Introduction
The liver has excellent regenerative capacities.
Liver cells are quiescent under normal conditions; however, they
enter the cell cycle when damaged and proliferate until the
original liver volume is restored (1). A number of conditions may alter liver
mass, including surgical resection, chemicals and pathogens
(2–4).
In the situation of liver cancer or hepatocirrhosis, surgical
resection is a standard medical therapy. Cirrhosis of the liver is
one of the leading causes of mortality, and liver cancer is the
third most common cancer and the second leading cause of
cancer-associated mortality worldwide (5,6).
Numerous signaling pathways are involved in liver
regeneration, including pathways involving hepatocyte growth
factor, epidermal growth factor, interleukin 6, tumor necrosis
factor-α and transforming growth factor (TGF)-α and β (2,7). Among
these, the TGF-β/Smad pathway is reported to suppress cellular
proliferation and to regulate numerous biological processes;
however, the responses to TGF-β differ according to cell or tissue
type and the microenvironment (8–10).
Activation of secreted TGF-β and assembly of TGF-β
receptor type 1 and 2 (TGF-β R1 and 2) in the cellular membrane is
the first step in the TGF-β/Smad pathway. Subsequently, TGF-β R2
phosphorylates and activates TGF-β R1, which in turn phosphorylates
cytoplasmic Smad2 and Smad3 [also termed receptor-Smads (R-Smads)].
Activated R-Smads bind Smad4 and move into the nucleus, and the
Smads complex, along with co-factors, positively or negatively
regulates the expression of target genes (11–14). In
cellular proliferation, cMyc and cyclin D1 genes or proteins are
downregulated, while the expression of p15, p21 and Smad2 genes is
upregulated following activation of the TGF-β/Smad pathway. In
non-Smad pathways, the TGF-β receptor activates other proteins,
including RAS, phosphoinositide 3-kinase and FAS (12,14–16).
As a target gene of TGF-β, KLF10 may regulate the
TGF-β/Smad pathway. KLF10 may enhance Smad2, p21 and plasminogen
activator inhibitor-1 expression, and repress the transcription of
the Smad7 gene. KLF10 also plays important roles in numerous
biological processes, and it has been reported to inhibit
proliferation and induce apoptosis in several cell types (10,17,18).
However, the role of KLF10 in various pathophysiological conditions
remains unclear.
Partial hepatectomy (PH), resulting in the removal
of ~70% of the liver, is widely utilized for studies of liver
regeneration, acute liver failure and the metastasis of liver
cancer (19,20). KLF10 is known as a potential
antiproliferative gene; however, to the best of our knowledge,
there are no reports on the role of KLF10 in liver regeneration
(10,21). In the present study, to elucidate the
role of the KLF10 gene in liver regeneration following tissue loss,
molecular and histopathological analyses were conducted using
KLF10-knockout (KO) mice following a PH that removed two-thirds of
the liver.
Materials and methods
Animals
All procedures were approved by the Institutional
Animal Care and Use Committee of Konkuk University (Seoul, South
Korea). Three pairs of 8-week-old KLF10-KO C57BL/6 J mice (age:
54–57 days, average 8 weeks; body weight: 23.1–24.9 g, average 24.1
g) were kindly provided by Professor Woon-Kyu Lee (Inha University,
Incheon, Korea) (22). and five pairs
of 6-week-old C57BL/6J mice (age: 32–35 days old, average 6 weeks
old; body weight: 21.9–25.1 g, average 23.7 g) were obtained from
the Korea Research Institute of Bioscience and Biotechnology
(Daejeon, Korea). All mice were bred in the laboratory animal
breeding room under specific pathogen-free conditions to produce
the KO and wild-type (WT) mice groups. For genotyping each mouse,
DNA samples were isolated from all mice tails using the Genomic DNA
extraction kit (Bioneer Corporation, Daejeon, Korea) and subjected
to polymerase chain reaction (PCR) using the AccuPower®
PCR PreMix (#K-2016; Bioneer Corporation). The DNA primers for
genotyping were: KLF10 forward, CCT TCC TGC CAA CAA CTC TC and
reverse, TCT GAG GAG TGA CCC TTG CT; and KLF10-KO forward, TCG CCT
TCT TGA CGA GTT CT (12) and reverse,
TCTGAGGAGTGACCCTTGCT. The cycling conditions were initial
denaturation for 5 min at 95°C, followed by 30 cycles of 1 min at
95°C, 1 min at annealing temperature, 1 min at 72°C and an
additional 10 min at 72°C for final elongation. After the reaction,
the PCR samples were electrophoresed on a 1.5% agarose gel. The
size of KLF10 KO gene is 658 base pairs (bp) and that of WT gene is
248 bp.
Experimental design and surgical
procedure
At 8 weeks of age, a two-thirds PH was performed.
Subsequent to anaesthetizing with Zoletil (Virbac Corporation, Fort
Worth, TX, USA) and Rompun (Bayer Korea, Ltd., Seoul, Korea), the
middle abdominal skin and linea alba were incised. The left lateral
and median liver lobes were excised and the peritoneum and the skin
were closed (20). From each group of
KLF10-KO and WT mice, 3–5 animals were sacrificed at 0, 24, 48 or
72 h post-PH using the CO2 euthanasia chamber (n=3, n=5,
n=5 and n=5, respectively). At 2 h prior to euthanasia, 100 mg/kg
5-bromo-2-deoxy-uridine (BrdU; Sigma-Aldrich; EMD Millipore,
Billerica, MA, USA) was injected intraperitoneally. At the time of
sacrifice, all mice were grossly examined and blood was collected
from the caudal vena cava. The liver was excised and weighed. The
upper right lateral lobe was fixed in 10% neutral-buffered formalin
for histopathological analysis and the other hepatic tissues were
frozen for subsequent analyses.
Histological and immunohistochemical
analysis
Fixed tissues were processed routinely for paraffin
sectioning. Liver tissues were then embedded in paraffin and cut
into 4-µm thick sections. The sections were deparaffinized,
rehydrated and stained with hematoxylin and eosin (H&E)
(Sigma-Aldrich; EMD Millipore). For immunohistochemistry, serial
sections were cut, deparaffinized, rehydrated and serial incubated
in 1.0% H2O2, 2 N HCl and 0.1% trypsin. The
slides were pre-incubated with normal blocking serum (Vectastain
ABC kit; #PK-6102; Vector Laboratories, Inc., Burlingame, CA, USA)
according to the manufacturer's protocol and then incubated with
anti-BrdU antibody (#B8434; dilution, 1:500; Sigma-Aldrich) for 2 h
at room temperature. Detection of the BrdU was performed using
biotinylated secondary antibodies (Vectastain ABC kit; #PK-6102;
dilution, 1:200; Vector Laboratories, Inc.;), avidin-coupled
peroxidase (Vectastain ABC kit; Vector Laboratories, Inc.), and
diaminobenzidine (DAB; DAB substrate kit; Vector Laboratories,
Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was prepared from frozen liver tissues
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and was reverse transcribed into cDNA using moloney-murine
leukemia virus reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc.). The cDNA was used as a template for
amplification in the PCR using the AccuPower® PCR PreMix
(#K-2016; Bioneer Corporation). Smad2, Smad3, Smad4, Smad7, p15,
p21, TGF-β R1, TGF-β R2, cMyc, cyclin D1 and β-actin mRNA
expression level were analyzed using the RT-qPCR method, as
described by Shiao (23), with the
previously described primers (12).
The cycling conditions were initial denaturation for 5 min at 95°C,
followed by 35 cycles of 1 min at 95°C, 1 min at annealing
temperature, 1 min at 72°C and an additional 10 min at 72°C for
final elongation. After the reaction, the PCR mixtures were
electrophoresed on a 1.5% agarose gel. Band intensities were
quantified using ImageQuant Software (Image Lab V4.0; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and were normalized to the
transcription levels of β-actin. Each sample was tested in
triplicate.
Western blot analysis
Protein was extracted from the liver using the
extraction solution (Pro-Prep™; Intron Biotechnology, Inc.,
Seongnam, Korea). The protein concentrations were determined using
the bicinchoninic kit (Pierce; Thermo Fisher Scientific, Inc.).
Subsequent to being transferred to nitrocellulose membranes, the
proteins were blocked with 5% skimmed milk and then incubated
overnight with specific antibodies against β-actin (#sc-47778;
dilution, 1:500), Smad4 (#sc-7966; dilution, 1:500), Smad7
(#sc-11392; dilution, 1:1,000), p15 (#sc-65223; dilution, 1:200),
p21 (#sc-817; dilution, 1:500), TGF-β R1 (#sc-398; dilution,
1:200), cMyc (#sc-56505; dilution, 1:200) (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), Smad2 (#3122; dilution,
1:1,000), Smad3 (#9513; dilution, 1:200), p27 (#2552; dilution,
1:500), TGF-β R2 (#11888, dilution, 1:500) and cyclin D1 (#2922;
dilution, 1:500; all from Cell Signaling Technology, Inc., Danvers,
MA, USA) at 4°C. Subsequently, the membranes were washed with TBS
with Tween-20 and incubated for 1 h with either horseradish
peroxidase-conjugated anti-rabbit (#sc-2054; dilution, 1:1,000) or
anti-mouse secondary antibodies (#sc-2031; dilution, 1:1,000; Santa
Cruz Biotechnology, Inc.) at room temperature. Specific antibodies
were detected with an electrochemiluminescence test kit (KPL, Inc.,
Gaithersburg, MD, USA). The band intensities were quantified using
Image Lab V4.0 (Bio-Rad Laboratories, Inc.) and were normalized to
β-actin expression (12).
Statistical analysis
Statistical analysis was performed using SPSS V14.0
software (SPSS, Inc., Chicago, IL, USA). Statistically significant
differences between the studied groups were evaluated using
Student's unpaired t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Gross findings
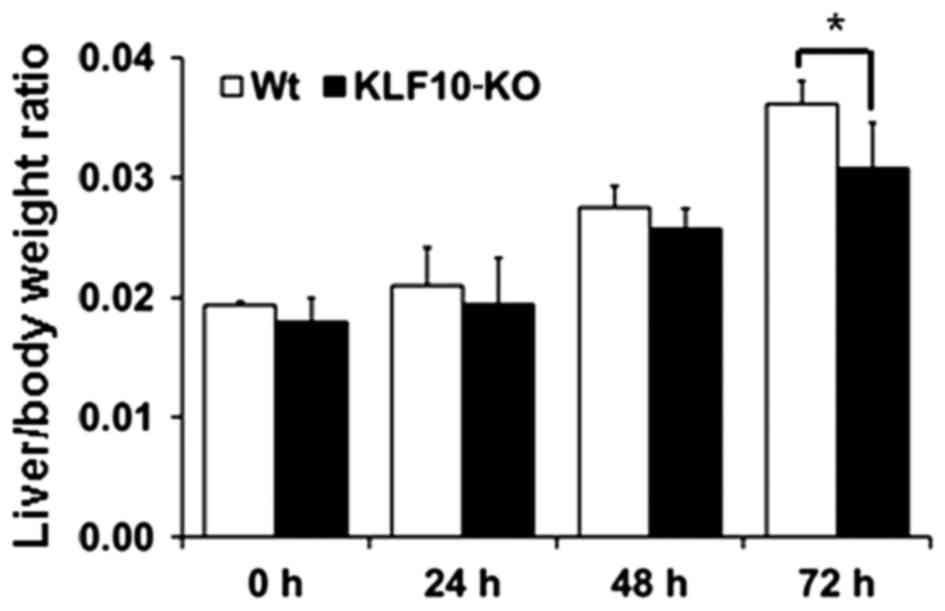
At 0, 24, 48 and 72 h post-PH, the body and liver
weights of all mice were measured. Macroscopically, the volume of
the remaining liver tissues increased from 48 h post-PH. The color
of the liver subsequent to PH was paler than that of normal liver
tissues from 48 h post-PH. At the time of sacrifice, the liver was
removed and weighed to examine the liver/body weight ratio. The
mean of the ratios at 0, 24, 48 and 72 h post-PH for the WT mice
vs. KLF10-KO mice was 0.0194 vs. 0.0180, 0.0210 vs. 0.0194, 0.0275
vs. 0.0257 and 0.0361 vs. 0.0307, respectively (Fig. 1). Overall, KLF10-KO mice exhibited
lower ratios compared with WT mice, and a significant difference
was observed at 72 h post-PH (P=0.028).
Histopathological analysis and BrdU
staining
H&E staining revealed no marked features at 0
and 24 h post-PH. Pale cytoplasm and hypertrophy of hepatocytes was
observed from 48 h post-PH (Fig. 2A).
Mitotic figures, mainly in the centrilobular area, were the most
frequent at 48 h post-PH, and these remained numerous at 72 h
post-PH. Until 24 h post-PH, few mitotic figures were observed
(Fig. 2A). Similarly, upon BrdU
staining to analyze the proliferative potential of cells, the
proportion of labeled nuclei was markedly increased at 48 h post-PH
(Fig. 2B). The number of
BrdU-positive nuclei was counted in an area of 0.1 mm2
under a light microscope at ×400 magnification. While calculating
the BrdU labeling indices, the number of mitotic figures was added
due to coarse staining of nuclei that were in the division stage
(Fig. 2B). The mean of the BrdU
labeling indices at 0, 24, 48 and 72 h post-PH for WT mice vs.
KLF10-KO mice was 0.60 vs. 0.87, 1.12 vs. 1.30, 72.85 vs. 59.45 and
40.1 vs. 26.56, respectively (Fig.
2C). Overall, KLF10-KO mice exhibited lower indices compared
with WT mice from 48 h post-PH, and a significant difference was
observed at 72 h post-PH (P=0.036).
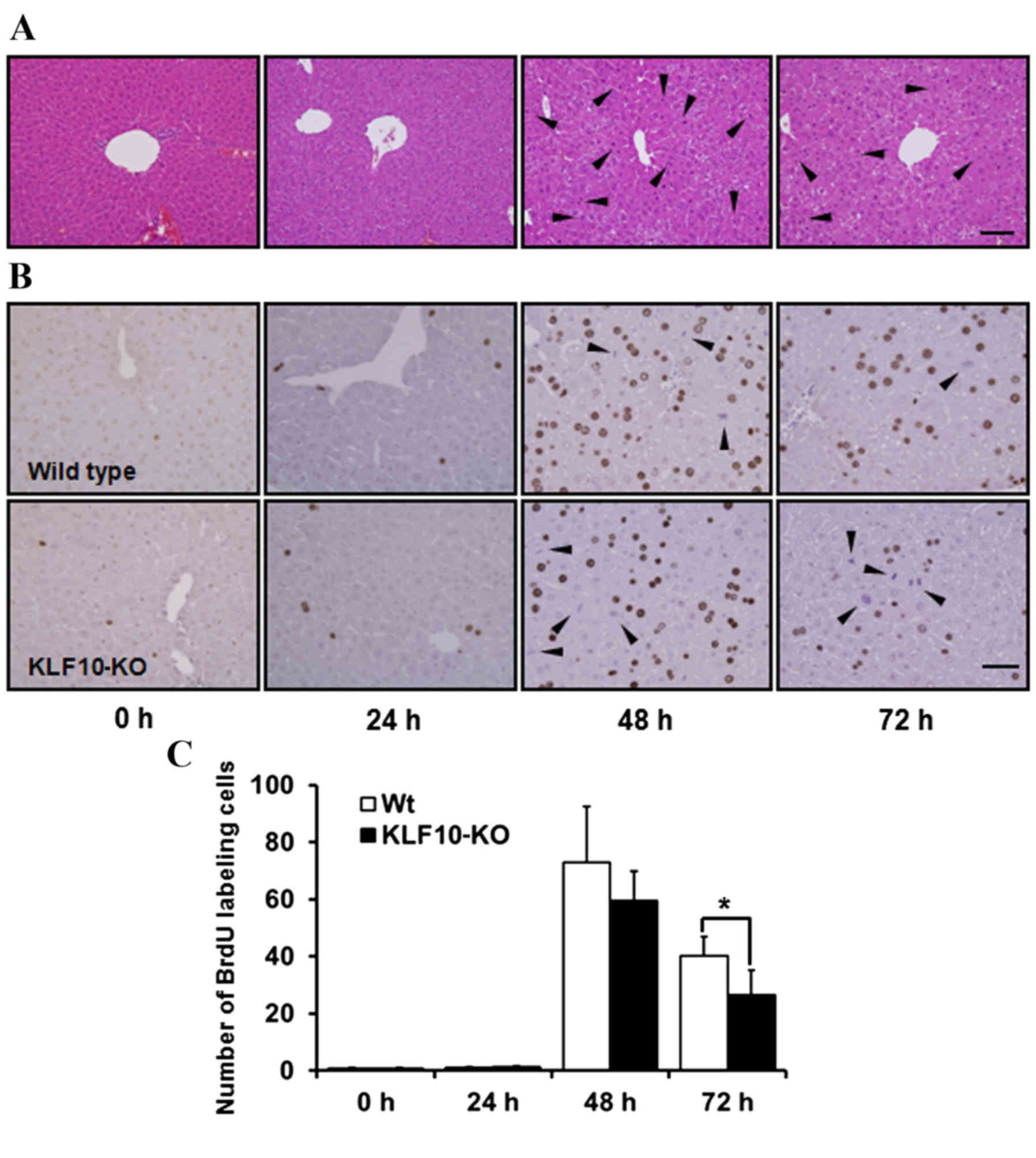 | Figure 2.Histopathological findings and BrdU
stains of the liver tissue following partial hepatectomy. (A)
Hematoxylin and eosin stains of 0, 24, 48 and 72 h post-PH. From 48
h post-PH, pale cytoplasm and hypertropy of hepatocytes was
observed. Mitotic figures, mainly in the centrilobular area, were
the most frequent at 48 h post-PH. Arrowheads indicate mitotic
figures. Bar, 100 µm. (B) Immunohistochemical staining of BrdU in
the liver tissue after PH. Nuclei of proliferating cells were
numerous in WT mice from 48 h post-PH. Mitotic cells were coarsely
stained (arrowheads). Bar, 50 µm. (C) BrdU indices of liver tissue.
BrdU-positive nuclei were counted in an area of 0.1 mm2
and the number of mitotic cells was added. KLF10-KO mice showed
lower indices compared with WT mice from 48 h post-PH, and there
was a significant difference at 72 h after PH. *P<0.05 by
unpaired Student's t-test. PH, partial hepatectomy; KLF10,
krüppel-like factor 10; KO, knockout; WT, wild-type; BrdU,
5-bromo-2-deoxy-uridine. |
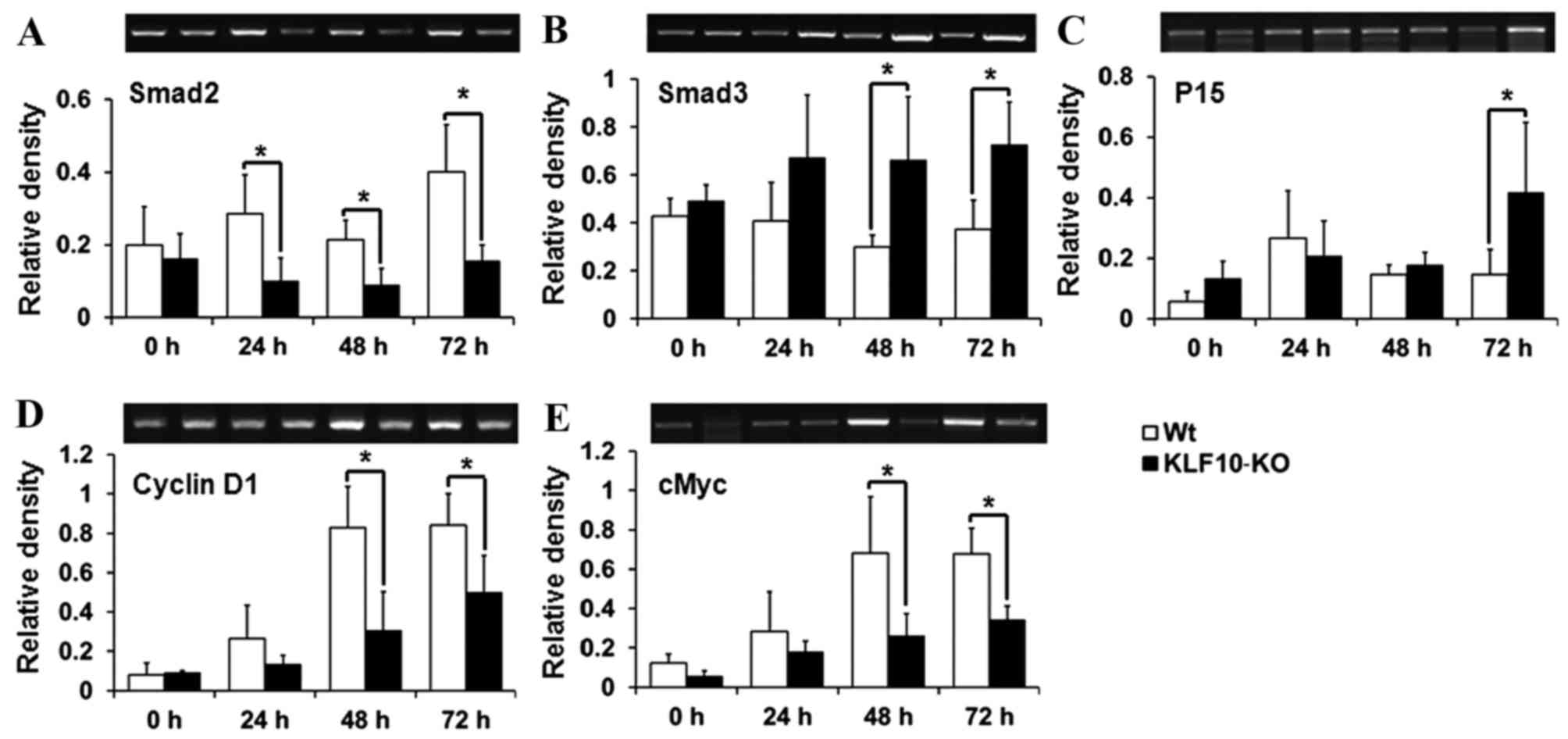
Expression of TGF-β/Smad pathway
genes
To determine the causes for the gross and
histological findings, the expression of the components of the
TGF-β/Smads signaling pathway, which is known to be regulated by
KLF10 (18), was analyzed. Smad2,
Smad7 and p21, which have previously been reported to be target
genes of KLF10 (10,17), and Smad3, Smad4, TGF-β1, TGF-βR1,
TGF-βR2, p15, p21, cMyc and cyclin D1, which are mediators or
target genes of the TGF-β/Smad pathway (14,15), were
analyzed by RT-qPCR. Smad2, one of the R-Smads, exhibited
significantly lower transcription levels in KLF10-KO mice compared
with WT mice from 24 h post-PH (24 h, P=0.034; 48 h, P=0.036; 72 h,
P=0.028; Fig. 3A). However, mRNA
levels of Smad3 (another R-Smad) were increased in KLF10-KO mice
compared with WT mice at all time points, and significant
differences were observed from 48 h post-PH (48 h, P=0.039; 72 h,
P=0.033; Fig. 3B). p15, a target gene
of the TGF-β/Smad pathway, exhibited significantly increased
expression in KLF10-KO mice compared with WT mice at 72 h post-PH
(P=0.049; Fig. 3C). By contrast, cMyc
and cyclin D1 genes, which are suppressed following activation of
the TGF-β/Smad pathway, exhibited significantly greater expression
in WT mice compared with KLF10-KO mice from 48 h post-PH (cMyc, 48
h, P=0.042; 72 h, P=0.011: cyclin D1, 48 h, P=0.036; 72 h, P=0.047;
Fig. 3D and E). No significant
differences were observed in the mRNA levels of other genes (data
not shown).
Expression of proteins involved in the
TGF-β/Smad pathway
The expression of proteins targeted by the TGF-β
pathway (Smad2, Smad3, Smad4, Smad7, p15, p21, p27, TGF-β1, TGF-β
R1, TGF-β R2 and cyclin D1) was then investigated. In RT-qPCR
analysis, significant differences were mostly observed at 48 and 72
h post-PH. The tissue lysates from these time points were analyzed
by western blotting. The results for Smad2, Smad3 and cyclin D1
were similar to those obtained by RT-qPCR. Smad2 expression was
significantly deacreased in KLF10-KO mice at 48 and 72 h post-PH
compared with that in WT mice (P=0.049 and P=0.043, respectively;
Fig. 4A). Protein levels of Smad3 and
p15 were increased in the KLF10-KO mice compared with those in the
WT mice, and significant differences were observed at 72 h post-PH
(P=0.019 and P=0.040, respectively; Fig.
4B and C). The cyclin D1 level was significantly lower at 48
and 72 h post-PH in the KLF10-KO mice compared with that in the WT
mice (P=0.045 and P=0.006, respectively; Fig. 4D). Additionally, the level of TGF-β1,
which is positively regulated by the TGF-β/Smad pathway (14), was increased in the KLF10-KO mice
compared with that in the WT mice, and significant differences were
observed at 72 h post-PH (P=0.012; Fig.
4E). Expression of TGF-β R1, which is involved in the early
steps of the TGF-β pathway (14), was
similar to that of TGF-β1 (Fig. 4F).
Other proteins did not show any significant differences (data not
shown).
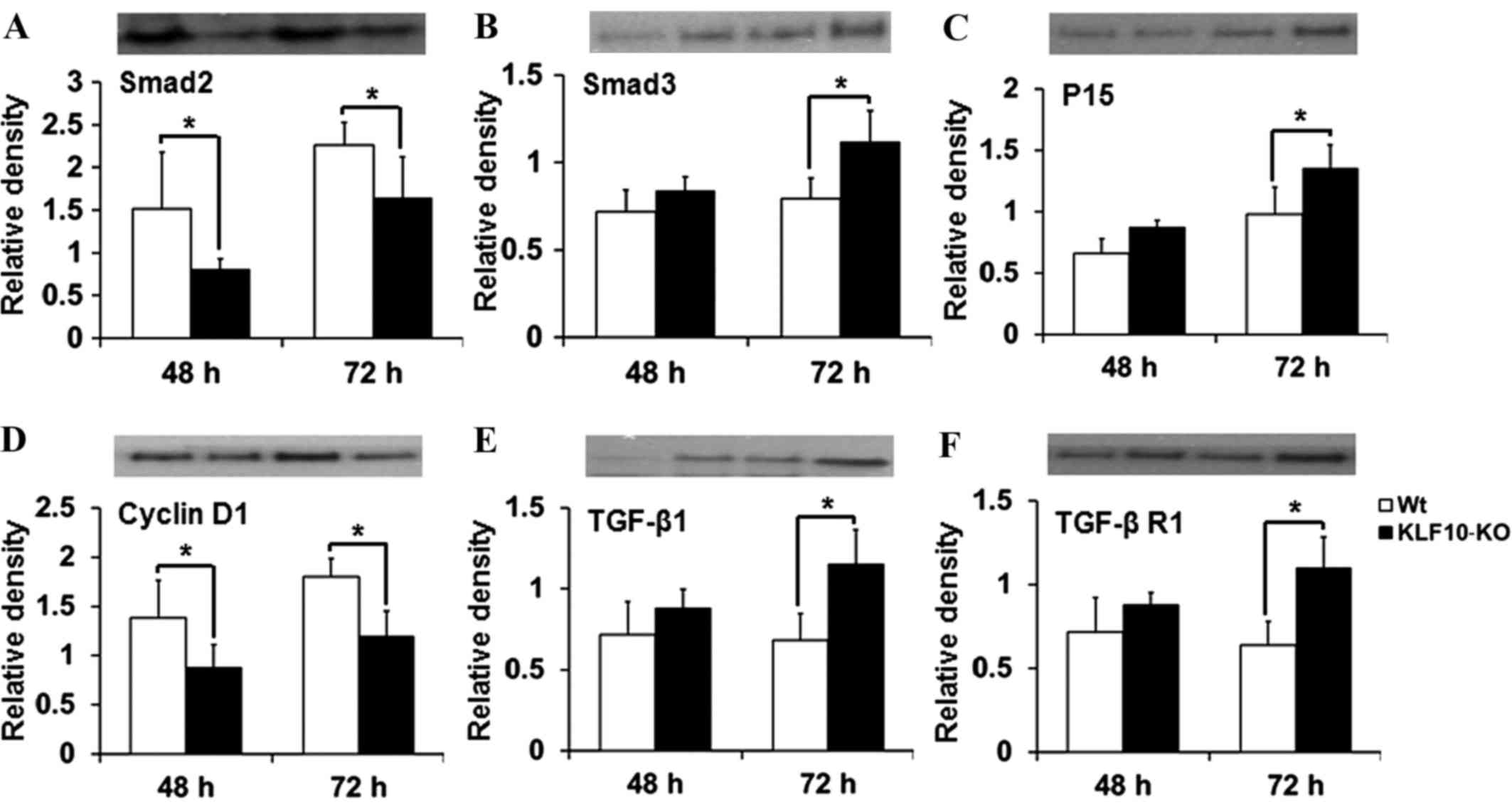 | Figure 4.Analysis of the levels of the proteins
(Smad2, Smad3, p15, cyclin D1, TGF-β1 and TGF-β R1) involved in the
TGF-β/Smad pathway. Considering reverse transcription-quantitative
polymerase chain reaction results, liver tissues at 48 and 72 h
post-partial hepatectomy were examined. (A) Smad2 levels were
lower, while (B) Smad3 and (C) p15 levels were higher in the
KLF10-KO mice compared with those in the WT mice. (D) Cyclin D1
levels were lower, while (E) TGF-β1 and (F) TGF-β R1 levels were
higher in the KLF10 KO mice compared with those in the WT mice.
Band intensities were quantified and normalized to the β-actin
level. *P<0.05 by unpaired Student's t-test. TGF-β1,
transforming growth factor-β1; TGF-β R1, transforming growth
factor-β receptor 1; KLF10, krüppel-like factor 10; KO, knockout;
WT, wild-type. |
Discussion
Numerous factors may alter liver mass, and a number
of signaling pathways are involved in liver regeneration (1,4,7). TGF-β is known to suppress cellular
proliferation, including hepatic regeneration (8,9). KLF10,
one of the target genes of TGF-β, enhances TGF-β-induced
anti-proliferative effects in certain cell types by inducing the
expression of several genes involved in TGF-β signaling, including
Smads, p15, p21 and TGF-β1 (24–26).
However, the exact function of KLF10 under various
pathophysiological conditions remains unclear, and to the best of
our knowledge, its role in liver regeneration has never been
examined.
In the present study, the role of KLF10 in liver
regeneration following tissue loss was investigated using KLF10-KO
mice. The liver/body weight ratio of the KLF10-KO mice was lower
than that of the WT mice at all examined time points, and a
significant difference was observed at 72 h post-PH (Fig. 1). KLF10-KO mice also exhibited lower
BrdU indices compared with WT mice from 48 h post-PH, and a
significant difference was observed at 72 h post-PH (Fig. 2C). Thus, cellular proliferation
appeared to be suppressed in KLF10-KO mice following PH (under
conditions that supported regeneration in WT mice). To determine
the causes for the decreased cellular proliferation in KLF10-KO
mice, the expression of genes involved in the TGF-β/Smad pathway
was examined by RT-qPCR. Under normal conditions and immediately
following PH, no difference was observed in the expression of the
examined genes. However, the mRNA levels of Smad2 decreased
significantly in KLF10-KO mice from 24 h post-PH (Fig. 3A). KLF10 has been reported to enhance
Smad2 expression (10), and this
decrease may be a cause for KLF10 gene ablation in the KO mice.
Unlike that of Smad2, the expression of Smad3 (a co-Smad) increased
from 48 h post-PH in the KLF10-KO mice (Fig. 3B). The expression of p15, whose
transcription is induced by the activated Smads complex, also
increased from 48 h post-PH in the KLF10-KO mice (Fig. 3C). Consequently, the cMyc and cyclin
D1 genes, which are downregulated by p15, showed significantly
lower expression in the KLF10-KO mice compared with the WT mice
from 48 h post-PH (Fig. 3D and E).
Western blot analysis results for Smad2, Smad3, p15 and cyclin D1
were similar to those of RT-qPCR. In addition, the levels of TGF-β1
and TGF-β R1, which are involved in the early phases of the TGF-β
pathway, were significantly increased at 72 h post-PH in the
KLF10-KO mice compared with the WT mice (Fig. 4).
These results indicated that the knockout of KLF10
led to repression of the transcription and function of the
proliferation-associated genes cMyc and cyclin D1 from 48 h
post-PH, thereby inhibiting liver regeneration. This repression may
be induced by the activation of the TGF-β/Smad pathway. Expression
of TGF-β R1, TGF-β1, Smad3 and p15, one of the genes positively
regulated by the TGF-β pathway, was increased in the KLF10-KO mice.
Thus, during liver regeneration following PH, lack of KLF10
suppresses the proliferation of hepatocytes by activation of the
TGF-β/Smad pathway, and the suppression of hepatocellular
proliferation can be observed in a delayed manner at 72 h
post-PH.
Cellular responses to TGF-β depend on the cell
types, the microenvironment and the tissue type (9,27). KLF10,
an early target of TGF-β, has been reported to exert effects
similar to those of TGF-β. However, the specific functions of KLF10
under various physiological conditions are not well characterized
(18,26,28,29). Our
previous study reported the tumor-suppressor effects against
chemically-induced liver tumorigenesis in KLF10-KO mice (12). Similarly, the anti-proliferative
effects on hepatocytes after PH were shown in the present study.
These results show that although there were no phenotypical changes
in the liver of KLF10-KO mice under normal physiological
conditions, the TGF-β/Smad pathway exhibited enhanced activation
and cellular proliferation was decreased under conditions that
promote cellular proliferative changes in the liver, including PH
and chemically-induced tumorigenesis.
The aforementioned results may be due to KLF10
transcription being induced by factors other than TGF-β, including
estrogen and epidermal growth factor (8,14,30,31).
Furthermore, certain intracellular proteins, including KLF11, were
recently reported to exhibit effects similar to those of KLF10
(26,32). Considering these findings, certain
compensatory mechanisms for KLF10 ablation may exist in non-Smad
TGF-β pathways or KLF10 signaling under proliferative conditions.
The detailed molecular mechanism underlying this compensatory
mechanism warrants additional study.
In conclusion, the hepatocytes of KLF10-KO mice
exhibited less proliferation compared with those of WT mice
following PH in the present study. This decrease was caused by
reinforcement of TGF-β/Smad signaling. TGF-β1, TGF-β R1 and Smad3
were upregulated, and this induced increased expression of p15.
These factors led to repressed expression of the
proliferation-associated genes, cMyc and cyclin D1. The present
findings demonstrated that knockout of KLF10 suppressed cellular
proliferation through reinforcement of the TGF-β/Smad pathway in
the presence of external stimuli that could induce hepatocyte
proliferation, including tumorigenesis and regeneration, following
tissue loss (12).
Acknowledgements
The present study was supported by the Konkuk
University in 2013.
References
|
1
|
Fausto N, Campbell JS and Riehle KJ: Liver
regeneration. J Hepatol. 57:692–694. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fausto N: Liver regeneration. J Hepatol.
32 1 Suppl:S19–S31. 2000. View Article : Google Scholar
|
|
3
|
Michalopoulos GK: Liver regeneration. J
Cell Physiol. 213:286–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Michalopoulos GK: Liver regeneration after
partial hepatectomy: Critical analysis of mechanistic dilemmas. Am
J Pathol. 176:2–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing and treating hepatocellular carcinoma. CA
Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mathers CD, Boerma T and Ma Fat D: Global
and regional causes of death. Br Med Bull. 92:7–32. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taub R: Liver regeneration: From myth to
mechanism. Nat Rev Mol Cell Biol. 5:836–847. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Itoh S and ten Dijke P: Negative
regulation of TGFbeta receptor/Smad signal transduction. Curr Opin
Cell Biol. 19:176–184. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Subramaniam M, Hawse JR, Rajamannan NM,
Ingle JN and Spelsberg TC: Functional role of KLF10 in multiple
disease processes. Biofactors. 36:8–18. 2010.PubMed/NCBI
|
|
11
|
Ellenrieder V: TGFbeta-regulated gene
expression by smads and Sp1/KLF-like transcription factors in
cancer. Anticancer Res. 28:1531–1539. 2008.PubMed/NCBI
|
|
12
|
Heo SH, Jeong ES, Lee KS, Seo JH, Lee WK
and Choi YK: Krüppel-like factor 10 null mice exhibit lower tumor
incidence and suppressed cellular proliferation activity following
chemically induced liver tumorigenesis. Oncol Rep. 33:2037–2044.
2015.PubMed/NCBI
|
|
13
|
Inman GJ: Switching TGFβ from a tumor
suppressor to a tumor promoter. Curr Opin Genet Dev. 21:93–99.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Massagué J: TGFbeta in Cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kaminska B, Wesolowska A and Danilkiewicz
M: TGF beta signalling and its role in tumour pathogenesis. Acta
Biochim Pol. 52:329–337. 2005.PubMed/NCBI
|
|
16
|
Kang JS, Liu C and Derynck R: New
regulatory mechanisms of TGF-beta receptor function. Trends Cell
Biol. 19:385–394. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Johnsen SA, Subramaniam M, Monroe DG,
Janknecht R and Spelsberg TC: Modulation of transforming growth
factor beta (TGF-beta)/Smad transcriptional responses through
targeted degradation of TGF-beta-inducible early gene-1 by human
seven in absentia homologue. J Biol Chem. 277:30754–30759. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Subramaniam M, Hawse JR, Johnsen SA and
Spelsberg TC: Role of TIEG1 in biological processes and disease
states. J Cell Biochem. 102:539–548. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Martins PN, Theruvath TP and Neuhaus P:
Rodent models of partial hepatectomies. Liver Int. 28:3–11. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mitchell C and Willenbring H: A
reproducible and well-tolerated method for 2/3 partial hepatectomy
in mice. Nat Protoc. 3:1167–201170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Breitkopf K, Weng H and Dooley S:
TGF-β/Smad-signaling in liver cells: Target genes and inhibitors of
two parallel pathways. Signal Transduction. 6:329–337. 2006.
View Article : Google Scholar
|
|
22
|
Song KD, Kim DJ, Lee JE, Yun CH and Lee
WK: KLF10, transforming growth factor-β-inducible early gene 1,
acts as a tumor suppressor. Biochem Biophys Res Commun.
419:388–394. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shiao YH: A new reverse
transcription-polymerase chain reaction method for accurate
quantification. BMC Biotechnol. 3:222003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Buenemann CL, Willy C, Buchmann A,
Schmiechen A and Schwarz M: Transforming growth
factor-beta1-induced Smad signaling, cell-cycle arrest and
apoptosis in hepatoma cells. Carcinogenesis. 22:447–452. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dooley S, Weng H and Mertens PR:
Hypotheses on the role of transforming growth factor-beta in the
onset and progression of hepatocellular carcinoma. Dig Dis.
27:93–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Spittau B and Krieglstein K: Klf10 and
Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue
Res. 347:65–72. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heldin CH and Miyazono K: Transforming
growth factor-beta. An interesting candidate for clinical use.
Lakartidningen. 92:1569–1572. 1995.(In Swedish). PubMed/NCBI
|
|
28
|
Bensamoun SF, Hawse JR, Subramaniam M,
Ilharreborde B, Bassillais A, Benhamou CL, Fraser DG, Oursler MJ,
Amadio PC, An KN and Spelsberg TC: TGF-beta inducible early gene-1
knockout mice display defects in bone strength and
microarchitecture. Bone. 39:1244–1251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bos JM, Subramaniam M, Hawse JR,
Christiaans I, Rajamannan NM, Maleszewski JJ, Edwards WD, Wilde AA,
Spelsberg TC and Ackerman MJ: TGFβ-inducible early gene-1 (TIEG1)
mutations in hypertrophic cardiomyopathy. J Cell Biochem.
113:1896–1903. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moustakas A and Heldin CH: Non-Smad
TGF-beta signals. J Cell Sci. 118:3573–3584. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsubone T, Moran SL, Subramaniam M, Amadio
PC, Spelsberg TC and An KN: Effect of TGF-beta inducible early gene
deficiency on flexor tendon healing. J Orthop Res. 24:569–575.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gohla G, Krieglstein K and Spittau B:
Tieg3/Klf11 induces apoptosis in OLI-neu cells and enhances the
TGF-beta signaling pathway by transcriptional repression of Smad7.
J Cell Biochem. 104:850–861. 2008. View Article : Google Scholar : PubMed/NCBI
|