Introduction
Lung cancer is the most common cause of
cancer-associated mortality in the UK for both males and females
(1), and >1/5 patients with cancer
succumb to this malignancy worldwide (2). Non-small cell lung carcinoma (NSCLC)
accounts for 80–85% of all cases of lung cancer, and develops
through the accumulation of molecular alterations, which may serve
as prognostic biomarkers for NSCLC outcome (3).
Mitotic spindle formation and the spindle checkpoint
are critical for the maintenance of cell division and chromosome
segregation (4). A number of mitotic
spindle-associated proteins have been implicated in multiple
malignancies, including lung cancer (5,6).
Overexpression and gene amplification have been reported to
contribute to the development and progression of malignant tumours
for a number of mitotic spindle genes, including those involved in
centrosome maturation [e.g., Aurora kinase (AURK)A,
microtubule nucleation factor TPX2 (TPX2) and kinesin-like
protein 11 (KIF11)] (7,8),
microtubule formation [e.g., AURKA, cytoskeleton-associated
protein 5 (CKAP5), tubulin β (TUBB) and TUBB3]
(9–11), and chromosomal alignment and
segregation [e.g., AURKA, AURKB, AURKC, discs
large-associated protein 5 (DLGAP5) and TTK protein kinase
(TTK)] (12–14). AURKA serves a central role in
recruiting other mitotic spindle members (5). A number of previous studies conducted in
lung cancer have investigated the prognostic value of various of
the aforementioned genes, including TPX2 (15), AURKA (16–18) and
AURKB (18–21); however, the prognostic value of AURKA
and AURKB remains a matter of debate. No information on the
potential prognostic significance in human NSCLC has yet been
provided for DLGAP5, CKAP5 or TTK.
Personalised medicine relies on the utilisation of
gene profiling (including expression, mutation and methylation) in
combination with clinicopathological characteristics to provide an
optimal management plan for the patient. Therefore, it is necessary
to expand our efforts in investigating the association of
particular molecular profiles with patient outcomes. The aim of the
present study was to acquire a comprehensive expression profile of
mitotic spindle-associated genes (AURKA, AURKB,
AURKC, CKAP5, DLGAP5, KIF11,
TPX2, TTK, TUBB and TUBB3) in NSCLC and
to investigate the potential associations with clinicopathological
characteristics and patient survival rates.
Materials and methods
Patients and samples
The present study was undertaken within the context
of the Liverpool Lung Project (22).
Appropriate ethical approval from the Liverpool Research Ethics
Committee, ref 157/97, was obtained and all patients provided
written informed consent. A total of 132 frozen surgical tumour
samples, collected between January 1999 and December 2005 at
Liverpool Heart and Chest Hospital (Liverpool, UK), were available
from patients with primary NSCLC, 56 from adenocarcinoma (AdC) and
76 from squamous cell carcinoma of the lung (SqCCL). In addition,
44 paired non-tumour surgical lung samples (20 from patients with
AdC and 24 from patients with SqCCL) were analysed. The median age
of the patients was 67 years (range, 45–82 years); 56 of the
patients were female and 77 were male. The majority of the
specimens were of the pathological tumour (pT)2 stage (n=101),
whereas the pT1 and pT3/4 groups comprised 19 and 12 patients,
respectively. The HBEC-3KT cell line (23) used as a calibrator was provided by
Professor John Minna and Professor Adi Gazdar.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from primary lung tumour
tissue (ten 20-µm thick sections per specimen) using a Direct-zol™
RNA MiniPrep kit (Zymo Research Corp., Irvine, CA, USA), according
to the manufacturer's protocol. The quality and quantity of RNA
were determined using a NanoDrop 2000 spectrophotometer (Thermo
Fisher Scientific, Inc., Wilmington, DE, USA) and 200 ng RNA was
reverse transcribed using a High-Capacity cDNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. Predesigned
6-carboxyfluorescein-labelled TaqMan Gene Expression Assays (Thermo
Fisher Scientific, Inc.) were employed, according to the
manufacturer's protocol, to analyse mRNA expression: AURKA,
Hs01582072_m1; AURKB, Hs00945858_g1; AURKC,
Hs00152930_m1; CKAP5, Hs01120723_m1; DLGAP5,
Hs00207323_m1, KIF11, Hs00189698_m1; TPX2,
Hs00201616_m1; TTK, Hs01009870_m1; TUBB,
Hs00962419_g1; and TUBB3, Hs00964962_g1, with a
4,7,2′-trichloro-7′-phenyl-6-carboxyfluorescein-labelled β-actin
(ACTB) TaqMan Gene Expression Assay (cat. no. 4326315E;
Thermo Fisher Scientific, Inc.) serving as an endogenous control.
RNA from human bronchial epithelial cells (HBEC-3KT) was used as
technical calibrator. Three technical replicates were performed for
every qPCR assay. Thermocycling conditions were 95°C for 10 min
(activation), 45 cycles of 95°C for 15 sec (denaturation), 60°C for
1 min (annealing and extension)] on a Life Technologies StepOnePlus
Real-Time PCR System. mRNA levels were expressed as relative
quantification (RQ) values, which were calculated as
RQ=2−ΔΔCq (24).
Quantification cycle (Cq) values were determined using StepOne
software (version 1.2; Thermo Fisher Scientific, Inc.) and
normalised to the corresponding Cq value for the endogenous control
ACTB, generating ΔCq values (ΔCq=Cq target-Cq ACTB). Sample ΔCq
values were further normalised against an immortalised bronchial
epithelial cell line HBEC-3KT (23)
calibrator using the formula: ΔΔCq=(ΔCq sample-ΔCq HBEC-3KT).
Statistical analysis
Gene expression in tumour and adjacent wild-type
tissues were compared using the Wilcoxon non-parametric test. The
study characteristics were examined using descriptive statistics.
Categorical variables were compared using a χ2 test and
continuous variables were examined using a Mann-Whitney U test.
Overall survival time was calculated from the date of surgery to
the date of mortality or last follow-up date. Overexpression for a
tumour sample was designated as >95% reference interval [mean ±
(2x standard deviation)] of wild-type tissues. Postoperative
univariate survival analysis was explored using Kaplan-Meier
estimator curves for all the categorical predictors. Tests of
equality across strata were also conducted to evaluate the
suitability of including potential predictors in the final
multivariate model. For the categorical variables, a log-rank test
of equality across strata was used, and a univariate Cox's
proportional hazard regression was used to analyse continuous
variables to examine the differences in survival rate. Variables
with P<0.25 in the univariate analysis were selected for
inclusion in the final multivariate model as previously suggested
(25). A multivariate Cox's
proportional hazard model was used to examine the association
between mRNA expression and other relevant prognostic factors. All
statistical analyses were performed using IBM®
SPSS® statistical software (version 22.0; IBM SPSS,
Armonk, NY, USA) and Stata® (version 13.1; StataCorp
LLC, College Station, TX, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Gene expression analysis
RT-qPCR analysis revealed that, with the exception
of AURKC, the mRNA expression levels of all the genes
examined in the present study (AURKA, AURKB,
AURKC, DLGAP5, CKAP5, KIF11,
TPX2, TTK, TUBB and TUBB3) were
significantly upregulated in NSCLC tissues compared with those in
wild-type adjacent lung tissues (P<0.0001; Fig. 1). Comparison between histology types
(Fig. 2) revealed that the mRNA
expression of seven genes was significantly increased in SqCCL
compared with that in AdC tissues (P<0.001 for AURKA,
AURKB, DLGAP5, TPX2, TTK and
TUBB; P=0.001 for KIF11).
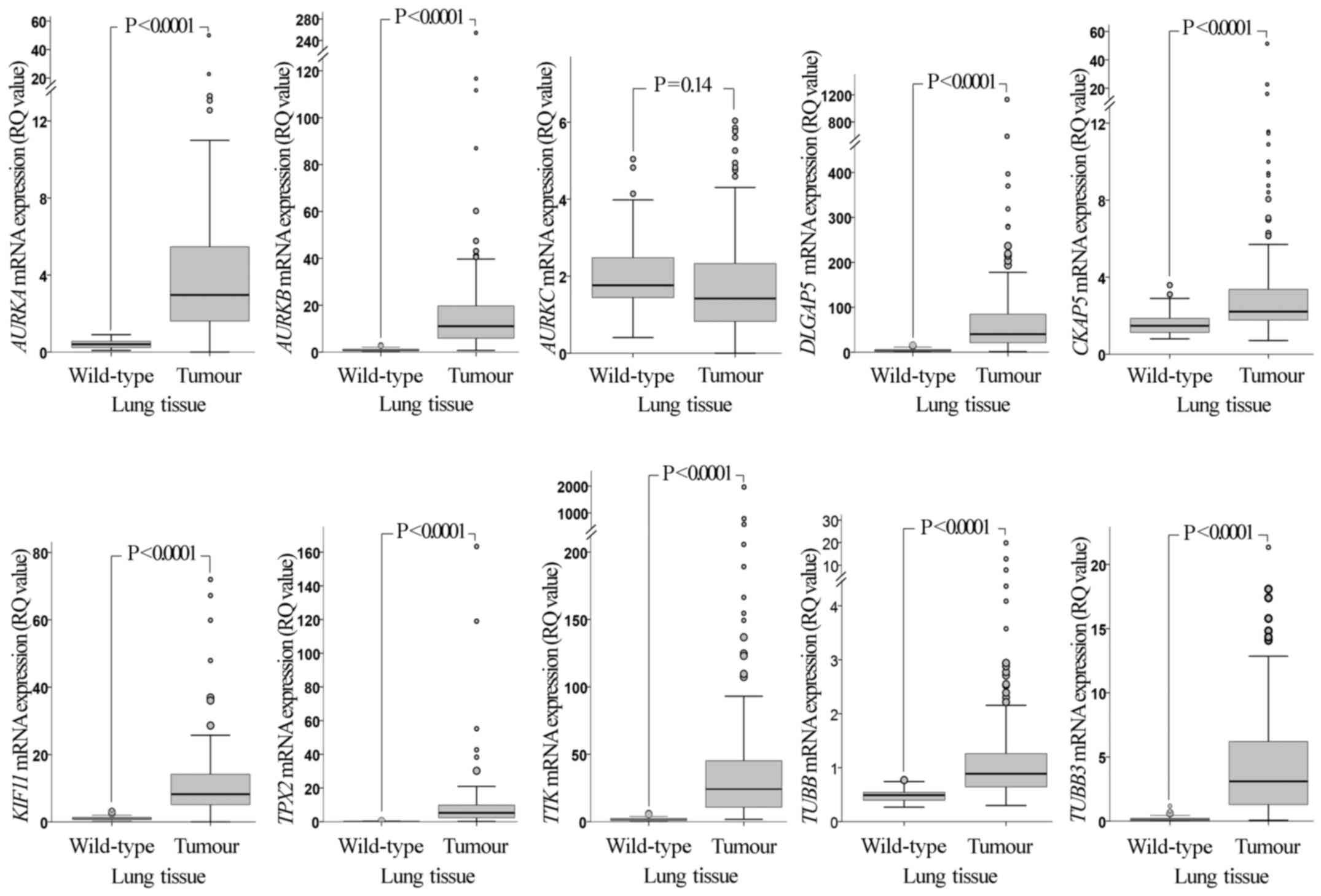 | Figure 1.Comparative mRNA expression of the
examined genes in NSCLC tissues and adjacent normal tissues. The
mRNA expression levels of the genes in NSCLC tumours are
significantly higher than those in adjacent normal tissues, with
the exception of AURKC. P-values were calculated using a
Mann-Whitney U test and adjusted for multiple comparisons by
Bonferroni correction. RQ values were calculated using RNA from the
non-tumorigenic immortalised human bronchial epithelial cell line
HBEC-3KT as a calibrator. Larger circles represent outlier values
(>1.5x interquartile range); smaller circles represent extreme
values (>3x interquartile range). NSCLC, non-small cell lung
cancer; RQ, relative quantification; AURK, Aurora kinase;
DLGAP5, discs large-associated protein 5; CKAP5,
cytoskeleton-associated protein 5; KIF11, kinesin-like
protein 11; TPX2, microtubule nucleation factor TPX2;
TTK, TTK protein kinase; TUBB, tubulin β. |
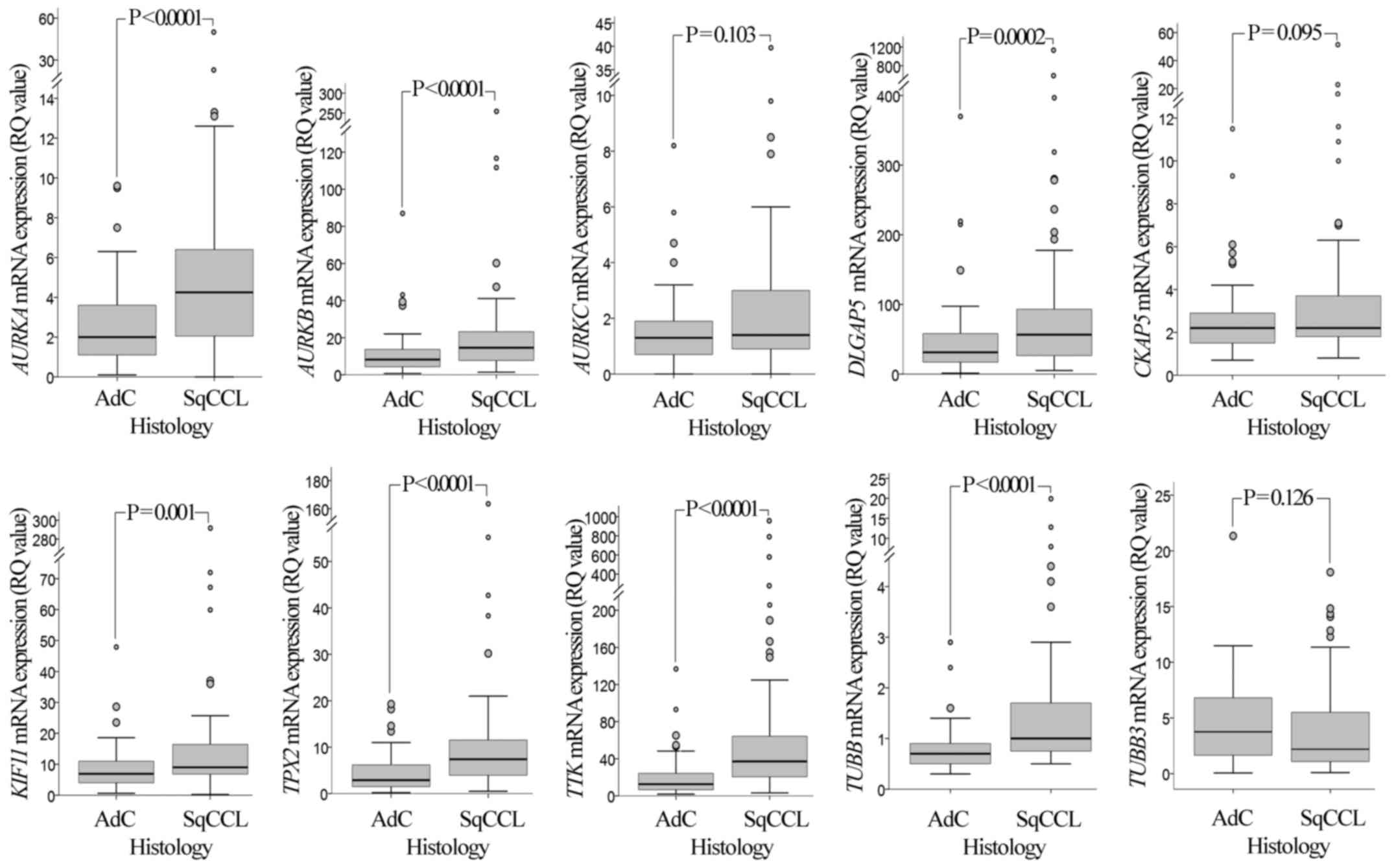 | Figure 2.mRNA expression of AURKA,
AURKB, AURKC, DLGAP5, CKAP5,
KIF11, TPX2, TTK, TUBB and TUBB3
genes in SqCCL and AdC of the lung. Comparison between histology
types demonstrated that the mRNA expression levels of the
AURKA, AURKB, DLGAP5, KIF11,
TPX2, TTK and TUBB genes in SqCCL tumours are
significantly higher than those in AdC tumours. P-values were
calculated using a Mann-Whitney U test and adjusted for multiple
comparisons by Bonferroni correction. RQ values were calculated
using RNA from the non-tumorigenic immortalised human bronchial
epithelial cell line HBEC-3KT as a calibrator. Larger circles
represent outlier values (>1.5x interquartile range); smaller
circles represent extreme values (>3x interquartile range).
AURK, Aurora kinase; DLGAP5, discs large-associated
protein 5; CKAP5, cytoskeleton-associated protein 5;
KIF11, kinesin-like protein 11; TPX2, microtubule
nucleation factor TPX2; TTK, TTK protein kinase;
TUBB, tubulin β; SqCCL, squamous cell carcinoma of the lung;
AdC, adenocarcinoma; RQ, relative quantification. |
Survival analysis
There was no association between the mRNA expression
of any of the genes evaluated with age, sex, pathological stage or
nodal status (Table I). Potential
associations between the expression level of the target genes and
overall survival rate were examined. In univariate analysis,
pathological stage, nodal status and AURKA mRNA expression
were predictors of overall survival rate (Table II). Most importantly, multivariate
analysis demonstrated that AURKA mRNA expression [hazard
ratio (HR), 1.81; 95% confidence interval (CI) 1.16–2.84; P=0.009]
independently predicts poor prognosis in patients with NSCLC upon
adjusting for age, pT2 and involvement of distal nodes
(pathological node stage 2) (Table
II). This observation was consistent with the Kaplan-Meier
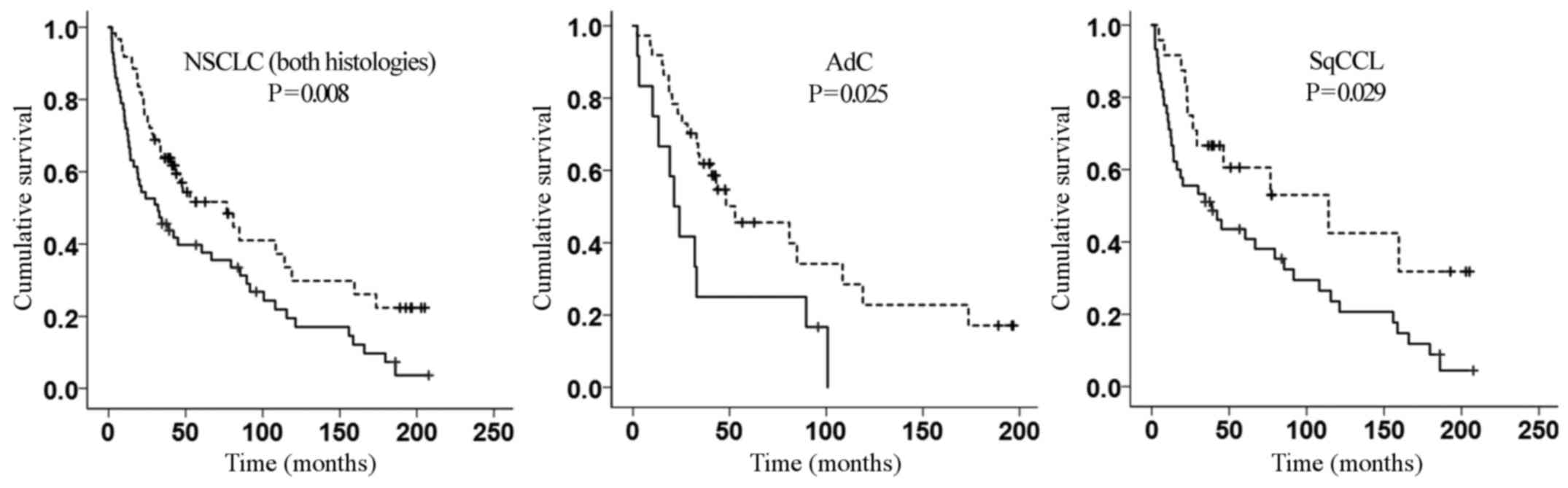
estimator curve (Fig. 3). The
association with prognosis remained significant even when SqCCL and
AdC tissues were tested separately (P=0.025 and P=0.029,
respectively; Fig. 3).
 | Table I.Clinicopathological characteristics
of the study patients in association with AURKA mRNA
expression profile. |
Table I.
Clinicopathological characteristics
of the study patients in association with AURKA mRNA
expression profile.
| Clinicopathological
characteristic | Total number of
patients (%) 124 (100) | High expression of
Aurora-A mRNA (n=59) | Low expression of
Aurora-A mRNA (n=65) | P-value |
|---|
| Mean age, years
(standard deviation) | 66.5 (8.5) | 65.9 (8.5) | 67.5 (8.5) | 0.223a |
| Gender |
|
|
| 0.180b |
|
Male | 70 (56.5) | 37 (52.9) | 33 (47.1) |
|
|
Female | 54 (43.5) | 22 (40.7) | 32 (59.3) |
|
| Histology |
|
|
|
<0.001b |
|
Adenocarcinoma | 52 (41.9) | 13 (25.0) | 39 (75.0) |
|
|
Squamous cell carcinoma | 72 (58.1) | 46 (63.9) | 26 (36.1) |
|
| Tumour stage |
|
|
| 0.513b |
| 1 | 19 (15.3) | 7 (36.8) | 12 (63.2) |
|
| 2 | 91 (73.3) | 45 (49.5) | 46 (50.6) |
|
| ≥3 | 12 (9.6) | 7 (58.3) | 5 (41.7) |
|
| Nodal status |
|
|
| 0.975b |
| 0 | 68 (54.8) | 32 (47.1) | 36 (52.9) |
|
| 1 | 38 (30.6) | 18 (47.4) | 20 (52.6) |
|
| 2 | 18 (14.6) | 9 (50.0) | 9 (50.0) |
|
 | Table II.Univariate and multivariate Cox's
proportional hazard regression analyses of potential predictors of
overall survival among the study patients. |
Table II.
Univariate and multivariate Cox's
proportional hazard regression analyses of potential predictors of
overall survival among the study patients.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Covariates | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| AURKA mRNA | 1.79
(1.16–2.77) | 0.009 | 1.81
(1.16–2.84) | 0.009 |
| Age | 1.02
(1.00–1.05) | 0.066 | 1.03
(1.00–1.06) | 0.020 |
| Tumour stage |
|
|
|
|
| 1 | Reference | Reference | Reference | Reference |
| 2 | 2.82
(1.35–5.86) | 0.006 | 2.43
(1.16–5.10) | 0.019 |
| ≥3 | 3.80
(1.42–10.15) | 0.008 | 1.39
(0.38–5.09) | 0.623 |
| Nodal status |
|
|
|
|
| 0 | Reference | Reference | Reference | Reference |
| 1 | 1.62
(1.03–2.55) | 0.037 | 1.45
(0.90–2.34) | 0.128 |
| 2 | 2.55
(1.35–4.84) | 0.004 | 3.14
(1.24–7.99) | 0.016 |
Discussion
Spindle formation is a key process for cell
proliferation (8). It is well known
that spindle assembly aberrations lead to aneuploidy and are
extensively involved in the development of cancer (26). Thus, it was hypothesised that the
expression of genes associated with this process may be indicative
of the aggressiveness of a tumour and therefore may exhibit
prognostic value.
In the present study, the mRNA expression of the
AURKA, AURKB, AURKC, CKAP5,
DLGAP5, KIF11, TPX2, TTK, TUBB
and TUBB3 genes was investigated in a large cohort of human
NSCLC tissues, and potential associations between expression
profiles and clinicopathological characteristics, including
survival rates, were evaluated. All genes, with the exception of
AURKC, were overexpressed in the malignant tissues in
comparison with adjacent wild-type tissues. These results possibly
reflect the requirement for increased mitotic spindle genes
expression to cope with the increased replication rate of cancer
cells (27,28). However, the important clinical
question is whether the overexpression of any of these genes is
able to confer a selective advantage on cancer cells and increase
their invasive properties. The results of the present study confirm
that up-regulation of mitotic spindle genes is a common abnormality
in NSCLC and further support a role for the maintenance of a
tumorigenic phenotype (5,29). The results of the present study
demonstrated that, of the 10 genes examined, only AURKA
overexpression was associated with poor prognosis, which suggests
that this gene has a particular contribution to a more aggressive
phenotype. It is notable that multivariate Cox's regression
analysis identified AURKA mRNA expression as an independent
predictor of poor prognosis in patients with NSCLC.
The overexpression of AURKA in NSCLC has been
demonstrated previously (16,17). Consistent with these previous studies,
it was observed in the present study that AURKA mRNA
overexpression was increased in SqCCL compared with that in AdC
tissue. However, the prognostic value of AURKA in lung
cancer has not yet been established. In contrast to the study of
Tang et al (30), the
prognostic significance of AURKA expression in the present
study appears to hold true for both histological subtypes. There is
a lack of consensus on this issue, with previous studies debating
on the prognostic significance of AURKA in SqCCL (17,18).
Furthermore, perimembrane immunohistochemical staining was
demonstrated to be a marked predictor of poor prognosis in patients
with SqCCL, but not in patients with AdC (16), whereas microarray data analysis
demonstrated that AURKA mRNA overexpression is associated
with poor prognosis in patients with AdC, but not in patients with
SqCCL (30). The reported differences
are possibly due to dissimilarities in the study design,
measurement of AURKA expression and the small study size,
which decreases statistical significance. It is imperative that a
large multicentre study is undertaken to determine a definitive
explanation of these discrepancies.
AURKA overexpression may serve an important
role in cancer aggressiveness through a range of underlying
molecular mechanisms. Elevated levels of AURKA perturb mitotic
spindle formation and therefore cytokinesis due to centrosome
amplification, leading to chromosomal instability and consequently
aneuploidy or polyploidy (5,31). AURKA overexpression also
inactivates several tumour-suppressor genes, including p53
(32). The association between
AURKA overexpression and p53 mutation, as well as
advanced tumour grade and advanced cancer stage, was also reported
in patients with hepatocellular carcinoma (33), and with clinically aggressive disease
and decreased survival rates in patients with ovarian cancer
(34). These AURKA-associated
events (the perturbation of spindle formation and inactivation of
tumour-suppressor genes by elevated AURKA expression) may
explain the association identified between up-regulated
AURKA expression and poor outcome of patients with NSCLC.
Nonetheless, the hypothesis that up-regulated AURKA
expression contributes to a poor survival outcome in lung cancer
has been debated, presumably because NSCLC represents a set of
heterogeneous malignancies, with various outcomes, even among those
with the same clinicopathological features (35). The results of the present study
provide evidence to support the prognostic role of AURKA
expression in patients with NSCLC and highlight the requirement for
a large multicentre clinical study which will take into
consideration further parameters, including therapeutic regimens.
Most importantly, the results of the present study suggest that
NSCLC patients may benefit from therapy with AURKA inhibitors and
this requires validation in a prospective clinical study.
Acknowledgements
The Liverpool Lung Project is funded by the Roy
Castle Lung Cancer Foundation (Liverpool, UK). The present study
was also supported through a PhD studentship awarded to A.S.K.
Al-Khafaji (grant no. SL25) by the University of Baghdad (Baghdad,
Iraq).
References
|
1
|
Field JK, Devaraj A, Duffy SW and Baldwin
DR: CT screening for lung cancer: Is the evidence strong enough?
Lung Cancer. 91:29–35. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lacroix L, Commo F and Soria JC: Gene
expression profiling of non-small-cell lung cancer. Expert Rev Mol
Diagn. 8:167–178. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wassmann K and Benezra R: Mitotic
checkpoints: From yeast to cancer. Curr Opin Genet Dev. 11:83–90.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fu J, Bian M, Jiang Q and Zhang C: Roles
of aurora kinases in mitosis and tumorigenesis. Mol Cancer Res.
5:1–10. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Martens-de Kemp SR, Nagel R, Stigter-van
Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ and
Brakenhoff RH: Functional genetic screens identify genes essential
for tumor cell survival in head and neck and lung cancer. Clin
Cancer Res. 19:1994–2003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sankaran S and Parvin JD: Centrosome
function in normal and tumor cells. J Cell Biochem. 99:1240–1250.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanenbaum ME and Medema RH: Mechanisms of
centrosome separation and bipolar spindle assembly. Dev Cell.
19:797–806. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Royle SJ: The role of clathrin in mitotic
spindle organisation. J Cell Sci. 125:19–28. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manning AL and Compton DA: Structural and
regulatory roles of nonmotor spindle proteins. Curr Opin Cell Biol.
20:101–106. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Leandro-Garcia LJ, Leskelä S, Landa I,
Montero-Conde C, López-Jiménez E, Letón R, Cascón A, Robledo M and
Rodríguez-Antona C: Tumoral and tissue-specific expression of the
major human beta-tubulin isotypes. Cytoskeleton (Hoboken).
67:214–223. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jelluma N, Brenkman AB, van den Broek NJ,
Cruijsen CW, van Osch MH, Lens SM, Medema RH and Kops GJ: Mps1
phosphorylates Borealin to control Aurora B activity and chromosome
alignment. Cell. 132:233–246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nguyen HG, Makitalo M, Yang D, Chinnappan
D, St Hilaire C and Ravid K: Deregulated Aurora-B induced
tetraploidy promotes tumorigenesis. FASEB J. 23:2741–2748. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Slattery SD, Mancini MA, Brinkley BR and
Hall RM: Aurora-C kinase supports mitotic progression in the
absence of Aurora-B. Cell Cycle. 8:2984–2994. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kadara H, Behrens C, Yuan P, Solis L, Liu
D, Gu X, Minna JD, Lee JJ, Kim E, Hong WK, et al: A five-gene and
corresponding protein signature for stage-I lung adenocarcinoma
prognosis. Clin Cancer Res. 17:1490–1501. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ogawa E, Takenaka K, Katakura H, Adachi M,
Otake Y, Toda Y, Kotani H, Manabe T, Wada H and Tanaka F:
Perimembrane Aurora-A expression is a significant prognostic factor
in correlation with proliferative activity in non-small-cell lung
cancer (NSCLC). Ann Surg Oncol. 15:547–554. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lo Iacono M, Monica V, Saviozzi S, Ceppi
P, Bracco E, Papotti M and Scagliotti GV: Aurora kinase A
expression is associated with lung cancer histological-subtypes and
with tumor de-differentiation. J Transl Med. 9:1002011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takeshita M, Koga T, Takayama K, Kouso H,
Nishimura-Ikeda Y, Yoshino I, Maehara Y, Nakanishi Y and Sueishi K:
CHFR expression is preferentially impaired in smoking-related
squamous cell carcinoma of the lung, and the diminished expression
significantly harms outcomes. Int J Cancer. 123:1623–1630. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vischioni B, Oudejans JJ, Vos W, Rodriguez
JA and Giaccone G: Frequent overexpression of aurora B kinase, a
novel drug target, in non-small cell lung carcinoma patients. Mol
Cancer Ther. 5:2905–2913. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smith SL, Bowers NL, Betticher DC,
Gautschi O, Ratschiller D, Hoban PR, Booton R, Santibáñez-Koref MF
and Heighway J: Overexpression of aurora B kinase (AURKB) in
primary non-small cell lung carcinoma is frequent, generally driven
from one allele, and correlates with the level of genetic
instability. Br J Cancer. 93:719–729. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Perumal D, Singh S, Yoder SJ, Bloom GC and
Chellappan SP: A novel five gene signature derived from stem-like
side population cells predicts overall and recurrence-free survival
in NSCLC. PLoS One. 7:e435892012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cassidy A, Myles JP, van Tongeren M, Page
RD, Liloglou T, Duffy SW and Field JK: The LLP risk model: An
individual risk prediction model for lung cancer. Br J Cancer.
98:270–276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sato M, Vaughan MB, Girard L, Peyton M,
Lee W, Shames DS, Ramirez RD, Sunaga N, Gazdar AF, Shay JW and
Minna JD: Multiple oncogenic changes (K-RAS(V12), p53 knockdown,
mutant EGFRs, p16 bypass, telomerase) are not sufficient to confer
a full malignant phenotype on human bronchial epithelial cells.
Cancer Res. 66:2116–2128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bursac Z, Gauss CH, Williams DK and Hosmer
DW: Purposeful selection of variables in logistic regression.
Source Code Biol Med. 3:172008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cimini D and Degrassi F: Aneuploidy: A
matter of bad connections. Trends Cell Biol. 15:442–451. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vader G and Lens SM: The aurora kinase
family in cell division and cancer. Biochim Biophys Acta.
1786:60–72. 2008.PubMed/NCBI
|
|
28
|
Garrido G and Vernos I: Non-centrosomal
TPX2-dependent regulation of the aurora a kinase: Functional
implications for healthy and pathological cell division. Front
Oncol. 6:882016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eymin B and Gazzeri S: Role of cell cycle
regulators in lung carcinogenesis. Cell Adh Migr. 4:114–123. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang H, Xiao G, Behrens C, Schiller J,
Allen J, Chow CW, Suraokar M, Corvalan A, Mao J, White MA, et al: A
12-gene set predicts survival benefits from adjuvant chemotherapy
in non-small cell lung cancer patients. Clin Cancer Res.
19:1577–1586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marumoto T, Zhang D and Saya H: Aurora-A-a
guardian of poles. Nat Rev Cancer. 5:42–50. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Katayama H, Sasai K, Kawai H, Yuan ZM,
Bondaruk J, Suzuki F, Fujii S, Arlinghaus RB, Czerniak BA and Sen
S: Phosphorylation by aurora kinase A induces Mdm2-mediated
destabilization and inhibition of p53. Nat Genet. 36:55–62. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jeng YM, Peng SY, Lin CY and Hsu HC:
Overexpression and amplification of Aurora-A in hepatocellular
carcinoma. Clin Cancer Res. 10:2065–2071. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lassmann S, Shen Y, Jütting U, Wiehle P,
Walch A, Gitsch G, Hasenburg A and Werner M: Predictive value of
Aurora-A/STK15 expression for late stage epithelial ovarian cancer
patients treated by adjuvant chemotherapy. Clin Cancer Res.
13:4083–4091. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Z, Fillmore CM, Hammerman PS, Kim CF
and Wong KK: Non-small-cell lung cancers: A heterogeneous set of
diseases. Nat Rev Cancer. 14:535–546. 2014. View Article : Google Scholar : PubMed/NCBI
|