Introduction
Breast cancer is the most common disease in women
worldwide. Cancer cells that have developed resistance to
chemotherapeutic agents are major clinical obstacles in the
successful treatment of breast cancer (1). When treated with one chemotherapeutic
agent, patients can develop either intrinsical or acquired
resistance to other drugs during the course of treatment (2). Chemotherapy drug resistance involves
various mechanisms, including overexpression of adenosine
triphosphate-binding cassette (ABC) transport proteins, changes in
the expression of glutathione, glutathione transferase and
topoisomerase, the enhancement of cancer cell DNA damage repair
mechanisms and programmed cell death pathway defects (3–6). At
present, acquired drug resistance and metastasis prevent breast
cancer from being successfully treated in every case (7). It is necessary to improve the clinical
management of cancer by increasing the sensitivity of tumor cells
to chemotherapeutic agents and via the prediction of the
effectiveness of chemotherapeutic agents, all without drug
resistance developing in individual patients.
ABC transporters contribute to the development of
the resistance of cancer cells to anticancer drugs via
ATP-dependent drug efflux (1). Drug
efflux in tumor cells is most frequently associated with
overexpression of one or more membrane-bound ABC transporters
(3). ABC sub-family B member 1
(ABCB1/P-gp) overexpression has been associated with poor drug
response in breast cancer patients (8). ATP-binding cassette sub-family C member
1 (MRP1/ABCC1) is involved in the multidrug resistance (MDR) of a
variety of solid tumors (9). ABC
sub-family G member 2 (ABCG2) encoding BCRP also belongs to the
frequently studied ABC transporters (10). However, the role of other ABC genes in
MDR is reported far less. Regulation of drug efflux is a key
mechanism involeved in drug resistance. The results of the study by
Pogribny et al (11)
determinded that cisplatin-resistance cells upregulated MRP1 when
compared with sensitive MCF-7 cells.
The eukaryotic initiation factor (eIF) 4F complex
consists of three proteins: cap-binding protein eIF4E, scaffolding
protein eIF4G and ATP-dependent RNA helicase eIF4A (12,13). All
three proteins converge to modulate the translation of specific
mRNAs. Generally, 4EBP1 inhibits the downstream mTOR pathway
through binding to eIF4E. Phosphorylation of 4EBP1 by mTOR results
in its dissociation from eIF4E and activation of cap-dependent mRNA
translation (14). The increased
amount of 4EBP1-bound eIF4E concomitantly decreases the amount of
eIF4G-bound eIF4E and vice versa. The correct functioning of
cellular processes, including drug resistance, is regulated by
controling gene expression at the mRNA translational level
(15,16). Recently, eIF4F complex formation was
found to be reduced in tumors responsive to anti-BRAF therapy, but
increased in resistant metastases, compared with tumors prior to
treatment. B-cell lymphoma-2 modifying factor (BMF) is a
pro-apoptotic gene that has previously been demonstrated to be
involved in the acquired resistance to PLX4720, a vemurafenib
analogue. BMF has also demonstrated involvment in the sensitivity
to vemurafenib by acting on the cleavage of eIF4G, which
consequently affects the formation of the eIF4F complex (17). This phenomenon is similar to the drug
resistance of breast cancer cells caused by high expression of the
ABC transporter family. Although drug resistance can be reversed by
disrupting the eIF4F complex, the association between eIF4F and ABC
transporters is not clear.
The present study investigated the potential role of
eIF4G in the resistance of MCF-7/ADR cells to anticancer drugs, and
unravelled the possible association between eIF4G and ABC
transporters.
Materials and methods
Cells and reagents
The human breast cancer MCF-7 cell line, maintained
at 37°C with 5% CO2 in a humidified atmosphere and grown
in Dulbecco's modified Eagle's medium (HyClone Laboratories; GE
Healthcare, Logan, UT, USA) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), was a gift from the Institute of Pharmacy and Pharmacology,
University of South China (Hengyang, Hunan, China). The MCF-7
Adriamycin (ADM)-resistant (MCF-7/ADR) cell line was alternately
fed with medium containing 1 µg/ml−1 ADM (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) and RPMI-1640 medium supplemented
with 10% FBS, and was regularly tested for maintenance of
drug-resistaince. The cells were treated with ADM (Sigma-Aldrich;
Merck KGaA), tamoxifen (TAM; Sigma-Aldrich; Merck KGaA) and taxol
(TAX; Sigma-Aldrich; Merck KGaA). All drugs were dissolved in
dimethyl sulfoxide (DMSO) for in vitro studies.
Target prediction
Several online Bioinformatics and Research software
programs, TargetScan (http://www.targetscan.org), miRanda (http://www.microrna.org/), Pictar (https://www.mdc-berlin.de/10440258/en/research/researchteams/systemsbiologyofgeneregulatoryelements/projects/pictar)
and miRBase (http://www.mirbase.org/search.shtml) were used to
predict conserved miRNA binding sites in the 3′-untranslated region
(3′UTR) of human eIF4G.
Transient transfection
miR-503 mimic, miRNA mimic control and miR-503
inhibitor (all GenePharma, Shanghai, China) marked with
carboxyfluorescein (FAM) in vitro were transfected into the
cells at a final concentration of 160 nM using Lipofectamine 2,000
(Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocols. After 6 h, fluorescence
microscopy was used to detect the percent of fluorescent cells.
After 48 h, western blot analysis was performed.
Western blot analysis
Western blot analysis was performed on cell extracts
of the indicated cell lines that had been transfected for 48 h with
the miRNA sequences. Immunoblots were performed from whole cell
lysate prepared using RIPA Buffer supplemented with PMSF, and
phosphatase inhibitors. Cell lysates were quantified for protein
content using a bicinchoninic acid protein assay kit. SDS PAGE was
used to separate the proteins, with 30 µg total protein per lane on
5% SDS-PAGE. Protein samples were transferred to 0.22-mm polyvinyl
difluoride membranes. After saturation in Tris-buffered saline with
Tween-20 and 5% powdered milk, the membranes were incubated with
anti-eIF4G (cat. no., 2617S; dilution, 1:1,000) and anti-β-actin
(cat. no., 8457S; dilution, 1:1,000) antibodies purchased from Cell
Signaling Technology, ABCB1 (cat. no., ab170904; dilution,
1:1,000), ABCC1 (cat. no., ab3368; dilution, 1:1,000) and ABCG2
(cat. no., ab108312; dilution, 1:1,000) purchased from Abcam,
overnight at 4°C. The membranes were then incubated with
horseradish peroxidase-conjugated secondary antibodies (cat. no.,
sc-2357; dilution, 1:5,000 Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) for 1 h at room temperature. The signals were visualized
using ECL Substrates (Tanon Ltd, shanghai, China) and quantified
using AlphaImager 2200 software.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells using the TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) chloroform extraction
method. The cDNA was prepared from 1 µg total RNA using reverse
transcriptase (Promega Corporation, Madison, WI, USA). Total
cellular RNA was quantified by qPCR analysis using gene-specific
primer pairs. RT-qPCR was performed by the SYBR-Green qPCR Master
Mix (Tiangen Biotech Co., Ltd., Beijing, China) on an ABI 7300 PCR
machine (Applied Biosystems, Inc., Foster, CA, USA). qPCR cycling
conditions were as follows: 95°C for 5 min, and then 95°C for 15
sec, 65°C for 15 sec and 72°C for 32 sec, for 40 cycles. The
melting curve was 60–95°C. The relative mRNA expression levels were
calculated as 2−∆∆Cq method (18). Oligonucleotides used for the analysis
were as follows: ABCB1 forward, 5′-CTAGAAGGTTCTGGGAAGAT and
reverse, 5′-GAGTTTCTGTATGGTACCTG; ABCC1 forward,
5′-GGCTCAAGGAGTATTCAGAG and reverse, 5′-CCATCGATGATGATCTCTCC; EIF4G
forward, 5′-CTCTGAGACCGTTGGGCAAA and reverse,
5′-GCGACGAATGCACCAATGT; ABCG2 forward, 5′-GCAGGTCAGAGTGTGGTTTC and
reverse, 5′-GACAGCCAAGATGCAATGGT; and 18S ribosomal RNA forward,
5′-CCTGGATACCGCAGCTAGGA and reverse, 5′-GCGGCGCAATACGAATGCCCC. Each
reaction was performed in triplicate and three independent
experiments were run.
MTT
Cells in the exponential growth phase were seeded in
96-well plates at a density of 1×104 per well. After
incubation for 24 h at 37°C with 5% CO2 in a humidified
atmosphere, the cells were transfected with miR-503 mimic, miR-503
inhibitor or miRNA mimic control (NC). At 24 h post-transfection,
various concentrations of ADM (0, 0.625, 12.5, 25, 50 and 100 µM),
TAX (0, 0.3125, 0.625, 1.25, 2.5 and 5 µM) and TAM (0, 3.75, 7.5,
15, 30 and 60 µM) were added. After an incubation period of 24 h,
20 µl MTT solutions with a concentration of 5 mg/ml were added to
each well for 4 h at 37°C. Next, the culture medium (medium
containing drug and MTT) was removed, and the insoluble formazan
crystals were dissolved in 150 ml DMSO. After shaking for 10 min,
the absorbance at 570 nm was optically monitored using a microplate
reader.
Luciferase reporter assay
The wild-type or mutant eIF4G 3′UTR-luciferase
reporter plasmids (Landbiology, Guangzhou, China) were
co-transfected with miR-503 mimics, miR-503 control (NC), miR-503
inhibitor and inhibitor control (NC inhibitor) into the MCF-7 and
MCF-7/ADR cells. At 48 h post-transfection, the cells were assayed
for luciferase activity using the Dual Luciferase assay system
(Promega Corporation) according to the manufacturer's instructions.
The Renilla luciferase activities were normalized to corresponding
firefly luciferase activities. For each transfection, the
luciferase activity was averaged from three replicates.
Assessment of cell cycle and
apoptosis
At 48 h post-transfection, the cells were washed
with phosphate-buffered saline and fixed with 70% ethanol
overnight. The cells were then stained with 50 µg/ml propidium
iodide, 100 µg/ml RNase A and 0.2% Triton X-100 (Nanjing KeyGen
Biotech, Co., Ltd.; KGA511), and kept out of the light for 30 min,
prior to undergoing cell cycle analysis using a flow cytometer. The
percentage of apoptotic cells was determined by flow cytometry (BD
calibur, USA) with the Annexin V staining kit (cat. no., KGA106;
Nanjing KeyGen Biotech, Co., Ltd.).
Statistical analysis
All experiments were performed at least three times
(n=3). Statistical analyses used the two-tailed Student's t-test,
with P<0.05 used to indicate a statistically significant
difference. Statistical analysis was performed using GraphPad Prism
5.00 (GraphPad, San Diego, CA, USA).
Results
ABCB1, ABCC1 and ABCG2 were
overexpressed in MCF-7/ADR cells
To analyze the alterations in ABCB1, ABCC1 and ABCG2
during the formation of breast cancer cellular drug resistance, the
MCF-7/ADR cell line, an ADM-resistant variant, was utilized and
confirmed to be more resistant compared with the MCF-7 cells
treated with ADM, TAM and TAX (Fig.
1A). It was identified that the ATP-dependent efflux genes
(ABCB1, ABCC1 and ABCG2) exhibited higher protein and mRNA levels
in the MCF-7/ADR cells compared to the parental MCF-7 cells
(Fig. 1B and C).
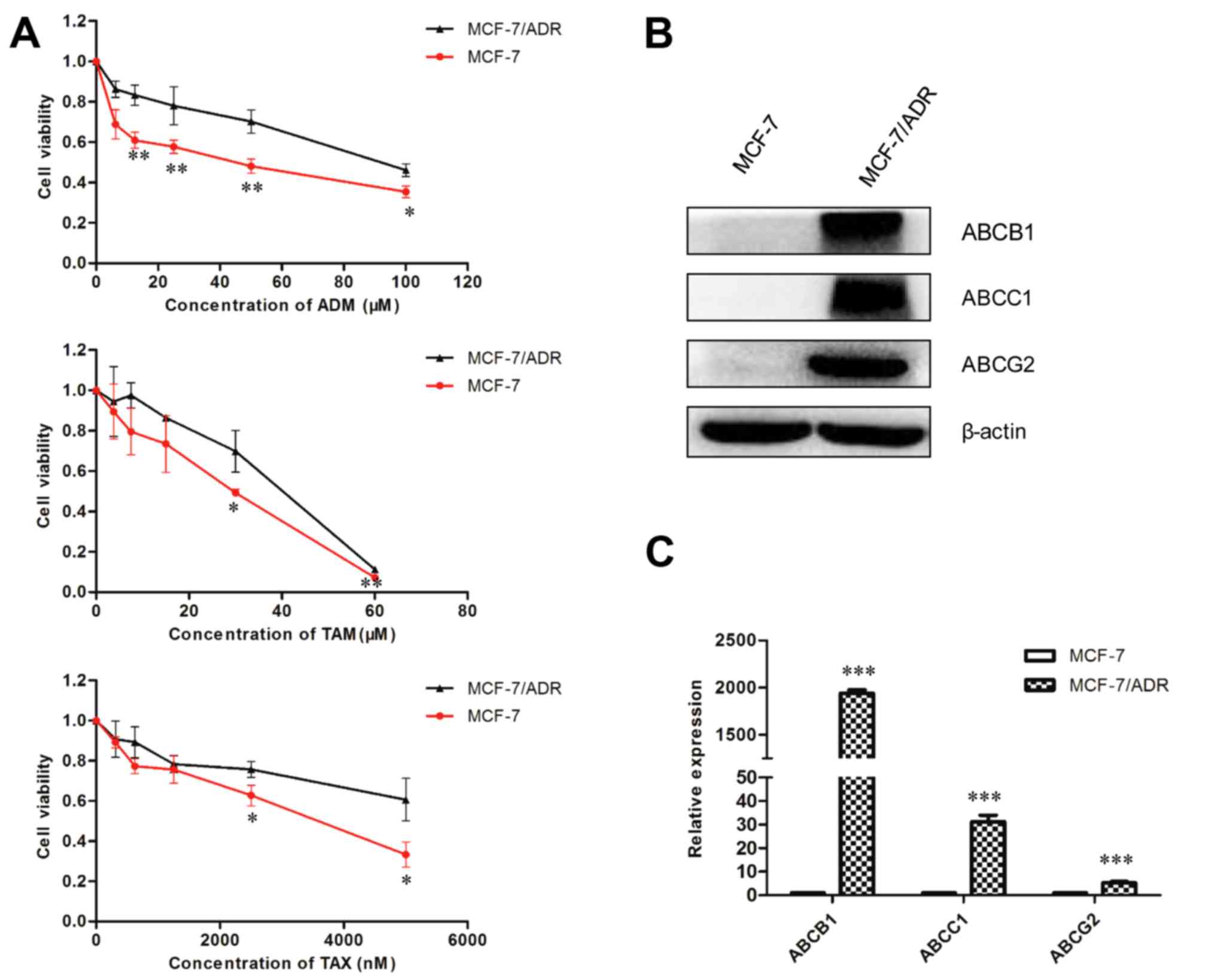 | Figure 1.Upregulation of ABCB1, ABCC1 and
ABCG2, and alterations in ADM, TAM and TAX sensitivity in MCF-7/ADR
cells. (A) MTT analyses showed that MCF-7 cells were sensitive to
ADM, TAM and TAX compared with the MCF-7/ADR cells (*P<0.05,
**P<0.01). (B) Western blotting analysis of ABCB1, ABCC1 and
ABCG2 in the indicated cell lines. β-actin was used as a loading
control. (C) Expression levels of ABCB1, ABCC1 and ABCG2 mRNA in
the MCF-7/ADR cells, as determined by reverse
transcription-quantitative polymerase chain reaction, were
upregulated in comparison with the MCF-7 cells (***P<0.001).
ABCB1, ABC sub-family B member 1; ABCC1, ABC sub-family C member 1;
ABCG2, ABC sub-family G member 2; ADM, Adriamycin; TAM, tamoxifen;
TAX, taxol. |
3′UTR of eIF4G gene contains conserved
miRNA binding sites for miR-503
miR-503 was predicted to bind the 3′UTR of human
eIF4G by bioinformatics and research computing programs, including
TargetScan, Pictar and miRanda (Fig.
2).
Putative miRNAs binding sites for
miR-503 were confirmed by luciferase reporter assays
To validate whether the 3′UTR of human eIF4G could
be recognized by miR-503, a luciferase reporter assay was utilized.
The luciferase activity in the cells transfected with the vector
containing the eIF4G 3′UTR fragment with the binding sequence of
miR-503 was inhibited by miR-503 transfection compared with the
activity of the cells that were transfected with the NC miRNA
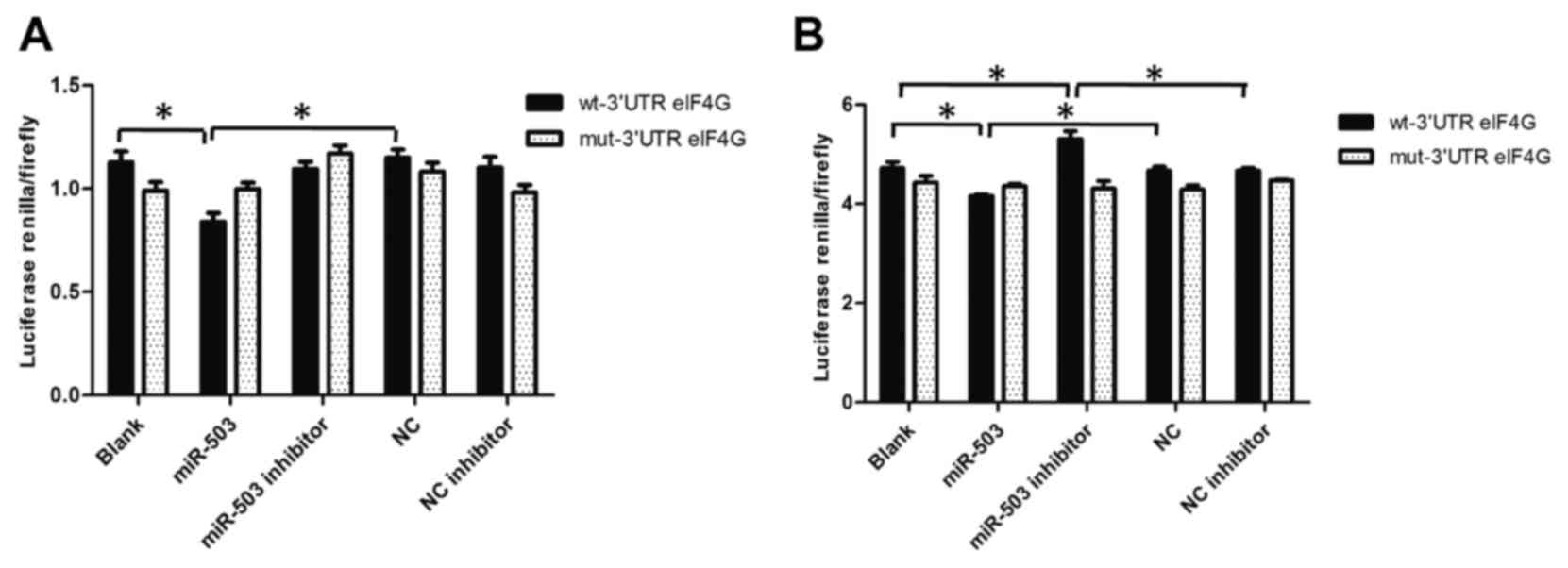
mimic, miR-503 inhibitor and the blank control (Fig. 3). From the results (Fig. 3A) in the MCF-7 group, it is apparent
that the 3′UTR eIF4G wild-type inhibition rate was 0.75 and that
the mut-3′UTR eIF4G inhibition rate was 1.01. Fig. 3B demonstrates that in MCF-7/ADR group,
the 3′UTR eIF4G wild-type inhibition rate was 0.88 and the
mut-3′UTR eIF4G inhibition rate was 0.98, suggesting that the
wild-type-3′UTR eIF4G serves a more important role in the binding
association than the mut-3′UTR eIF4G.
Elevated levels of miR-133 decreased
eIF4G mRNA expression
The MCF-7/ADR cells were transfected with
Lipofectamine 2000 (mock), miRNA control (NC), miR-503 and miR-503
inhibitor marked with FAM in vitro (Fig. 4A). After 48 h, fluorescence microscopy
was used to detect the transfection efficiency. The data (Fig. 4B) showed that miR-503 did not
significantly affect the expression of ABCB1, ABCC1 or ABCG2 mRNA,
but that it significantly downregulated the expression of eIF4G.
These results demonstrated that ABCB1, ABCC1 and ABCG2 were not the
target genes of miR-503, but that miR-503 could downregulate eIF4G
mRNA expression.
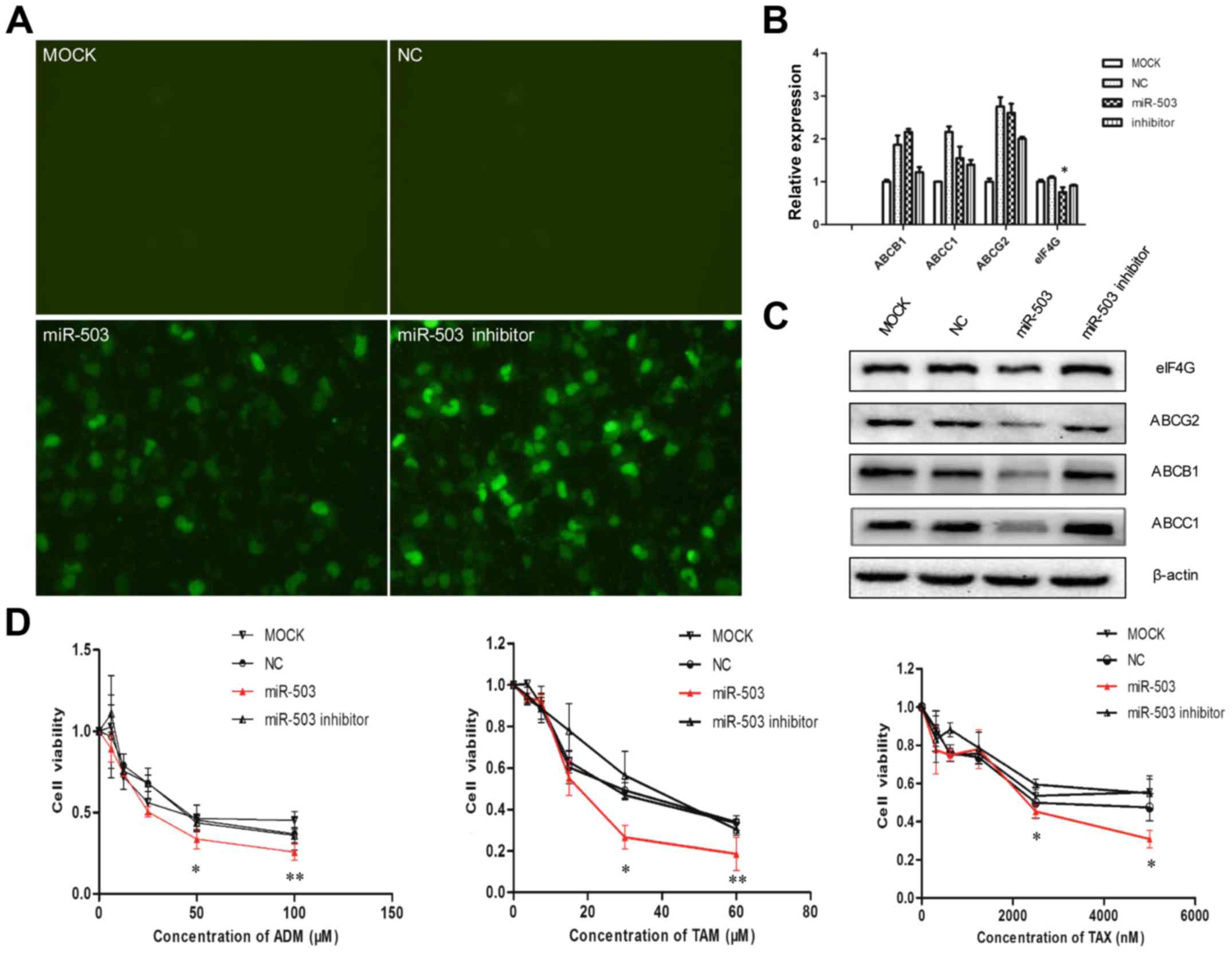 | Figure 4.miR-503 negatively regulates the
expression of ABCB1, ABCC1 and ABCG2, and increases MCF-7/ADR
sensitivity to ADM, TAX and TAM. (A) After 48 h, fluorescence
microscopy was used to detect transfection efficiency, and the
transfection efficiency of the miR-503 and miR-503 inhibitor was
>35%. (B) The expression of eIF4G mRNA was downregulated, while
the expression levels of ABCB1, ABCC1 and ABCG2 mRNA in the miR-503
mimic-transfected MCF-7/ADR cells were not changed in comparison
with the control cells and untreated cells, as determined by
reverse transcription-quantitative polymerase chain reaction
(**P<0.01 vs. negative microRNA or mock group). (C) miR-503
negatively regulated the expression of eIF4G, ABCB1, ABCC1 and
ABCG2, as determined by western blot assay. (D) miR-503 inhibited
drug resistance, as determined by MTT assay (*P<0.05,
**P<0.01). (E) Cell cycle distributions were analyzed with flow
cytometry, MCF-7/ADR cells transfected with miR-503 were arrested
in G0/G1 phases. (F) miR-503 increased the percentage of apoptosis
cells treated with anticancer drugs (20 µM ADM, 10 µM TAM and 5 µM
TAX). ABCB1, ABC sub-family B member 1; ABCC1, ABC sub-family C
member 1; ABCG2, ABC sub-family G member 2; ADM, Adriamycin; TAM,
tamoxifen; TAX, taxol; miR, microRNA; eIF4G, eukaryotic translation
initiation factor 4-γ 1; NC, negative control. |
miR-503 downregulates the expression
of eIF4 G, ABCB1, ABCC1 and ABCG2 proteins, and increases MCF-7/ADR
sensitivity to ADM, TAM and TAX
The present study assessed the function of miR-503
in human breast cancer, with a particular focus on MCF-7/ADR cells.
It was revealed that miR-503 may cause the downregulation of eIF4G,
ABCB1, ABCC1 and ABCG2 (Fig. 4C), and
the sensitivity of the MCF-7/ADR cells to ADM, TAM and TAX was
demonstrated to be increased by miR-503 (Fig. 4D). In addition, miR-503 was identified
to be able to block the cell cycle at G0/G1
(Fig. 4E) and lead to the occurrence
of apoptosis in MCF-7/ADR cells (Fig.
4F). These results suggested that miR-503 may decrease the
chemoresistance of breast cancer cells by the downregulation of
eIF4G, ABCB1, ABCC1 and ABCG2.
eIF4G siRNA negatively regulates the
expression of ABCB1, ABCC1 and ABCG2, and increases MCF-7/ADR cell
sensitivity to ADM, TAM and TAX
In transient cells transfected with eIF4G siRNA,
after 6 h the transfection efficiency was >35%, as detected by
fluorescence microscopy (Fig. 5A).
Using RT-qPCR, the difference in eIF4G expression in the MCF-7/ADR
cells transfected with siR-eIF4G was examined and was revealed to
be significantly downregulated in comparison with that of the
control cells and untreated cells (Fig.
5C). eIF4G siRNA negatively regulated the protein expression of
ABCB1, ABCC1 and ABCG2, as determined by western blot assay
(Fig. 5B), but exhibited no effect on
the mRNA expression (Fig. 5C). The
results (Fig. 5D) demonstrated that
eIF4G siRNA reduced cell viability compared with siRNA NC or
untransfected cultures, as assessed by MTT assay. In addition,
eIF4G siRNA was identified to be able to block the cell cycle at
G0/G1 (Fig. 5E)
and lead to the occurrence of apoptosis in MCF-7/ADR cells
(Fig. 5F).
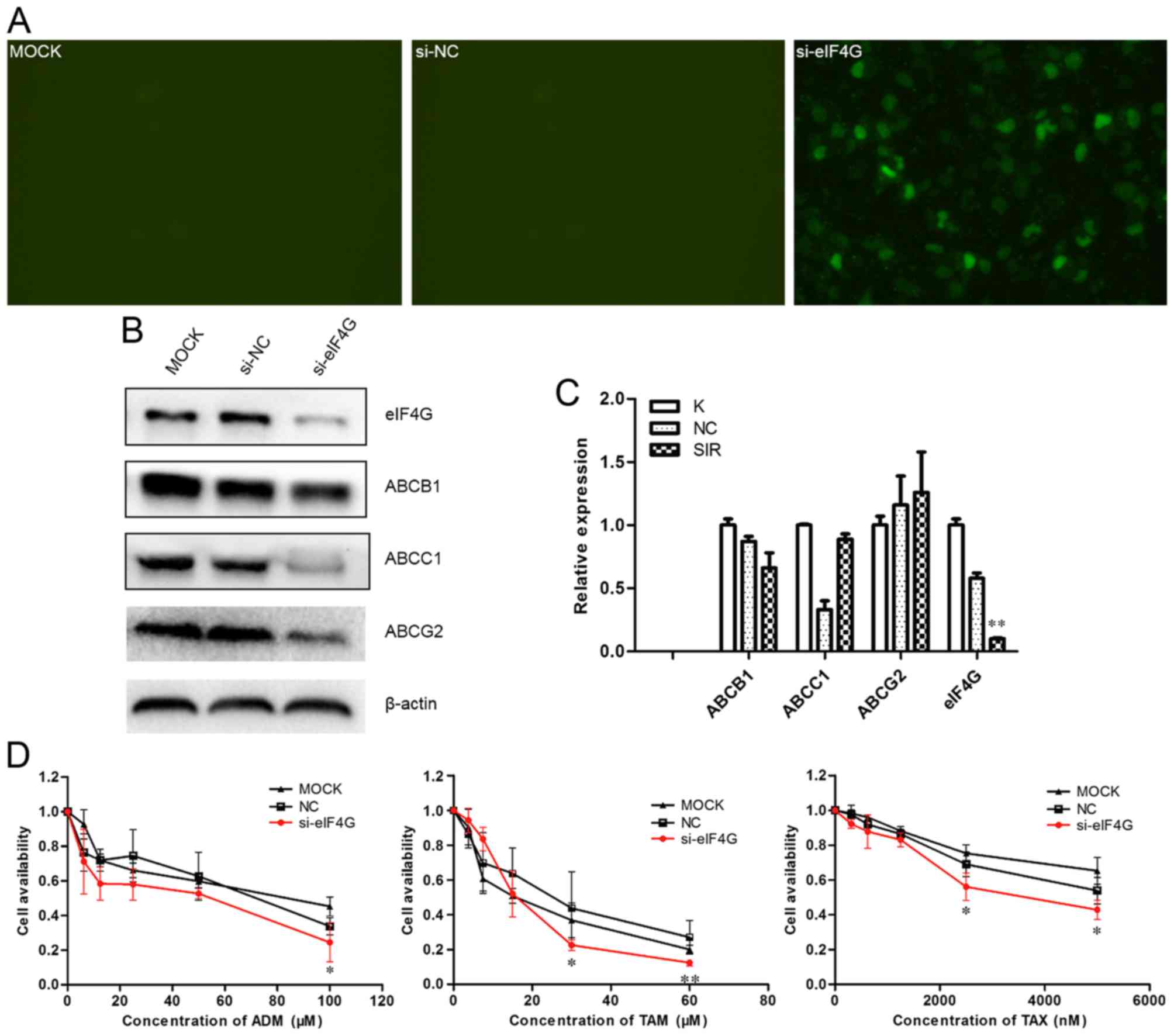 | Figure 5.eIF4G siRNA negatively regulated the
expression of ABCB1, ABCC1 and ABCG2, and increased MCF-7/ADR
sensitivity to ADM, TAX and TAM. (A) Transfection efficiency of
eIF4G siRNA and NC siRNA. After 48 h, fluorescence microscopy was
used to detect transfection efficiency, and the transfection
efficiency was >35%. (B) eIF4G siRNA negatively regulated the
expression of ABCB1, ABCC1 and ABCG2, as assessed by western blot
assay. (C) The effect of eIF4G siRNA on mRNA. (D) eIF4G siRNA
reduced cell viability compared with siRNA NC or untransfected
cultures, as determined by MTT assay (*P<0.05, **P<0.01). (E)
Cell cycle distributions were analyzed by flow cytometry, and
MCF-7/ADR cells transfected with eIF4G siRNA were arrested in G0/G1
phases. (F) eIF4G siRNA increased the percentage of apoptotic cells
treated with anticancer drugs (20 µM ADM, 10 µM TAM and 5 µM TAX).
eIF4G, eukaryotic translation initiation factor 4-γ 1; siRNA, small
interfering RNA; ABCB1, ABC sub-family B member 1; ABCC1, ABC
sub-family C member 1; ABCG2, ABC sub-family G member 2; ADM,
Adriamycin; TAM, tamoxifen; TAX, taxol; NC, negative control. |
Discussion
Several studies previously reported that ABCB1,
ABCC1 and ABCG2 were upregulated in pre- and post-treatment breast
tumors (1,21). Despite notable progress in research,
MDR remains the main obstacle to successful cancer treatment. ABC
transporters are one of the major factors leading to
chemoresistance owing to their ability to efflux drugs outside of
tumor cells, thus diminishing the therapeutic effect (22). In the present study, an ADM-resistant
breast cancer cell line was used as a model to address the role of
ABC transport proteins in tumor cells. ABCB1, ABCC1 and ABCG2 were
found to be overexpressed in the MCF-7/ADR cells, indicating that
MCF-7/ADR tumor cells may develop MDR to a variety of chemotherapy
drugs (23,24). When treated with three types of drugs
whose structures and mechanisms of action are completely different,
the MCF-7/ADR cells were shown to be resistant to ADM, TAX and TAM
when compared with the MCF-7 cells.
The molecular mechanism of eIF4G, a component of the
translation initiation complex, awaits further clarification.
Previous studies identified that eIF4G is a positive regulator of
tumor growth via regulation of translation initiation activity.
Accordingly, the corresponding inhibition of translation and
induction of senescence by miRNAs and/or small molecule inhibitors
targeting eIF4G are potential novel strategies for cancer treatment
(25,26). Increasing evidence shows that the
abnormal function of the cellular eIF4F complex, which is caused by
increased expression of initiation factors, serves a key role in
tumorigenesis. It is a widely held belief that the targeting of the
molecules of this complex may be used as a cancer therapy.
A number of studies have demonstrated that miRNAs
are associated with numerous critical cellular pathways and that
they are able to function together with other genes that are
crucial for the control of cellular growth (20,27).
Accordingly, the results of our earlier studies provided an
experimental basis for using these small molecules in a therapeutic
approach (28), and miR-133a and
miR-326 have been established as personalized therapy for HCC
treatment. It is vital that the gene/pathway that these small
regulatory molecules regulate is identified. We predicted that the
3′UTR of eIF4G contained one potential binding site for miR-503,
and the present study confirmed the binding of miR-503 to the 3′UTR
of eIF4G in breast cancer cells. Elevated levels of miR-503 were
confirmed to lead to the inhibition of eIF4G gene translation and
cell proliferation, and to promote senescence in MCF-7/ADR cells. A
number of studies have demonstrated that disruption of the
translational machinery one of the main contributors to the
development and progression of cancer (29,30). A
large body of research has become focused upon investigating the
mechanisms of translational regulation by miRNAs (31,32). The
present results provided important evidence of the regulation of
translational initiation complex by the targeting of eIF4G by
miRNA, and thereby the modulation of drug sensitivity in cancer
cells.
In summary, the present study demonstated that
miR-503 could increase the sensitivity of human MCF-7/ADR cells to
ADM, TAX and TAM via targeting eIF4G, leading to the downregulation
of ABCB1, ABCC1 and ABCG2 transport proteins. Targeting miR-503
would be a potential therapeutic strategy for the treatment of
breast cancer with MDR.
Acknowledgements
This study was supported by the grants from the
National Natural Science Foundation of China (no. 81372579), the
Hunan Provincial Innovation Foundation For Postgraduate (no.
2015SCX29) and the Hunan Province Cooperative Innovation Center for
Molecular Target New Drug Study.
References
|
1
|
Hlaváč V, Brynychová V, Václavíková R,
Ehrlichová M, Vrána D, Pecha V, Koževnikovová R, Trnková M, Gatěk
J, Kopperová D, et al: The expression profile of ATP-binding
cassette transporter genes in breast carcinoma. Pharmacogenomics.
14:515–529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gottesman MM, Hrycyna CA, Schoenlein PV,
Germann UA and Pastan I: Genetic analysis of the multidrug
transporter. Annu Rev Genet. 29:607–649. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lo YL and Liu Y: Reversing multidrug
resistance in Caco-2 by silencing MDR1, MRP1, MRP2, and
BCL-2/BCL-xL using liposomal antisense oligonucleotides. PLoS One.
9:e901802014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Deeley RG, Westlake C and Cole SP:
Transmembrane transport of endo- and xenobiotics by mammalian
ATP-binding cassette multidrug resistance proteins. Physiol Rev.
86:849–899. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krishnamurthy P and Schuetz JD: Role of
ABCG2/BCRP in biology and medicine. Annu Rev Pharmacol Toxicol.
46:381–410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gong MQ, Wu JL, Chen B, Zhuo RX and Cheng
SX: Self-assembled polymer/inorganic hybrid nanovesicles for
multiple drug delivery to overcome drug resistance in cancer
chemotherapy. Langmuir. 31:5115–5122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bao L, Hazari S, Mehra S, Kaushal D, Moroz
K and Dash S: Increased expression of P-glycoprotein and
doxorubicin chemoresistance of metastatic breast cancer is
regulated by miR-298. Am J Pathol. 180:2490–2503. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Berger W, Hauptmann E, Elbling L,
Vetterlein M, Kokoschka EM and Micksche M: Possible role of the
multidrug resistance-associated protein (MRP) in chemoresistance of
human melanoma cells. Int J Cancer. 71:108–115. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Padmanabhan R, Chen KG, Gillet JP, Handley
M, Mallon BS, Hamilton RS, Park K, Varma S, Mehaffey MG, Robey PG,
et al: Regulation and expression of the ATP-binding cassette
transporter ABCG2 in human embryonic stem cells. Stem Cells.
30:2175–2187. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pogribny IP, Filkowski JN, Tryndyak VP,
Golubov A, Shpyleva SI and Kovalchuk O: Alterations of microRNAs
and their targets are associated with acquired resistance of MCF-7
breast cancer cells to cisplatin. Int J Cancer. 127:1785–1794.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Silvera D, Formenti SC and Schneider RJ:
Translational control in cancer. Nat Rev Cancer. 10:254–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li BD, McDonald JC, Nassar R and De
Benedetti A: Clinical outcome in stage I to III breast carcinoma
and eIF4E overexpression. Ann Surg. 227:756–761; 761–763. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Demosthenous C, Han JJ, Stenson MJ, Maurer
MJ, Wellik LE, Link B, Hege K, Dogan A, Sotomayor E, Witzig T and
Gupta M: Translation initiation complex eIF4F is a therapeutic
target for dual mTOR kinase inhibitors in non-Hodgkin lymphoma.
Oncotarget. 6:9488–9501. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan S, Li Y, Yue P, Khuri FR and Sun SY:
The eIF4E/eIF4G interaction inhibitor 4EGI-1 augments
TRAIL-mediated apoptosis through c-FLIP Down-regulation and DR5
induction independent of inhibition of cap-dependent protein
translation. Neoplasia. 12:346–356. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
She QB, Halilovic E, Ye Q, Zhen W,
Shirasawa S, Sasazuki T, Solit DB and Rosen N: 4E-BP1 is a key
effector of the oncogenic activation of the AKT and ERK signaling
pathways that integrates their function in tumors. Cancer Cell.
18:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boussemart L, Malka-Mahieu H, Girault I,
Allard D, Hemmingsson O, Tomasic G, Thomas M, Basmadjian C, Ribeiro
N, Thuaud F, et al: eIF4F is a nexus of resistance to anti-BRAF and
anti-MEK cancer therapies. Nature. 513:105–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Karthikeyan S and Hoti SL: Development of
fourth generation ABC inhibitors from natural products: A novel
approach to overcome cancer multidrug resistance. Anticancer Agents
Med Chem. 15:605–615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ween MP, Armstrong MA, Oehler MK and
Ricciardelli C: The role of ABC transporters in ovarian cancer
progression and chemoresistance. Crit Rev Oncol Hematol.
96:220–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Szakács G, Paterson JK, Ludwig JA,
Booth-Genthe C and Gottesman MM: Targeting multidrug resistance in
cancer. Nat Rev Drug Discov. 5:219–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baguley BC: Multiple drug resistance
mechanisms in cancer. Mol Biotechnol. 46:308–316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu P, Cheng H, Roberts TM and Zhao JJ:
Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev
Drug Discov. 8:627–644. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mazan-Mamczarz K, Zhao XF, Dai B,
Steinhardt JJ, Peroutka RJ, Berk KL, Landon AL, Sadowska M, Zhang
Y, Lehrmann E, et al: Down-regulation of eIF4GII by miR-520c-3p
represses diffuse large B cell lymphoma development. PLoS Genet.
10:e10041052014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS,
Hong QS, Singh S, Kao CL, Chen HY, Su KY, et al:
Enterovirus-induced miR-141 contributes to shutoff of host protein
translation by targeting the translation initiation factor eIF4E.
Cell Host Microbe. 9:58–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma J, Wang T, Guo R, Yang X, Yin J, Yu J,
Xiang Q, Pan X, Tang H and Lei X: Involvement of miR-133a and
miR-326 in ADM resistance of HepG2 through modulating expression of
ABCC1. J Drug Target. 23:519–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Zhao J, Zhang JW, Huang QY, Huang
JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ and Ma WM: MicroRNA-217,
down-regulated in clear cell renal cell carcinoma and associated
with lower survival, suppresses cell proliferation and migration.
Neoplasma. 60:511–515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang H, Zhang P, Xiang Q, Yin J, Yu J,
Yang X and Lei X: Let-7 g microRNA sensitizes
fluorouracil-resistant human hepatoma cells. Pharmazie. 69:287–292.
2014.PubMed/NCBI
|
|
31
|
Borel F, Han R, Visser A, Petry H, van
Deventer SJ, Jansen PL and Konstantinova P: Réseau Centre de
Ressources Biologiques Foie (French Liver Biobanks Network),
France: Adenosine triphosphate-binding cassette transporter genes
up-regulation in untreated hepatocellular carcinoma is mediated by
cellular microRNAs. Hepatology. 55:821–832. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z,
Liu J, Cui Y, Bian X, Bie P and Qian C: MicroRNA-122 sensitizes HCC
cancer cells to adriamycin and vincristine through modulating
expression of MDR and inducing cell cycle arrest. Cancer Lett.
310:160–169. 2011.PubMed/NCBI
|