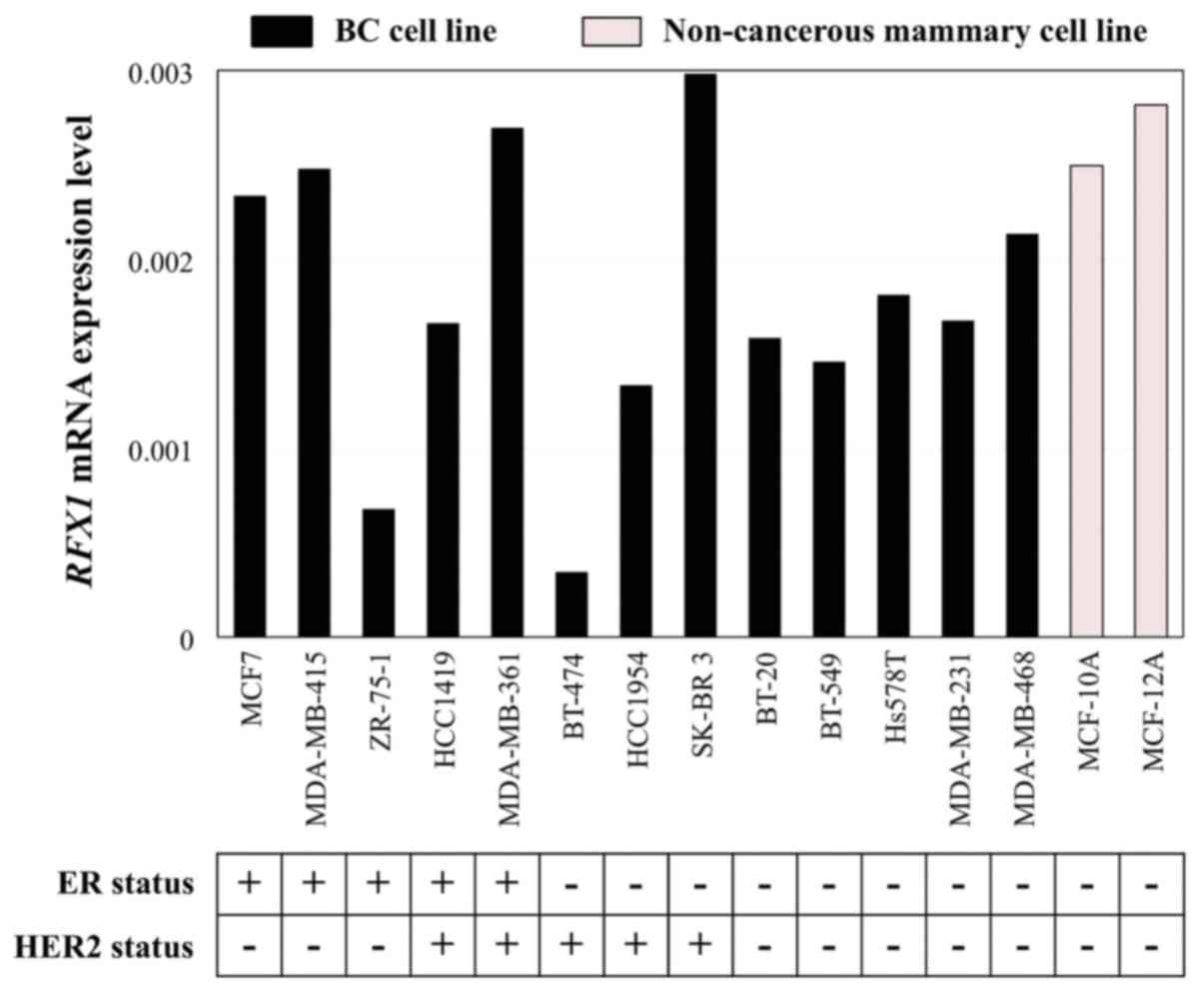
RFX1 mRNA expression levels were
heterogeneous among the 13 BC cell lines and two non-cancerous
mammary cell lines (Fig. 1). We
accepted the reports of each cell line's immunostaining status from
previous studies (16,17).
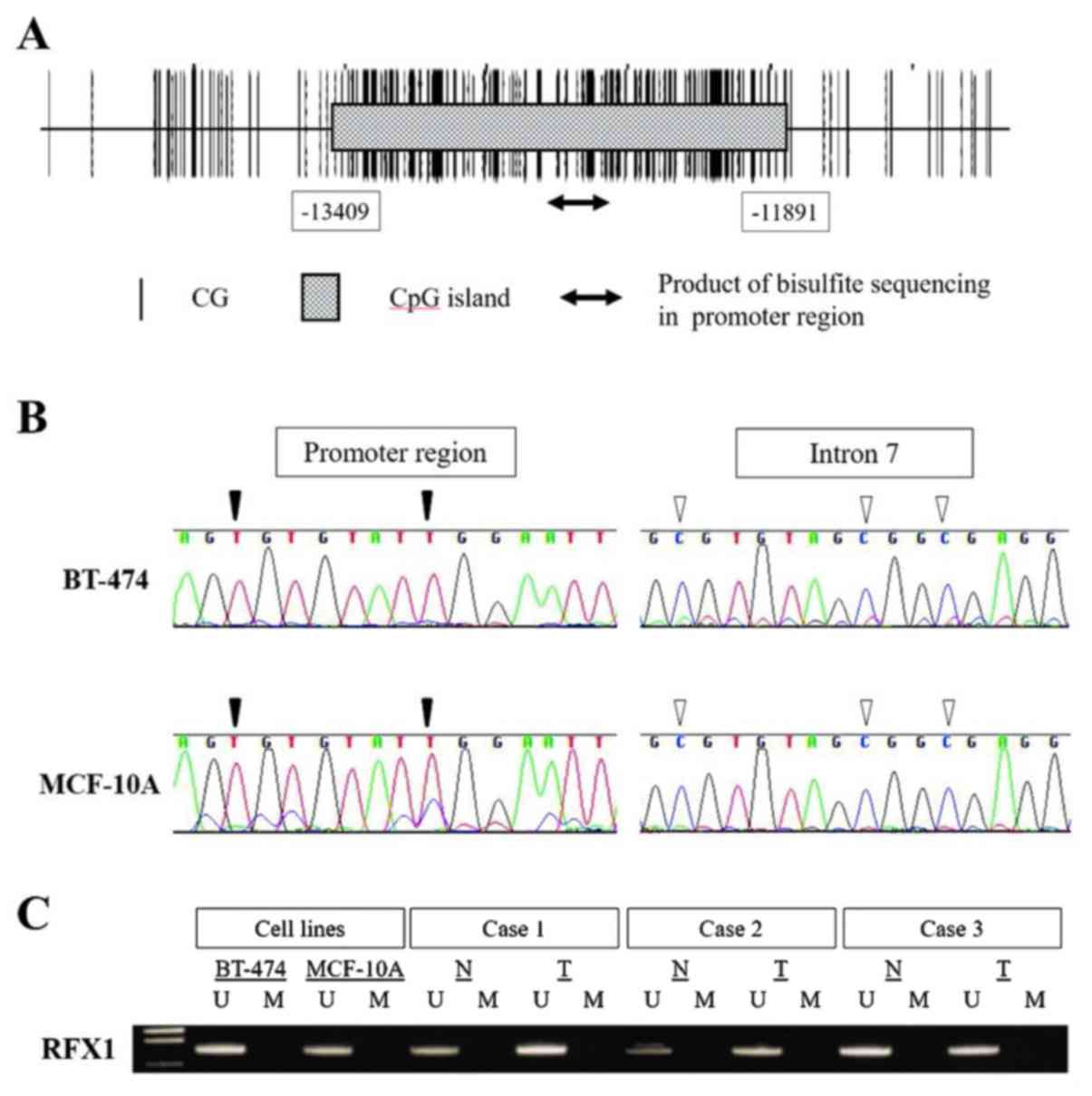
A CpG island was found at upstream of the
transcription initiation site of RFX1 (length 1,519 bases;
Fig. 2A). We examined the methylation
status of the 15 cell lines using bisulfite sequence analysis,
which detected no CpG methylation in the RFX1 promoter
regions of all cell lines, whereas CpG hypermethylation in intron 7
was detected in all cell lines including the non-cancerous mammary
cell lines (Fig. 2B). BT-474,
MCF-10A, and three representative clinical BC and corresponding
non-cancerous tissues were subjected to MSP. We found absence of
methylation in the promoter region of RFX1 gene both in cell
lines and clinical samples (Fig.
2C).
Patient characteristics
All 167 patients were women. Their mean age was
54.4±11.6 years (mean ± standard deviation; range, 26–78 years).
Their disease stage (by UICC staging system) were stage 0, 7; stage
I, 47; stage II, 78; stage III, 34; and stage IV, 1. The median
duration of patient follow-up was 100.0 months (range, 8–155
months) or until death. The immunostaining status of primary tumors
were ER positive, n=127; negative, n=40; progesterone receptor
(PgR) positive, n=115; negative, n=52; and HER2 positive, n=39;
negative, n=119.
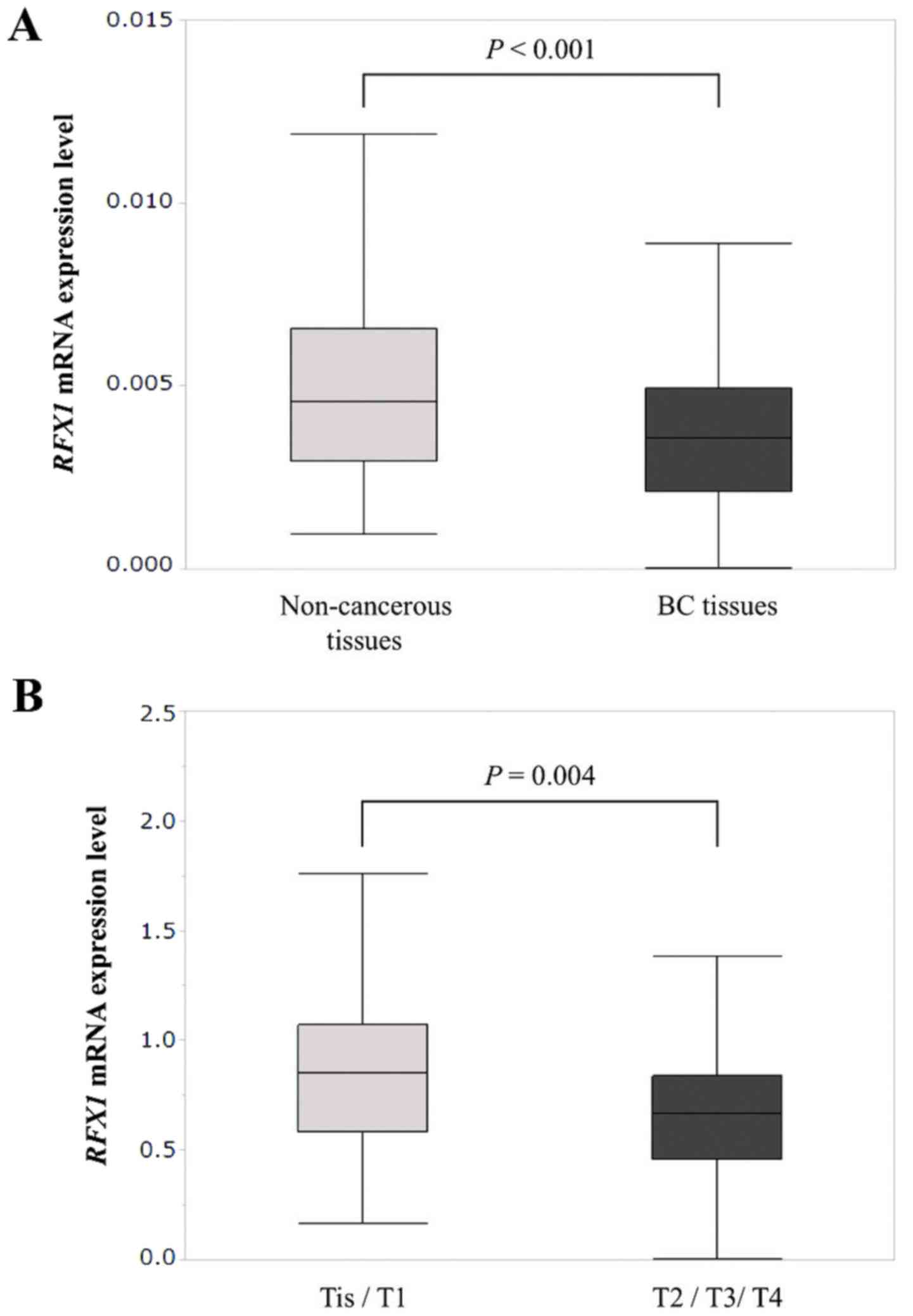
Mean expression levels of RFX1 mRNA in BC
tissues was significantly lower than those in corresponding
non-cancerous tissues (P<0.001; Fig.
3A). In 43 (26%) of the 167 patients, RFX1 mRNA
expression levels in BC tissues were higher than in adjacent normal
tissues. We calculated the ratio of RFX1 mRNA expression
levels between BC and adjacent non-cancerous tissues. Patients
whose RFX1 mRNA levels were lower in BC tissue than in
adjacent tissues were designated as ‘the low RFX1 group’
(n=124), and patients with higher RFX1 expression were
designated as ‘the high RFX1 group’ (n=43) in the following
analyses.
Clinical significance of RFX1 mRNA
levels
The high RFX1 group was significantly
associated with less advanced UICC T factor (P=0.028) and lower
pathological stage (P=0.015). The high RFX1 group correlated
significantly with ER positive (P=0.005), PgR positive (P=0.011),
HER2 negative status (P=0.001; Table
II).
 | Table II.Associations between expression of
RFX1 mRNA and clinicopathological parameters in 167 patients
with breast cancer. |
Table II.
Associations between expression of
RFX1 mRNA and clinicopathological parameters in 167 patients
with breast cancer.
| Clinicopathological
parameter | The low RFX1
group (n=124) | The high
RFX1 group (n=43) | P-value |
|---|
| Age |
|
|
|
| ≤60
year | 78 | 30 |
|
| >60
year | 46 | 13 | 0.413 |
| Histology |
|
|
|
|
DCIS | 4 | 3 |
|
|
IDC | 110 | 38 |
|
|
ILC | 5 | 2 |
|
|
Others | 5 | 0 | 0.265 |
| UICC T factor |
|
|
|
|
Tis/T1 | 51 | 26 |
|
|
T2/T3/T4 | 73 | 17 |
0.028a |
| Node status |
|
|
|
|
Negative | 62 | 23 |
|
|
Positive | 62 | 20 | 0.693 |
| UICC pathological
stage |
|
|
|
|
0/I/II | 92 | 39 |
|
|
III/IV | 32 | 4 | 0.015a |
| ER status |
|
|
|
|
Positive | 88 | 39 |
|
|
Negative | 36 | 4 |
0.005a |
| PgR status |
|
|
|
|
Positive | 79 | 36 |
|
|
Negative | 45 | 7 |
0.011a |
| HER2 status |
|
|
|
|
Positive | 37 | 2 |
|
|
Negative | 82 | 37 |
|
|
Unknown | 5 | 4 |
0.001a |
|
Triple-negative |
|
|
|
|
True | 15 | 3 |
|
|
False | 108 | 40 |
|
|
Unknown | 1 | 0 | 0.323 |
| Adjuvant
therapy |
|
|
|
|
Endocrine therapy alone | 35 | 22 |
|
|
Chemotherapy alone | 27 | 3 |
|
|
Endocrine and
chemotherapy | 48 | 16 |
|
|
None | 14 | 2 |
0.012a |
Mean RFX1 mRNA expression in T2/T3/T4
specimens (n=90) was significantly lower than in the Tis (carcinoma
in situ)/T1 group specimens (n=77; P=0.004; Fig. 3B). RFX1 mRNA levels did not
significantly differ between patients with negative and positive
lymph node metastases (n=85 and 82 respectively, P=0.181), or
between UICC stage 0/I/II specimens (n=131) and stage III/IV (n=36;
P=0.069).
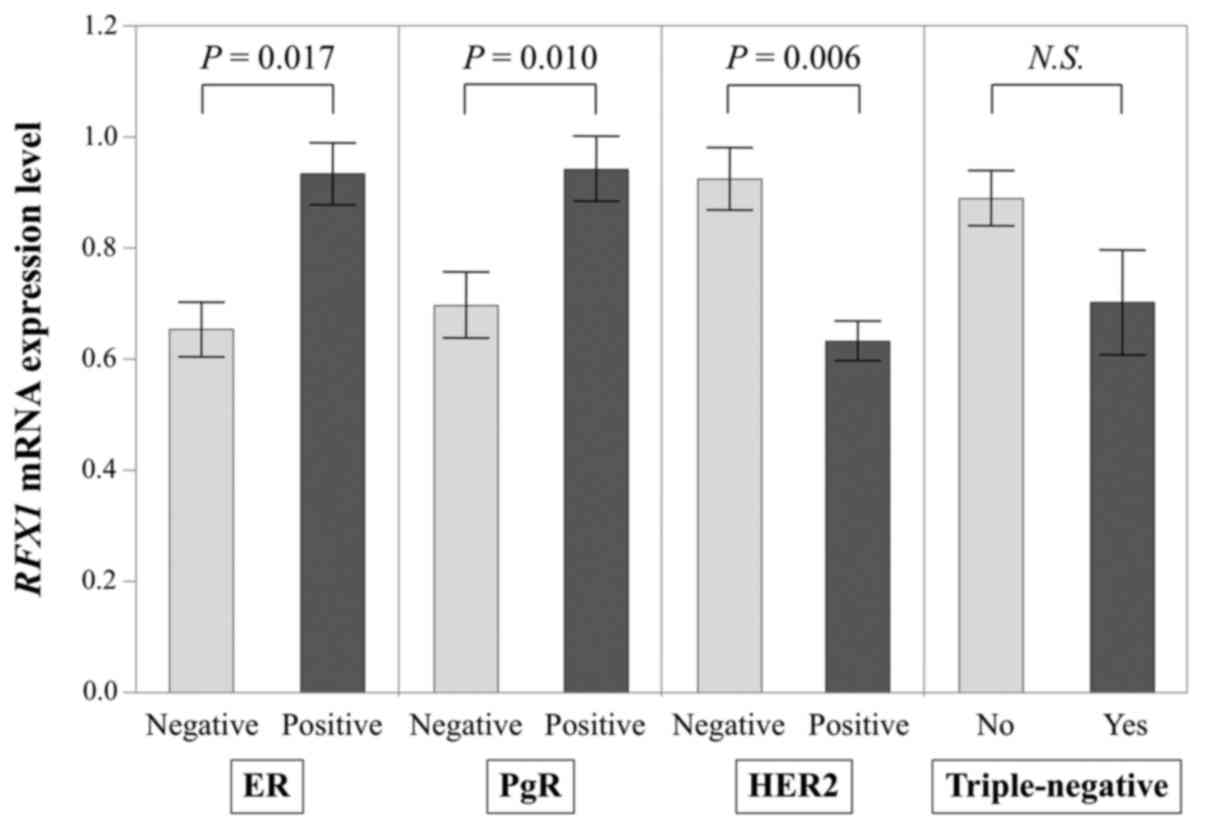
With regard to relevance to the conventional
biomarkers (Fig. 4), RFX1 mRNA
levels in ER positive specimens (n=127) were expressed
significantly higher than in ER negative specimens (n=40; P=0.017);
PgR positive specimens' RFX1 mRNA level (n=115) was also
significantly higher than that of PgR negative specimens (n=52;
P=0.010); HER2 negative specimens (n=119) expressed significantly
more RFX1 mRNA than HER2 positive specimens (n=39; P=0.006;
missing HER2 data for 1 patient). RFX1 mRNA levels did not
significantly differ between triple-negative (n=18) and other BC
subtypes (n=148; P=0.363; missing data for 1 patient).
Prognostic impact of RFX1 mRNA
expression
The high RFX1 group experienced significantly
longer DFS than the low RFX1 group (5-year DFS rates, high
RFX1 group: 95%; low RFX1 group: 77%; P=0.007;
Fig. 5A). Among 43 patients of the
high RFX1 group, only two suffered recurrences. The OS in
the high RFX1 group was also significantly longer than in
the low RFX1 group (5-year OS rates, high RFX1 group:
98%; low RFX1 group: 90%; P=0.013; Fig. 5B). Multivariate analysis of DFS
identified UICC pathological stage III/IV [hazard ratio (HR): 4.80;
95% confidence interval (CI): 1.80–14.9, P=0.001] and the low
RFX1 group (HR, 4.77; 95% CI: 1.32–30.9, P=0.014) as
independent prognostic factors (Table
III).
 | Table III.Prognostic factors for disease-free
survival in 167 patients with breast cancer. |
Table III.
Prognostic factors for disease-free
survival in 167 patients with breast cancer.
| Variables | n | Univariate Hazard
ratio | 95% CI | P | Multivariate Hazard
ratio | 95% CI | P |
|---|
| Age (>60) | 59 | 1.06 | 0.49–2.17 | 0.879 |
|
|
|
| UICC T factor
(T2-4) | 90 | 3.96 | 1.73–10.7 |
<0.001a | 1.85 | 0.67–5.65 | 0.239 |
| Lymph node
metastasis (positive) | 82 | 4.98 | 2.18–13.4 |
<0.001a | 2.84 | 0.79–10.0 | 0.107 |
| UICC pathological
stage (III/IV) | 36 | 7.73 | 3.78–16.4 |
<0.001a | 4.80 | 1.80–14.9 |
0.001a |
| ER status
(negative) | 40 | 2.02 | 0.93–4.14 | 0.073 |
|
|
|
| PgR status
(negative) | 52 | 1.81 | 0.87–3.67 | 0.112 |
|
|
|
| HER2 status
(positive) | 39 | 2.31 | 1.09–4.76 |
0.031a | 0.94 | 0.42–2.08 | 0.883 |
| Triple-negative
(yes) | 18 | 2.37 | 0.88–5.40 | 0.084 |
|
|
|
| Adjuvant
chemotherapy (yes) | 94 | 2.37 | 1.10–5.65 |
0.026a | 0.32 | 0.09–1.07 | 0.064 |
| The low RFX1
group | 124 | 5.69 | 1.72–35.2 |
0.002a | 4.77 | 1.32–30.9 |
0.014a |
Discussion
Our data showed that higher RFX1 expression
was associated with less advanced T factor and lower disease stage,
and patients with higher RFX1 expression had excellent
prognoses.
The RFX1 gene is cytogenetically located at
19p13.1, and belongs to the RFX family genes which share the
conserved DNA-binding domain named ‘winged helix’ (4,18). The
RFX family codes transcription factors that contain a highly
conserved 76-amino acid DNA binding domain (4). RFX1 forms a homodimer or heterodimers
with the RFX2 or RFX3 proteins through a dimerization domain
(19,20). RFX1-4 can bind methylated DNA
sequences preferentially within a sequence-specific, 14-bp
consensus sequence (21). As RFX1
downregulates the proto-oncogene c-myc (8), it is considered to be a tumor
suppressor. In hepatocellular carcinoma (HCC), RFX1 regulates
SC-2001-mediated SHP1 transcription in HCC cell lines, and
SC-2001 inhibits tumor growth (7).
RFX1 downregulation in tumors has been also verified in
glioma (5), neuroblastoma (6), and esophageal adenocarcinoma (22).
We investigated the mRNA expression level and
methylation status of RFX1 using BC cell lines and
non-cancerous mammary gland epithelial cell lines. The RFX1
mRNA expression levels were heterogeneous across cell lines. There
were no reports on hypermethylation of the promoter region of
RFX1 gene in non-neoplastic tissues (5). In this study, we found no CpG
methylation within the promoter region in cell lines and some
clinical tissues. Reportedly, CpGs in intron 7 were hypermethylated
in glioma cell lines and tissue samples, although CpG methylation
in the promoter region was not seen (5). BC cells were also reported to have
hypermethylated CpGs in intron 7 (23). CpGs in gene bodies are sometimes
methylated in a tissue-specific manner. Gene body methylation has
been shown to increase gene expression contrary to methylation in
the promoter region (24). In the
current study, we found CpG hypermethylation in intron 7 of all BC
cell lines and the non-cancerous mammary cell lines. These results
suggest that methylation status is not associated with
downregulation of RFX1 mRNA in BC cells.
This study demonstrated that RFX1 mRNA
expression has a significant relationship with patients'
clinicopathological factors and prognosis. The most striking
finding was that the high RFX1 group experienced excellent
prognosis (5-year DFS rates, high RFX1 group: 95%; low RFX1
group: 77%). Notably, only two of 43 patients suffered recurrences
in the high RFX1 group. Multivariate analysis of DFS
identified UICC pathological stage and the low RFX1 group as
independent prognostic factors. These results emphasize the
usefulness of RFX1 as a prognostic marker. RFX1 mRNA
level was indicated as a potent prognostic factor which was
independent of ER, PgR or HER2 status. Considering these findings,
if a patient's BC specimen's RFX1 mRNA expression is high,
adjuvant chemotherapy could be tailored, thus reducing the adverse
events and expense associated with chemotherapy.
Breast cancers are commonly treated according to
their subtypes (3), which are
determined by investigating their immunostaining status for ER, PgR
and HER2. The subtype reflects each tumor's character, prognosis
and sensitivity to medication. In this study, the high RFX1
group correlated significantly with ER positive, PgR positive and
HER2 negative. This result corresponds to our understanding that BC
with such phenotypes are less aggressive. Although triple-negative
BC is thought to be a more malignant phenotype with a poorer
prognosis, our results showed no significant differences between
triple-negative BC and other types. This might be because of the
small sample size of the triple-negative group.
This is the first study to show the relationship
between RFX1 mRNA expression and BC prognosis; and our
findings may have several possible clinical applications. For
example, RFX1 levels in resected samples might help evaluate
each patient's prognosis and need for adjuvant chemotherapy.
Multigene expression assays might be refined by adding RFX1
expression. In addition, RFX1 expression can be analyzed by
needle biopsy samples before operation, thus aiding in treatment
decisions for neoadjuvant therapy.
This study has some limitations. First, the function
of RFX1 in BC cells is unknown. Basic functional analyses in which
RFX1 expression is inhibited are needed. Second, the
mechanism of RFX1 downregulation is still unclear. The
methylation status of two regions (the promoter and intron 7) in
the RFX1 gene was determined in the present study.
Therefore, it remains formally possible that additional intragenic
or intergenic regions may have undergone methylation in breast
cancer tissues. Mechanisms other than methylation (such as histone
modification and micro RNAs) should also be investigated for this
gene and its product. Further, this study was conducted
retrospectively. Therefore, therapeutic intervention might affect
significance of RFX1 mRNA level. Thus study for larger
series of patients or prospective study is warranted.
In conclusion, our data indicate that higher
expression of RFX1 mRNA level in BC tissue than adjacent
normal site predicts better prognosis. RFX1 could become a
useful prognostic marker for BC.
References
|
1
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coates AS, Winer EP, Goldhirsch A, Gelber
RD, Gnant M, Piccart-Gebhart M, Thürlimann B and Senn HJ: Panel
Members: Tailoring therapies-improving the management of early
breast cancer: St Gallen international expert consensus on the
primary therapy of early breast cancer 2015. Ann Oncol.
26:1533–1546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Emery P, Durand B, Mach B and Reith W: RFX
proteins, a novel family of DNA binding proteins conserved in the
eukaryotic kingdom. Nucleic Acids Res. 24:803–807. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ohashi Y, Ueda M, Kawase T, Kawakami Y and
Toda M: Identification of an epigenetically silenced gene, RFX1, in
human glioma cells using restriction landmark genomic scanning.
Oncogene. 23:7772–7779. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng C, Zhang Y, Yin J, Li J, Abounader R
and Zuo Z: Regulatory factor X1 is a new tumor suppressive
transcription factor that acts via direct downregulation of CD44 in
glioblastoma. Neuro Oncol. 16:1078–1085. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Su JC, Tseng PH, Hsu CY, Tai WT, Huang JW,
Ko CH, Lin MW, Liu CY, Chen KF and Shiau CW: RFX1-dependent
activation of SHP-1 induces autophagy by a novel obatoclax
derivative in hepatocellular carcinoma cells. Oncotarget.
5:4909–4919. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen L, Smith L, Johnson MR, Wang K,
Diasio RB and Smith JB: Activation of protein kinase C induces
nuclear translocation of RFX1 and down-regulates c-myc via an
intron 1 X box in undifferentiated leukemia HL-60 cells. J Biol
Chem. 275:32227–32233. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanda M, Shimizu D, Fujii T, Tanaka H,
Shibata M, Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, et
al: Protein arginine methyltransferase 5 is associated with
malignant phenotype and peritoneal metastasis in gastric cancer.
Int J Oncol. 49:1195–1202. 2016.PubMed/NCBI
|
|
10
|
Kanda M, Shimizu D, Nomoto S, Takami H,
Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, et
al: Prognostic impact of expression and methylation status of
DENN/MADD domain-containing protein 2D in gastric cancer. Gastric
Cancer. 18:288–296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kanda M, Nomoto S, Oya H, Takami H,
Shimizu D, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: The expression of melanoma-associated antigen D2 both in
surgically resected and serum samples serves as clinically relevant
biomarker of gastric cancer progression. Ann Surg Oncol. 23:(Suppl
2). S214–S221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanda M, Shimizu D, Fujii T, Sueoka S,
Tanaka Y, Ezaka K, Takami H, Tanaka H, Hashimoto R, Iwata N, et al:
Function and diagnostic value of Anosmin-1 in gastric cancer
progression. Int J Cancer. 138:721–730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takai D and Jones PA: Comprehensive
analysis of CpG islands in human chromosomes 21 and 22. Proc Natl
Acad Sci USA. 99:3740–3745. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanda M, Shimizu D, Tanaka H, Shibata M,
Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, Fujii T, et
al: Metastatic pathway-specific transcriptome analysis identifies
MFSD4 as a putative tumor suppressor and biomarker for hepatic
metastasis in patients with gastric cancer. Oncotarget.
7:13667–13679. 2016.PubMed/NCBI
|
|
16
|
Finn RS, Dering J, Conklin D, Kalous O,
Cohen DJ, Desai AJ, Ginther C, Atefi M, Chen I, Fowst C, et al: PD
0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially
inhibits proliferation of luminal estrogen receptor-positive human
breast cancer cell lines in vitro. Breast Cancer Res. 11:R772009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Subik K, Lee JF, Baxter L, Strzepek T,
Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG and
Tang P: The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67
and AR by immunohistochemical analysis in breast cancer cell lines.
Breast Cancer (Auckl). 4:35–41. 2010.PubMed/NCBI
|
|
18
|
Gajiwala KS, Chen H, Cornille F, Roques
BP, Reith W, Mach B and Burley SK: Structure of the winged-helix
protein hRFX1 reveals a new mode of DNA binding. Nature.
403:916–921. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reith W, Ucla C, Barras E, Gaud A, Durand
B, Herrero-Sanchez C, Kobr M and Mach B: Rfx1, A transactivator of
hepatitis-B virus enhancer-I, belongs to a novel family of
homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol.
14:1230–1244. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Katan-Khaykovich Y, Spiegel I and Shaul Y:
The dimerization/repression domain of RFX1 is related to a
conserved region of its yeast homologues Crt1 and Sak1: A new
function for an ancient motif. J Mol Biol. 294:121–137. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sengupta PK, Smith EM, Kim K, Murnane MJ
and Smith BD: DNA hypermethylation near the transcription start
site of collagen alpha2(I) gene occurs in both cancer cell lines
and primary colorectal cancers. Cancer Res. 63:1789–1797.
2003.PubMed/NCBI
|
|
22
|
Watts JA, Zhang C, Klein-Szanto AJ,
Kormish JD, Fu J, Zhang MQ and Zaret KS: Study of FoxA pioneer
factor at silent genes reveals Rfx-repressed enhancer at Cdx2 and a
potential indicator of esophageal adenocarcinoma development. PLoS
Genet. 7:e10022772011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rauscher GH, Kresovich JK, Poulin M, Yan
L, Macias V, Mahmoud AM, Al-Alem U, Kajdacsy-Balla A, Wiley EL,
Tonetti D and Ehrlich M: Exploring DNA methylation changes in
promoter, intragenic, and intergenic regions as early and late
events in breast cancer formation. BMC Cancer. 15:8162015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang X, Han H, De Carvalho DD, Lay FD,
Jones PA and Liang G: Gene body methylation can alter gene
expression and is a therapeutic target in cancer. Cancer Cell.
26:577–590. 2014. View Article : Google Scholar : PubMed/NCBI
|