Introduction
It is well known that Schiff bases are important in
the development of coordination chemistry due to their ease of
synthesis and structural tenability, and their ability to form a
wide variety of complexes of chemical, biological and industrial
importance (1–5). Aroyl hydrazones are members of the
Schiff bases family, which are built with aromatic acid hydrazides
(aroyl hydrazides) and carbonyl compounds (1). Hydrazone complexes have been studied for
numerous years due to their antimicrobial and antitumor activities
(6–11). Hydrazone ligands can offer a
combination of amide oxygen and imine nitrogen as donor atoms, with
the imine nitrogen being involved in the formation of an N-N bond
reminiscent of a doubly reduced azo functionality. Due to the short
N-N bond length, the hydrazone ligands act mostly as tridentate
moieties, although they have the potential to behave as bridging
tetradentate ligands (12).
The chemistry of the trivalent lanthanide (Ln) ions
has attracted significant attention in the last 40 years (13–15). In
addition to their magnetic and photophysical properties, the
bioactivities of Lns, including their antimicrobial, antitumor,
antivirus and anticoagulant action, as well as their ability to
prevent arteriosclerosis, have been explored in recent decades
(16–19).
The present study synthesized the hydrogen ligand
(HL)(E)-N'-[1-(2-pyridinyl)ethylidene]isonicotinohydrazone and
prepared its two Ln(III) complexes. Furthermore, the antitumor
effects and potential mechanisms of the two Ln(III) complexes with
HL were explored in the human lung cancer cell line A549 and in the
human gastric cancer cell lines BGC823 and SGC7901.
Materials and methods
Materials and measurements
All starting materials were obtained commercially
and used as received. All the rear earth salts were purchased from
Beijing Chemical Works (Beijing, China). The A549 human lung
cancer, and the BGC823 and SGC7901 human gastric cancer cell lines,
were obtained from the Cell Culture Center of the Basic Institute
of Medical Sciences, Peking Union Medical College (Beijing, China).
Cell culture reagents were purchased from Gibco (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). MTT and dimethyl sulfoxide
(DMSO) were purchased from Sigma-Aldrich (Merck KGaA, Darmstadt,
Germany). Polyvinylidene fluoride (PVDF) membranes and non-fat dry
milk were obtained from EMD Millipore (Billerica, MA, USA).
The elemental analyses were performed in the
microanalytical laboratory of the Department of Chemistry, Lanzhou
University (Lanzhou, China). The infrared (IR) spectra (KBr pellet)
were recorded using an FTS 165 Fourier transform (FT)-IR
spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
in the range of 4,000–400 cm−1. Cell lines were cultured
in an incubator (Thermo Fisher Scientific, Inc.) under the
condition of 100% humidity, 5% CO2 at 37°C. RPMI-1640
culture medium was purchased from HycClone (GE Healthcare Life
Sciences, Chalfont, UK). CKX51 microscope (Olympus Corporation,
Tokyo, Japan) and ToupCam FMA050 (ToupTek Photonics, Hangzhou,
China) were used to obtain images of the cells. The MTT assay was
performed using a microplate reader (Infinite F50; Tecan Schweiz
AG, Männedorf, Switzerland). The flow cytometer Accuri C6 was
purchased from BD Biosciences (Franklin Lakes, NJ, USA).
Preparations of the ligand and
complexes
The ligand HL was prepared as described previously
(20). In brief, HL was obtained by
condensation of 2-acetylpyridine (0.1 mmol) and
isonicotinohydrazone (0.1 mmol) in ethanol solution with the
addition of 0.1 ml concentrated acetic acid. The parameters of the
IR spectrum of HL were as follows: IR (KBr) cm−1: ν(NH),
3,194; ν(C=O), 1,679; and ν(C=N), 1,586. Complex 1 was prepared as
follows: Ce(NO3)3·6H2O (217.1 mg,
0.5 mmol; Beijing Chemical Works) in ethanol (20 ml) was added to a
solution containing the ligand HL (240 mg, 1 mmol) in methanol (10
ml) with 0.05 ml triethylamine. The mixture was stirred for 12 h at
25°C to allow the formation of the precipitate of the Ce(III)
complex. The precipitate was separated by centrifugation at 2,000 ×
g for 10 min at 25°C, and washed three times with ethanol and one
time with ether, and finally dried in vacuo. Slow evaporation of
the mixed methanol/ethanol solution containing Ce(III) complex 1
led to a yellow single crystal that was suitable for X-ray
diffraction (XRD). The calculated parameters for
C29H34CeN9O8 were as
follows: C, 44.84; H, 4.41; and N, 16.23. The determined parameters
for C29H34CeN9O8 were
as follows: C, 44.51; H, 4.82; and N, 15.98. IR (KBr)
cm−1: ν(NH), 3,074; ν(C=O), 1,586; and ν(C=N), 1,519.
The La(III) complex 2 was synthesized as described earlier. The
calculated parameters for
C28H34LaN9O9 were as
follows: C, 43.14; H, 4.40; and N, 16.17, while the measured
parameters where as follows: C, 43.35; H, 4.77; and N, 15.98. IR
(KBr) cm−1: ν(NH), 3,081; ν(C=O), 1,578; and ν(C=N),
1,518. The complexes are soluble at room temperature in ethanol,
methanol, dimethylformamide and DMSO.
X-ray crystallography
The XRD measurements for the two complexes were
performed on a SMART APEX II CCD diffractometer (Bruker
Corporation, Billerica, MA, USA) equipped with a graphite
monochromatized Mo-Ka radiation (λ=0.071073 nm) by using φ-ω scan
mode. Mo-Ka radiation is the standard operation used in structure
elucidation, where Mo-Ka indicates the molybdenum-kalium target
radiography and λ is the X-Ray diffraction wavelength.
Semi-empirical absorption correction was applied to the intensity
data using the SADABS program version 2.03 (Bruker Corporation)
(21). The structures were solved by
direct methods and refined by full matrix least-square on
F2 using the SHELXTL version 97 program (Bruker
Corporation) (22). The non-hydrogen
atoms of the disordered methanol solvent molecules were refined
isotropically. However, the H atoms on the solvent molecules were
not added, and were directly included in the molecular formula. The
other non-hydrogen atoms were refined anisotropically. H atoms for
coordinated solvent molecules were located from difference Fourier
maps and refined with restraints in bond length and thermal
parameters. All the other H atoms were positioned geometrically and
refined using a riding model. Details of the crystal parameters,
data collection and refinements for complexes 1 and 2 are
summarized in Table I. The selected
bond lengths and angles are listed in Tables II and III, respectively.
 | Table I.Crystal data and structure refinement
for complexes 1 and 2. |
Table I.
Crystal data and structure refinement
for complexes 1 and 2.
|
Characteristicsa | Complex 1 | Complex 2 |
|---|
| Empirical
formula |
C29H34N9O8La |
C29H34N9O8Ce |
| Molecular weight,
Da | 775.56 | 776.77 |
| Temperature, K | 296(2) | 296(2) |
| Wavelength, nm | 0.071073 | 0.071073 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | Pn | Pn |
| Z | 4 | 4 |
| a, nm | 1.15714(6) | 1.16586(6) |
| b, nm | 1.13776(6) | 1.14190(6) |
| c, nm | 1.30471(7) | 1.29827(7) |
| β, ° | 99.4880(10) | 100.0540(10) |
| V,
nm3 | 1.69421(15) | 1.70184(16) |
| Dcalc,
g/cm3 | 1.520 | 1.516 |
| µ,
mm−1 | 1.321 | 1.397 |
| F(000) | 784 | 786 |
| Crystal size,
mm | 0.20×0.18×0.16 | 0.22×0.15×0.14 |
| θ range for data
collection, ° | 2.18–25.00 | 1.78–25.00 |
| Reflections
collected | 8,446 | 8,517 |
| Independent
reflections, Rint | 4,956(0.0153) | 4,547(0.0170) |
| Final GooF | 1.060 | 1.091 |
|
R1,
wR2 [I>2σ(I)] |
R1=0.0305,
wR2=0.0805 |
R1=0.0300,
wR2=0.0871 |
|
R1,
wR2 (all data) |
R1=0.0323,
wR2=0.0830 |
R1=0.0329,
wR2=0.0901 |
 | Table II.Selected bond lengths in the
complexes. |
Table II.
Selected bond lengths in the
complexes.
| A, Complex 1 |
|---|
|
|---|
| Bond | Lengtha, nm |
|---|
| La1-O1 | 0.2463(7) |
| La1-O6 | 0.2628(7) |
| La1-O3 | 0.2692(10) |
| La1-N2 | 0.2809(5) |
| La1-O2 | 0.2465(7) |
| La1-O7 | 0.2561(8) |
| La1-N1 | 0.2786(6) |
| La1-N6 | 0.2671(7) |
| La1-O4 | 0.2633(8) |
| La1-N5 | 0.2693(3) |
|
| B, Complex 2 |
|
| Bond | Lengtha, Å |
|
| Ce1-O2 | 0.2430(9) |
| Ce1-O6 | 0.2583(9) |
| Ce1-N6 | 0.2676(7) |
| Ce1-N2 | 0.2788(5) |
| Ce1-O1 | 0.2433(7) |
| Ce1-O4 | 0.2623(11) |
| Ce1-N5 | 0.2685(3) |
| Ce1-O7 | 0.2573(9) |
| Ce1-O3 | 0.2661(11) |
| Ce1-N1 | 0.2761(5) |
 | Table III.Selected bond angles in the
complexes. |
Table III.
Selected bond angles in the
complexes.
| A, Complex 1 |
|---|
|
|---|
| Angle | Sizea, ° |
|---|
| O1-La1-O2 | 147.8(2) |
| O1-La1-O6 |
70.2(2) |
| O1-La1-O4 |
85.0(3) |
| O6-La1-O4 | 140.8(3) |
| O7-La1-N6 | 119.4(2) |
| O1-La1-O3 | 131.3(3) |
| O6-La1-O3 | 135.6(3) |
| O1-La1-N5 |
73.9(2) |
| O6-La1-N5 |
72.4(5) |
| O3-La1-N5 |
78.0(2) |
| O7-La1-N1 |
75.0(2) |
| N6-La1-N1 | 117.8(2) |
| O1-La1-N2 |
58.1(2) |
| O1-La1-O7 |
81.9(2) |
| O2-La1-O6 |
84.0(2) |
| O2-La1-O4 | 126.8(3) |
| O1-La1-N6 | 123.7(2) |
| O6-La1-N6 |
69.2(2) |
| O2-La1-O3 |
80.7(3) |
| O4-La1-O3 |
48.2(2) |
| O2-La1-N5 | 116.8(5) |
| O4-La1-N5 |
71.7(2) |
| O1-La1-N1 | 118.0(2) |
| O6-La1-N1 | 144.6(2) |
| O3-La1-N1 |
67.6(2) |
| O2-La1-N2 | 125.8(2) |
| O4-La1-N2 |
65.9(2) |
| N5-La1-N2 | 116.8(2) |
| O2-La1-O7 |
72.0(2) |
| O7-La1-O6 |
72.2(2) |
| O7-La1-O4 | 135.0(3) |
| O2-La1-N6 |
59.2(2) |
| O4-La1-N6 | 103.6(2) |
| O7-La1-O3 | 138.6(3) |
| N6-La1-O3 |
67.1(3) |
| O7-La1-N5 | 142.2(3) |
| N6-La1-N5 |
57.6(4) |
| O2-La1-N1 |
73.4(2) |
| O4-La1-N1 |
73.8(2) |
| N5-La1-N1 | 142.2(4) |
| O7-La1-N2 |
70.6(2) |
| N6-La1-N2 | 169.5(2) |
| N1-La1-N2 |
59.9(2) |
|
| B, Complex 2 |
|
| Angle | Sizea, ° |
|
| O2-Ce1-O1 | 147.5(2) |
| O2-Ce1-O6 |
83.3(3) |
| O2-Ce1-O4 | 128.5(3) |
| O6-Ce1-O4 | 140.2(3) |
| O7-Ce1-O3 | 140.3(3) |
| O2-Ce1-N6 |
60.1(2) |
| O6-Ce1-N6 |
69.2(2) |
| O2-Ce1-N5 | 118.1(2) |
| O6-Ce1-N5 |
72.9(2) |
| N6-Ce1-N5 |
58.1(2) |
| O7-Ce1-N1 |
74.9(2) |
| O3-Ce1-N1 |
68.7(2) |
| O2-Ce1-N2 | 125.9(2) |
| O6-Ce1-N2 | 119.4(2) |
| N6-Ce1-N2 | 168.6(2) |
| O2-Ce1-O7 |
71.8(3) |
| O1-Ce1-O6 |
70.8(3) |
| O1-Ce1-O4 |
83.5(3) |
| O2-Ce1-O3 |
83.0(3) |
| O6-Ce1-O3 | 134.6(3) |
| O1-Ce1-N6 | 123.5(2) |
| O4-Ce1-N6 | 103.6(3) |
| O1-Ce1-N5 |
73.1(2) |
| O4-Ce1-N5 |
70.8(2) |
| O2-Ce1-N1 |
73.7(2) |
| O6-Ce1-N1 | 145.2(2) |
| N6-Ce1-N1 | 117.7(2) |
| O1-Ce1-N2 |
57.8(2) |
| O4-Ce1-N2 |
65.1(3) |
| N5-Ce1-N2 | 115.5(3) |
| O1-Ce1-O7 |
82.1(3) |
| O7-Ce1-O6 |
73.3(3) |
| O7-Ce1-O4 | 133.7(3) |
| O1-Ce1-O3 | 129.1(3) |
| O4-Ce1-O3 |
48.1(2) |
| O7-Ce1-N6 | 121.0(2) |
| O3-Ce1-N6 |
66.4(2) |
| O7-Ce1-N5 | 143.1(2) |
| O3-Ce1-N5 |
76.1(2) |
| O1-Ce1-N1 | 118.0(2) |
| O4-Ce1-N1 |
73.8(2) |
| N5-Ce1-N1 | 141.2(2) |
| O7-Ce1-N2 |
70.0(2) |
| N1-Ce1-N2 |
60.3(2) |
| O3-Ce1-N2 | 103.6(2) |
Cytotoxic activity assay
The effects of the DMSO-soluble HL and the complexes
1 and 2 on the proliferation of the human lung cancer cell line
A549 and the human gastric cancer lines BGC823 and SGC7901 were
explored by MTT assay. The half maximal inhibitory concentration
(IC50) was further calculated according to the results
of the MTT assay. Briefly, the three types of tumor cell lines
evaluated in the present study were plated in 96-well plates in
triplicate and grown to 75% confluence. Following treatment with
different concentrations of the complexes 1 and 2 for 72 h, the
media were replaced with 10 µl of fresh medium containing 0.45
mg/ml MTT reagent. The cells were incubated for 1 h at 37°C in the
presence of 5% CO2 to allow the formation of formazan
crystals. The formazan crystals were dissolved by addition of 100
µl DMSO during a 4-h incubation period at 37°C and 5%
CO2. The colorimetric change was measured on a
spectrophotometer at an absorbance wavelength of 570 nm. Data were
expressed as the percentage of viability relative to vehicle. At
least three independent experiments were performed.
Flow cytometry analysis
Apoptotic rates were determined by flow cytometry
using the FITC Annexin V Apoptosis Detection kit I, which contains
propidum iodide (PI) for red fluorescence detection (cat. no.
556547; BD Biosciences). Briefly, the cancer cells (A549) were
seeded at a density of 1×106 cells/well in 6-well plates
overnight, and then treated with 5, 10 and 15 µmol/l complex 2 for
48 h. Cells (1×106) were collected by centrifugation 200
× g for 5 min at 4°C and washed twice with cold PBS. Staining was
performed according to the manufacturer's protocol, and the cells
were analyzed using a flow cytometer (Accuri C6; BD Biosciences)
with 488 nm excitation wavelength and 530 nm emission wavelength.
The green fluorescence of Annexin V and the red fluorescence of PI
was detected by flow cytometry. The results were analyzed using
Cell Quest software version 6.0 (BD Biosciences). At least three
independent experiments were performed.
Western blotting
The aforementioned three types of tumor cells were
seeded in 6-well plates overnight. Cells were treated with 5 µmol/l
complex 2 for 48 h. Upon treatment, all the cell samples were
washed twice with PBS and lysed in RIPA Lysis Buffer (cat. no.
P0013C; Beyotime Institute of Biotechnology, Haimen, China). The
protein concentration of each sample was determined with a
bicinchoninic acid assay kit (cat. no. P0009; Beyotime Institute of
Biotechnology). The samples were separated by 12% SDS-PAGE,
transferred to PVDF membranes by electroblotting and blocked using
5% dried skimmed milk overnight at 4°C. Next, the membranes were
probed with polyclonal antibodies against caspase 3 (1:1,000; cat.
no. 3004; BioVision, Inc., Milpitas, CA, USA), the apoptosis
regulators B cell lymphoma (Bcl)-2-associated X protein (Bax)
(1:1,000; cat. no. 3032; BioVision, Inc.) and Bcl-2 (1:1,000; cat.
no. 3195; BioVision, Inc.) and GAPDH (1:2,000; cat. no. E7EUT5;
Abmart, Inc., Shanghai, China) diluted in 5% bovine serum albumin
(cat. no. 10711454001; Roche Applied Science, Penzberg, Germany).
The membranes were next incubated for 2 h at room temperature with
secondary alkaline phosphatase (AP)-conjugated anti-mouse
immunoglobulin (Ig)G (cat. no. A0258; Beyotime Institute of
Biotechnology) and AP-conjugated anti-rabbit IgG (cat. no. A0239;
Beyotime Institute of Biotechnology) antibodies, which were diluted
1:1,000 in PBS, followed by incubation with an enhanced
chemiluminescent reagent (Lumi-Phos™ WB Chemiluminescent Substrate;
Thermo Fisher Scientific, Inc.) and visualization on an
autoradiography film. Membranes were scanned using an ScanJet G4010
Photo Scanner (HP, Inc., Palo Alto, CA, USA) Densitometry values
were determined using UN-SCAN-IT version 6.0 software (Silk
Scientific, Inc., Orem, UT, USA).
Statistical analysis
All data were analyzed by SAS 6.12 software, and the
results were expressed as the mean + standard deviation. To compare
the differences between the groups, the statistical significance
was analyzed using a one-way analysis of variance followed by post
hoc comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results and Discussion
Crystal structures of complexes 1 and
2
As revealed by single-crystal XRD, complexes 1 and 2
are isostructural and crystallize in the monoclinic Pn space group
(Table I). The structure of complex 1
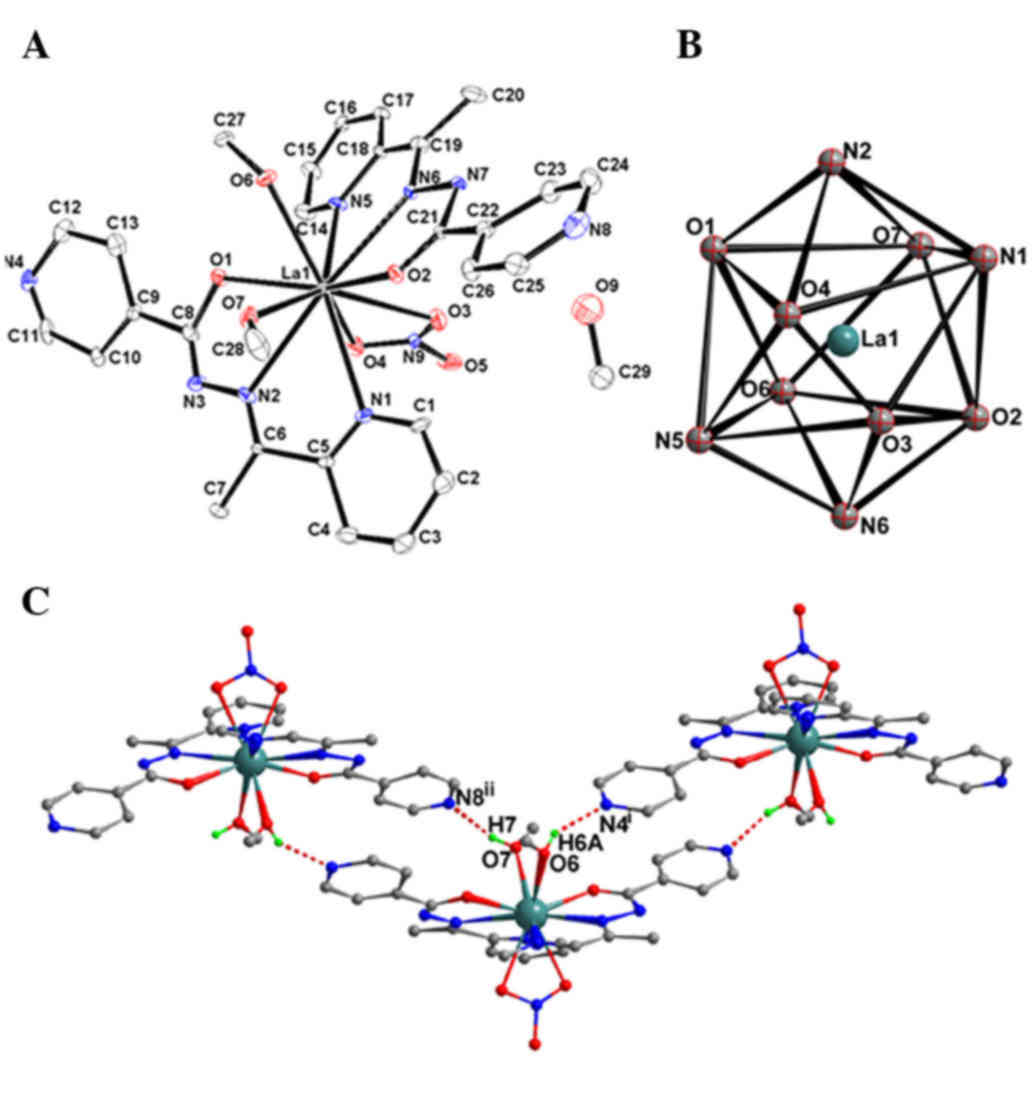
will be discussed in detail as a representative example. As shown
in Fig. 1A, once coordinated with a
metal ion, the acylhydrazone ligand is deprotonated and acts as
tridentate mono-basic ligand. The 10-coordinated La(III) ion with
bicapped square antiprism coordination geometry (Fig. 1B) is surrounded by two L−
anions with N2O donor sets, one bidentate nitrate anion
and two methanol molecules. The Ln-O (N) bond lengths are in the
range of 0.2463(7) to 0.2809(5) nm in complex 1, slightly longer than
those in complex 2 [0.2430(9)-0.2788(5)
nm] and the reported complex
[Pr(L)2(NO3)(CH3OH)2]·CH3CH2OH
[0.2414(2)-0.2707(2) nm] (20),
which is probably due to the Ln contraction. The numbers between
parentheses in the above lenghts indicate the standard deviation of
the bond distances to the third decimal place. In the crystal
structure of complex 1, intramolecular O–H···N hydrogen bonds
[O6–H6A···N4i and O7–H7···N8ii, with D···A
distances of 0.267(4) and
0.2752(8) nm, and D–H···A angles of
142.9 and 163.2°, respectively] link the complex molecules into a
chain-like structure (Fig. 1C). This
is similar in the crystal structure of complex 2, with D···A
distances of 0.272(5) and
0.2719(9) nm, and D–H···A angles of
142.1 and 165.9°, respectively.
IR spectra
The solid-state FT-IR spectra of the complexes,
recorded on a FT-IR spectrophotometer in the range 4,000–200
cm−1, were fully consistent with their single crystal
structures. The samples were studied as powder dispersed in KBr
pellets. The spectral region for all the complexes was somewhat
similar due to the similarity in the coordination modes of the
ligands with the metal centre. The ν(C=O) of the free ligand is
1,679 cm−1, while for complexes 1 and 2, this peak
shifted to 1,586 and 1,578 cm−1, respectively. In
addition, the ν(ligand-complexes) is ~100 cm−1, thus
indicating that the O atom of the C=O bond participates in the
coordination to the metal ion. The stretching frequency of the
azomethine (CH=N) group in the hydrazone ligand was observed to be
~1,586 cm−1, which shifted to lower frequency values
upon complexation with the metal by 70 cm−1, indicating
that the N atom of the C=N bond participates in the coordination to
the metal ion. The vibrations ν(N-H) disappear in the spectra of
the two complexes, indicating that the ligand is deprotonated. The
new bands at ~550 cm−1 for complexes 1 and 2 are
assigned to ν(M-O), and the weak peaks at ~470 cm−1 are
assigned to ν(M-N).
Antitumor activities
It has been reported that arylhydrazone and their
metal complexes display particularly effective antitumor
activities, possibly due to their NO bidentate systems (23). In consequence, in vitro cytotoxicity
assays were explored in the human lung cancer cell line A549 and in
the human gastric cancer cell lines BGC823 and SGC7901. The two
metal complexes displayed a concentration-dependent cytotoxic
profile in all cell lines, and their IC50 values are
shown in Table IV. The
IC50 values of complexes 1 and 2 were significantly
lower than that of the metal-free ligand in all the tested cell
lines, which suggested that the coordinated Ln(III) ion served a
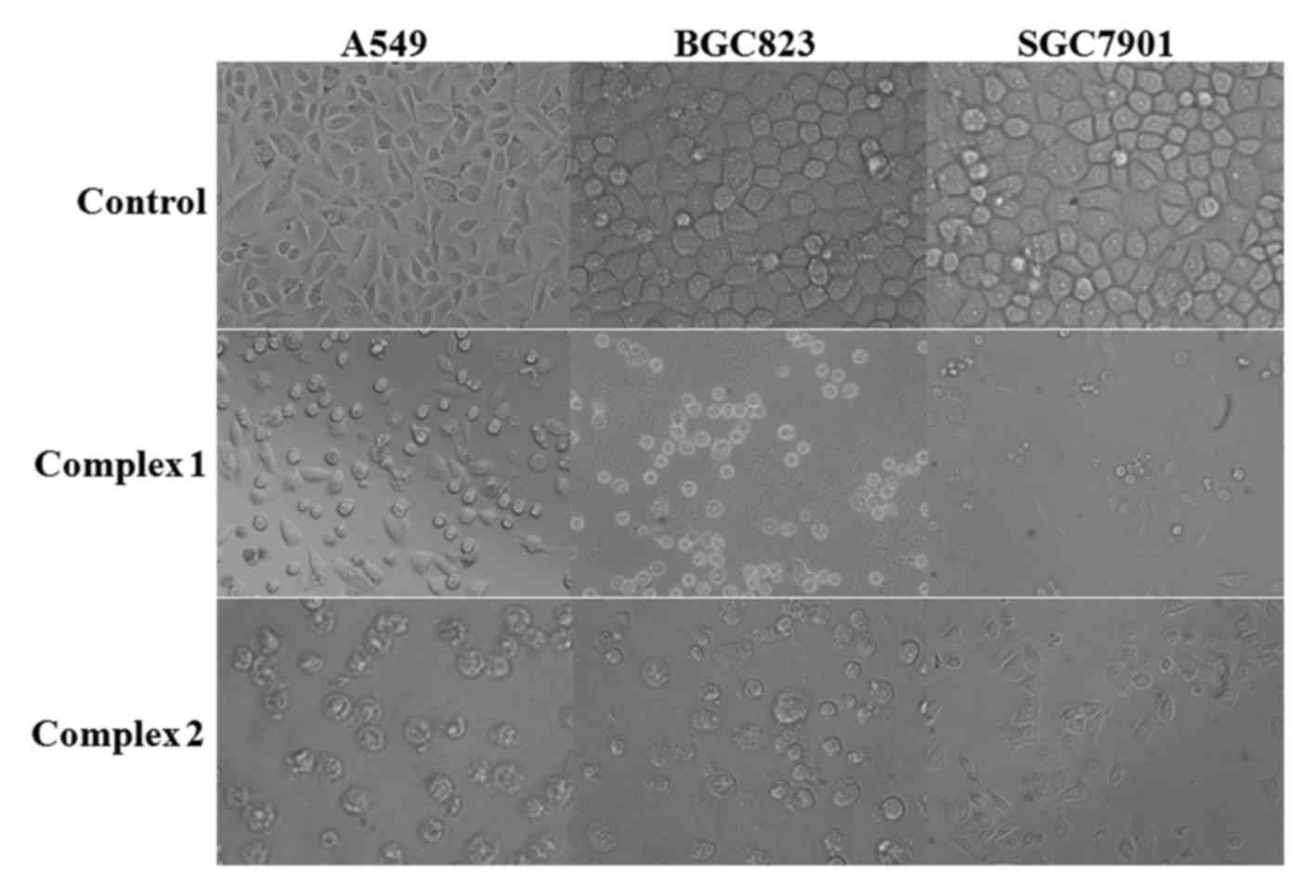
major role in mediating the potency of the complexes. Morphological
examinations also revealed that the proliferation of the cells was
significantly inhibited, and that the cells exhibited morphological
changes such as cell shrinkage and detachment (Fig. 2).
 | Table IV.IC50 (µmol/l) values of
the ligand and complexes 1 and 2 against the human lung cancer A549
and the human gastric cancer BGC823 and SGC7901 cell lines after
incubation for 72 h. |
Table IV.
IC50 (µmol/l) values of
the ligand and complexes 1 and 2 against the human lung cancer A549
and the human gastric cancer BGC823 and SGC7901 cell lines after
incubation for 72 h.
|
| IC50
(µmol/l) |
|---|
|
|
|
|---|
| Tested
compounds | A549 | BGC823 | SGC7901 |
|---|
|
Ce(NO3)3·6H2O | >150.0 | >150.0 | >150.0 |
|
La(NO3)3·6H2O | >150.0 | >150.0 | >150.0 |
| Ligand | 41.8 | 39.2 | 43.5 |
| Complex 1 | 11.5 | 13.3 | 15.6 |
| Complex 2 | 10.9 | 12.6 | 14.8 |
Cell apoptosis
MTT assay demonstrated that complexes 1 and 2
exhibited growth inhibition abilities on the three human cancer
cell lines evaluated, and that complex 2 exhibited relatively
better cytotoxic activities than complex 1 on the human lung cancer
cell line A549. For this reason, complex 2 and the human lung
cancer cell line A549 were employed in the subsequent apoptosis and
western blot analyses. To determine whether the proliferation
inhibition induced by complex 2 on A549 cells was attributed to the
induction of apoptosis, Annexin V/PI staining and flow cytometry
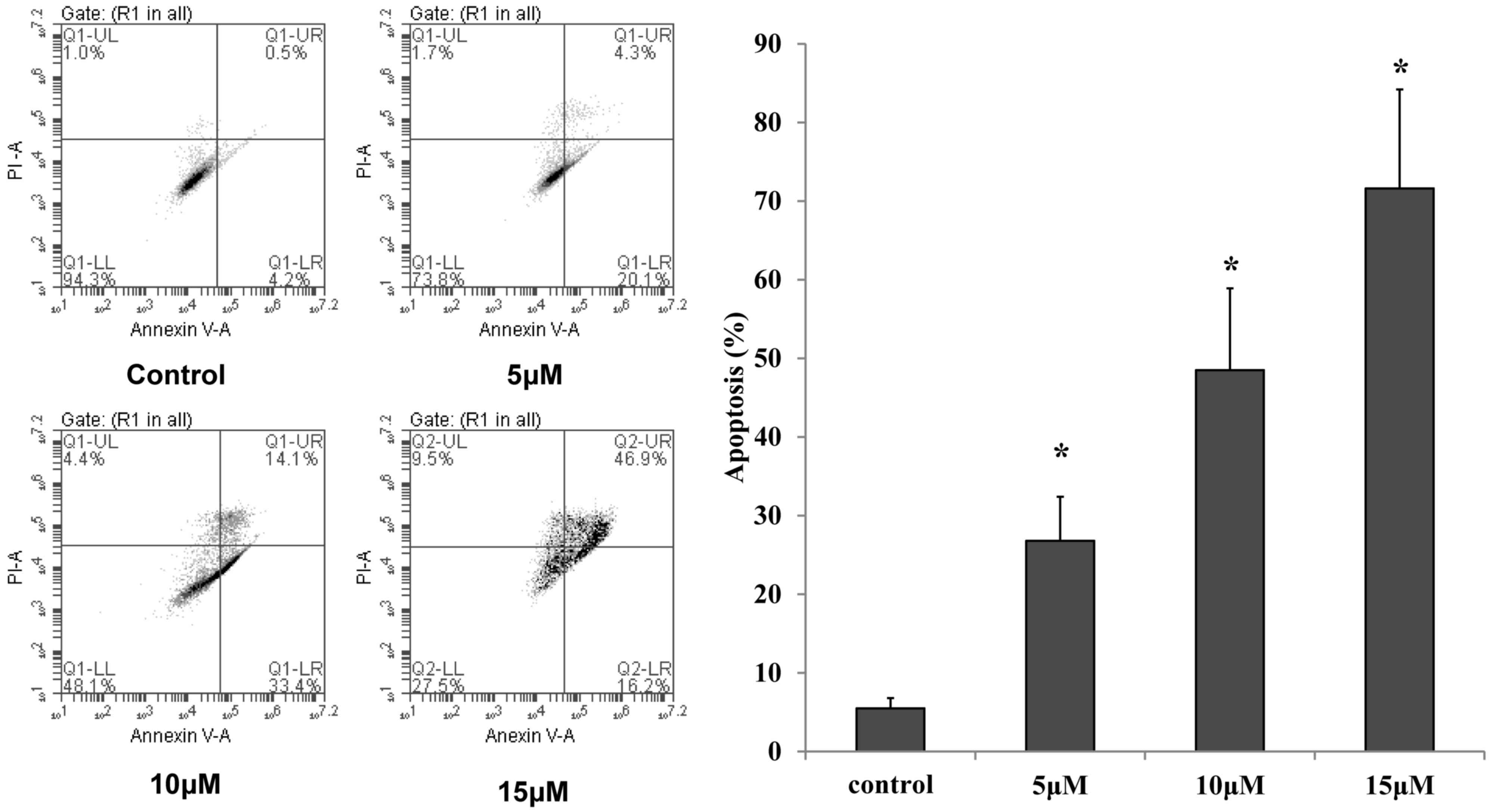
were applied to quantify the percentage of apoptosis. As shown in
Fig. 3, the number of apoptotic A549
cells increased in a dose-dependent manner following incubation
with 5, 10 and 15 µmol/l complex 2 for 48 h (P<0.05).
Caspase 3, Bax and Bcl-2 protein
expression
As previously reported, caspase 3 and Bcl-2 family
proteins are key regulators of the apoptotic pathway. When caspase
3 and Bax expression is high, the cells proceed into apoptosis,
while when Bcl-2 is produced in excess, cells are protected from
apoptosis (24,25). In the present study, western blotting
was applied to detect the protein expression of caspase 3, Bax and
Bcl-2 in the human lung cancer cell line A549 treated with
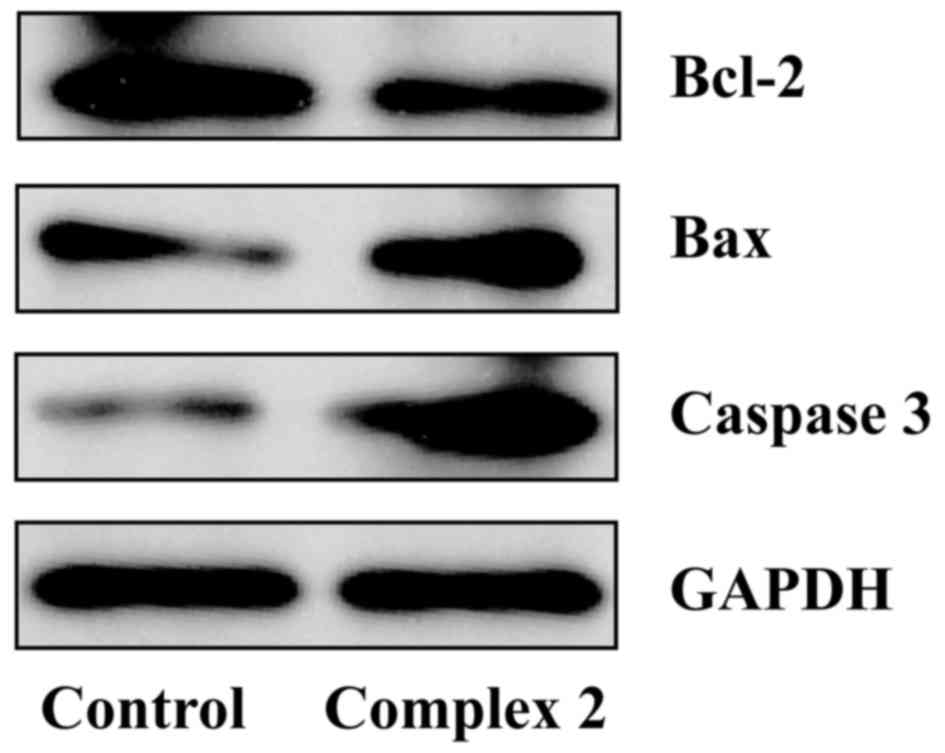
different concentrations of complex 2 for 48 h. The results
revealed that caspase 3 and Bax protein expression significantly
increased, while Bcl-2 protein expression was remarkably
downregulated upon incubation with 5 µmol/l complex 2 for 48 h
(Fig. 4). These results suggested
that the possible mechanism of complex 2 responsible for its
ability to induce A549 cell apoptosis may be associated with its
ability to increase the expression of caspase 3 and Bax, and to
decrease the expression of Bcl-2. These results may improve the
understanding of the pharmacological mechanisms of novel Ln(III)
complexes in cancer treatment.
In summary, two isostructural Ln(III) complexes with
isonicotinohydrazone ligand have been isolated and characterized by
elemental analyses, IR spectra and single-crystal XRD analyses in
the present study. The results indicate that the two complexes
exert considerable cytotoxic activity against three cancer cell
lines (human lung cancer cell line A549, and human gastric cancer
cell lines BGC823 and SGC7901), and that the IC50 values
of complexes 1 and 2 are lower than that of the ligand. Annexin
V/PI staining and western blotting further suggested that Ln(III)
complexes can induce apoptosis in A549 cells. Those findings may
potentially be useful for biomedical applications of novel Ln(III)
complexes, particularly in the human cancer therapeutic field.
Acknowledgements
The present study was supported in part by the
National Natural Science Foundation of China (grant nos. 81201908
and 21404033), Fund of the Natural Science Foundation of Jiangsu
(grant no. BK20141122), and Development Fund of Clinical Science
and Technology of Jiangsu University (grant no. JLY20140065).
References
|
1
|
Tsuchida E and Oyaizu K: Oxovanadium
(III–V) mononuclear complexes and their linear assemblies bearing
tetradentate Schiff base ligands: Structure and reactivity as
multielectron redox catalysts. Coord Chem Rev. 237:213–228. 2003.
View Article : Google Scholar
|
|
2
|
Sherrington DC: Utilisation of homogeneous
and supported chiral metal (salen) complexes in asymmetric
catalysis. Chem Soc Rev. 28:85–93. 1999. View Article : Google Scholar
|
|
3
|
Shongwe MS, Al-Rahbi SH, Al-Azani MA,
Al-Muharbi AA, Al-Mjeni F, Matoga D, Gismelseed A, Al-Omari IA,
Yousif A, Adams H, et al: Coordination versatility of tridentate
pyridyl aroylhydrazones towards iron: Tracking down the elusive
aroylhydrazono-based ferric spin-crossover molecular materials.
Dalton Trans. 41:2500–2514. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krishnamoorthy P, Sathyadevi P,
Senthilkumar K, et al: Copper (I) hydrazone complexes: Synthesis,
structure, DNA binding, radical scavenging and computational
studies. Inorg Chem Commun. 14:1318–1322. 2011. View Article : Google Scholar
|
|
5
|
Ni WX, Li M, Zhan SZ, Hou JZ and Li D: In
situ immobilization of metalloligands: A synthetic route to
homometallic mixed-valence coordination polymers. Inorg Chem.
48:1433–1441. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tamasi G, Chiasserini L, Savini L, Sega A
and Cini R: Structural study of ribonucleotide reductase inhibitor
hydrazones. Synthesis and X-ray diffraction analysis of a copper
(II)-benzoylpyridine-2-quinolinyl hydrazone complex. J Inorg
Biochem. 99:1347–1359. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song YM, Li WJ and Yang ML: Study on
synthesis, characterization and antitumor activity of rare earth
metal complexes with all trans retinoic acid and arginine acid.
Chinese J Inorg Chem. 30:1087–1096. 2014.
|
|
8
|
Bernhardt PV, Chin P, Sharpe PC, Wang JY
and Richardson DR: Novel diaroylhydrazine ligands as iron
chelators: Coordination chemistry and biological activity. J Biol
Inorg Chem. 10:761–777. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kalinowski DS, Sharpe PC, Bernhardt PV and
Richardson DR: Structure-activity relationships of novel iron
chelators for the treatment of iron overload disease: The methyl
pyrazinylketone isonicotinoyl hydrazone series. J Med Chem.
51:331–344. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xie QF, Gao PZ and Chen YM: Synthesis,
crystal structure and anti-tumor activity in vitro of a hydrazone
Schiff base and Its Cd(II) complex. Chinese J Inorg Chem.
30:2382–2388. 2014.
|
|
11
|
Sridhar SK, Saravanan M and Ramesh A:
Synthesis and antibacterial screening of hydrazones, Schiff and
Mannich bases of isatin derivatives. Eur J Med Chem. 36:615–625.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Banerjee S, Ray A, Sen S, et al:
Pseudohalide-induced structural variations in hydrazone-based metal
complexes: Syntheses, electrochemical studies and structural
aspects. Inorganica Chim Acta. 361:2692–2700. 2008. View Article : Google Scholar
|
|
13
|
Albrecht M, Osetska O and Fröhlich R:
2-[(8-Hydroxyquinolinyl)methylene]hydrazinecarboxamide: Expanding
the coordination sphere of 8-hydroxyquinoline for coordination of
rare-earth metal(III) ions. Dalton Trans. 3757–3762. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Parker D, Dickins RS, Puschmann H,
Crossland C and Howard JA: Being excited by lanthanide coordination
complexes: Aqua species, chirality, excited-state chemistry, and
exchange dynamics. Chem Rev. 102:1977–2010. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hunter RB and Walker W: Anticoagulant
action of neodymium 3-sulpho-isonicotinate. Nature. 178:471956.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kramsch DM, Aspen AJ and Rozler LJ:
Atherosclerosis: Prevention by agents not affecting abnormal levels
of blood lipids. Science. 213:1511–1512. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hodnett EM and Dunn WJ III: Cobalt
derivatives of Schiff bases of aliphatic amines as antitumor
agents. J Med Chem. 15:3391972. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hodnett EM and Mooney PD: Antitumor
activities of some Schiff bases. J Med Chem. 13:7861970. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y and Yang ZY: Synthesis,
characterization and DNA-binding properties of three 3d transition
metal complexes of the Schiff base derived from diethenetriamine
with PMBP. Transit Metal Chem. 30:902–906. 2005. View Article : Google Scholar
|
|
20
|
Hao ZY, Liu QW, Xu J, Jia L and Li SB:
Synthesis, characterization, antioxidant, activities and
DNA-binding studies of
(E)-N'-[1-(pyridin-2-yl)ethylidene]isonicotinohydrazide and its
Pr(III) and Nd(III) complexes. Chem Pharm Bull (Tokyo).
58:1306–1312. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sheldrick GM: SADABS, program for
empirical absorption correction of area detector data. University
of Göttingen; Germany: 1996
|
|
22
|
Sheldrick GM: SHELX-97, program for the
solution and refinement of crystal structures. University of
Göttingen; Germany: 1997
|
|
23
|
Cui Z, Li Y, Ling Y, Huang J, Cui J, Wang
R and Yang X: New class of potent antitumor acylhydrazone
derivatives containing furan. Eur J Med Chem. 45:5576–5584. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
May P and May E: Twenty years of p53
research: Structural and functional aspects of the p53 protein.
Oncogene. 18:7621–7636. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cory S and Adams JM: The Bcl2 family:
Regulators of the cellular life-or-death switch. Nat Rev Cancer.
2:647–656. 2002. View
Article : Google Scholar : PubMed/NCBI
|