Introduction
Circulating tumor cells (CTCs) were first described
in 1869 (1). CTCs, which are shed
from primary tumor tissue (2),
circulate in the bloodstream and promote metastasis (3,4). CTCs have
molecular characteristics that are also exhibited by the primary
tumor tissue (5,6); therefore, it may be possible to evaluate
drug sensitivity and resistance and predict patient prognosis
following therapy using the CTCs obtained by liquid biopsy
(7,8).
Breast cancer mortality is the fifth highest among
all cancer types and is the highest within the forms of cancer that
affect only females (9). Although
effective therapies targeting hormone receptors and human epidermal
growth factor receptor 2 expression have improved survival rates,
tumor recurrence and metastasis occur in a number of patients
(10,11). Recurrent tumors and metastases have
genetic characteristics that differ from those of the original
tumor and, therefore, alternative therapies may be required for
these tumors (12). CTCs from
patients with breast cancer may be able to indicate tumor
recurrence and metastasis (13),
predict survival rate (14) and
predict which therapy may be optimal (15,16).
A previous study demonstrated a correlation between
the number of CTCs and breast cancer recurrence or survival rate
(17). Another previous study
indicated that the number of CTCs detected during therapy may be a
predictive tool for progression-free and overall survival rate
(18). Additionally, genomic profiles
of CTCs may be used to predict therapeutic prognoses, identify an
optimal therapy and analyze the molecular variation of the tumor
during treatment (19). However,
genomic analyses of CTCs are challenging to perform due to the
rarity of these cells (20). In the
present study, live, intact CTCs were isolated by size and were
subsequently cultured to obtain sufficient quantities of cells for
genomic analysis.
Materials and methods
Clinical information of patients
A total of six patients with breast cancer from the
Asan Medical Center (Seoul, Korea) were included in the present
study. The median age was 44 years (range, 37–47). The stages of
the cancer were evaluated using the Tumor, Node and Metastasis
(TNM) system based on the recommendations of the 7th American Joint
Committee on Cancer (21). All blood
samples, tumor tissues and medical data used were anonymous, to
ensure patient confidentiality. The protocol that was used for the
current study was ethically approved by the institutional review
board of ASAN Medical Center (clearance no. 2013-1048).
Blood collection and CTC enrichment
process
Blood (10 ml) from each patient was obtained, stored
in acid citrate dextrose tubes and processed within 4 h. The CTC
culture kit (#CIKC10; Cytogen, Inc., Seoul, Korea) was used to
isolate CTCs from blood samples for culture. Briefly, density
gradient centrifugation was performed at 400 × g for 30 min at room
temperature using the blood samples, and the fraction containing
peripheral blood mononuclear cells was diluted with a dilution
buffer from the kit. Diluted cell suspensions were filtered using a
high-density microporous (HDM) chip (Cytogen, Inc.) (22) and the cells retrieved from the HDM
chip were cultured.
Primary culture of CTCs
The CTCs that were isolated were washed with PBS and
cultured in 6-well Costar® Ultra-Low Attachment plates
(Costar®; Corning Korea Company, Ltd., Seoul, Korea)
containing mesenchymal stem cell growth medium (MSCGM™, human
Mesenchymal Stem Cell Growth BulletKit™ Medium and Supplements;
Lonza Group, Basel, Switzerland) at 37°C, in an atmosphere
containing 5% CO2. Following 16–18 days of culture,
cells were fixed in 4% paraformaldehyde on microscope slides, to be
used in the immunofluorescence staining protocol. Cell images were
taken every other day under light microscopy (Eclipse TS 100; Nikon
Corporation, Tokyo, Japan). Cell pellets were stored at −80°C prior
to cancer gene panel analysis.
Immunofluorescence staining
The fixed cells on microscope slides were incubated
with 0.2% Triton X-100 in PBS for 10 min at room temperature and
subsequently treated with 0.3% hydrogen peroxide for 30 min at room
temperature. Following blocking with 1% bovine serum albumin (cat.
no. SH30574.02; GE Healthcare Life Sciences, Chalfont, UK) in PBS
for 30 min, the cells were incubated with mouse anti-epithelial
cell adhesion molecule (EpCAM) antibody (dilution, 1:200, cat. no.
2929; CST Biological Reagents Company Limited, Shanghai, China) at
room temperature for 1 h. EpCAM signals were amplified with the
Tyramide Signal Amplification™ kit (cat. no. T20922; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. The slides were mounted using Fluoroshield™ with DAPI
(ImmunoBioScience Corp., Mukilteo, WA, USA). Stained cells were
captured on a Nikon Eclipse Ti fluorescence microscope equipped
with a 200X objective.
Whole genome amplification
The cellular DNA was obtained from cell pellets that
were stored at −80°C and were amplified using the REPLI-g Single
Cell kit (Qiagen GmbH, Hilden, Germany) according to the
manufacturer's protocol. Briefly, cell pellets were resuspended
with the denaturation buffer and incubated at 65°C for 10 min.
Following the addition of the stop solution, denatured DNA samples
were added to REPLI-g single cell DNA polymerase and the reaction
buffer. This mixture was incubated at 30°C for 8 h and subsequently
at 65°C for 3 min.
Genomic DNA extraction from primary
tumor tissues
The genomic DNA was extracted from 5-µm sections of
formalin-fixed, paraffin-embedded (FFPE) primary tumor tissues.
H&E-stained FFPE slides were initially examined by a
pathologist (Asan Medical Center) to validate the presence of tumor
cells. DNA that was present in the tumor cells was extracted using
the Gentra® Puregene® DNA Isolation kit
(Qiagen GmbH) according to the manufacturer's protocol.
Ion AmpliSeq™ Cancer Panel
analysis
Genomic mutations were detected using the Ion
AmpliSeq™ Cancer Hotspot Panel v2 (Thermo Fisher Scientific, Inc.).
Briefly, the genomic DNA was amplified using the REPLI-g
Amplification kit (Qiagen GmbH) and the amplicons were purified
using the Agencourt AM-Pure XP kit (Beckman Coulter, Inc., Brea,
CA, USA), followed by end repair and ligation using the Ion Xpress™
Barcode Adapters kit (cat. no. 4471250; Thermo Fisher Scientific,
Inc.). Subsequent end-repair and ligation was performed with Ion
Xpress Barcode Adapters (Thermo Fisher Scientific, Inc.). The
median fragment size and the concentration of the final library
were determined using a BioAnalyzer equipped with a
High-Sensitivity Chip (Agilent Technologies GmbH, Waldbronn,
Germany). Subsequently the library was diluted to 10 pM with TE
using a low-Tris EDTA buffer, 5 µl of the library was used for
emulsion polymerase chain reaction (PCR) using the Onetouch™
reagent kit (Invitrogen; Thermo Fisher Scientific, Inc.). The
following cycling conditions were used: 80°C for 3 min; 18 cycles
of 99°C for 20 sec, 58°C for 30 sec, 72°C for 60 sec, 99°C for 20
sec, 56°C for 30 sec, and 70°C for 60 sec; and 10 cycles of 99°C
for 20 sec, and 58°C for elongated duration from 3 min to 20 min,
with the thermocycler lid heated to 85°C. The products of these
emulsion PCR reactions were enriched using Dynabeads®
MyOne™ Streptavidin C1 beads (Invitrogen; Thermo Fisher Scientific,
Inc.). The final enriched ion spheres were mixed with sequencing
primers and polymerase and loaded onto five Ion 316 chips. Base
calls were generated using Torrent Suite 3.0 software (Thermo
Fisher Scientific, Inc.) with tmap-f3 and maintained on the Ion
Torrent server for further analysis. Base calling was generated
using Torrent Suite software (version 3.0; Thermo Fisher
Scientific, Inc.) with tmap-f3 indexing. BAM and FASTQ alignment
files were generated based on the base calling results and were
used for variant calling, including single nucleotide polymorphisms
and insertions/deletions.
Results
Expansion of CTCs via cell
culture
CTCs from six patients with breast cancer were
cultured to obtained optimal numbers of cells for characterization.
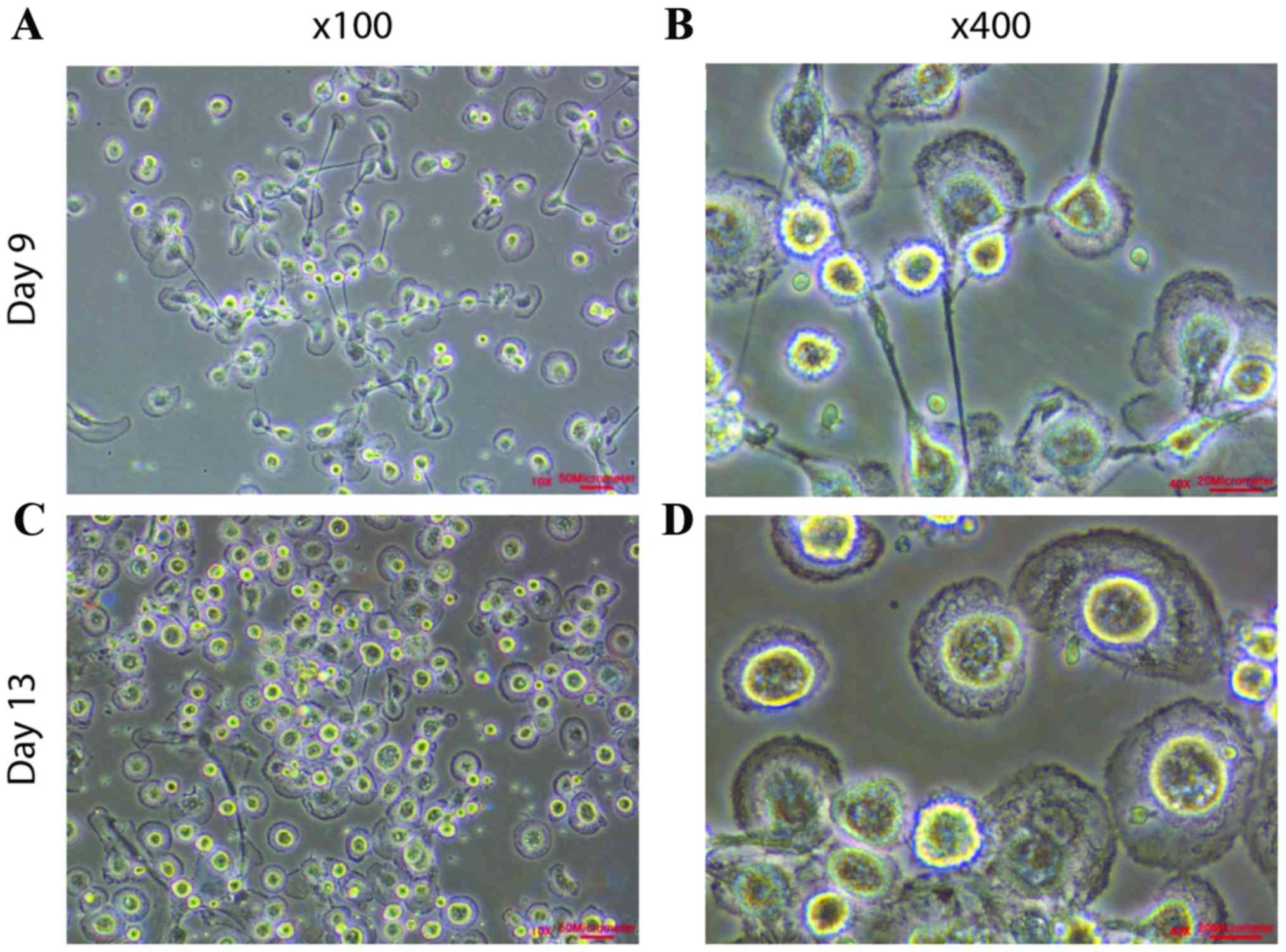
During the first nine days of culture, cells were attached or
suspended as single cells (Fig. 1A and
B). The cells were cultured until there were between
4×105 and 8×105 cells (Fig. 1C; Table
I) and the attached cells exhibited cell membrane ruffling
(Fig. 1D). The presence of cell
membrane ruffling demonstrated the selective expansion of
epithelial cells and improved cell motility.
 | Table I.Clinical characteristics of patients
with breast cancer, including immunofluorescence staining analysis
of EpCAM-positive cells. |
Table I.
Clinical characteristics of patients
with breast cancer, including immunofluorescence staining analysis
of EpCAM-positive cells.
|
|
|
| No. of cultured
cells |
|---|
|
|
|
|
|
|---|
| Patient ID | Age, years | AJCC/TNM stage | Total no. of
cells | No. of EpCAM+ cells,
% |
|---|
| AMC-15–01 | 47 | IIA |
4.0×105 | 34.92 |
| AMC-15–02 | 38 | IIA |
5.0×105 | 53.74 |
| AMC-15–03 | 43 | IIA |
5.0×105 | 53.76 |
| AMC-15-04 | 51 | IIB |
5.2×105 | 41.20 |
| AMC-15-05 | 37 | IIIC |
8.3×105 | 86.54 |
| AMC-15-06 | 46 | IIB |
4.5×105 | 86.14 |
CTC characterization
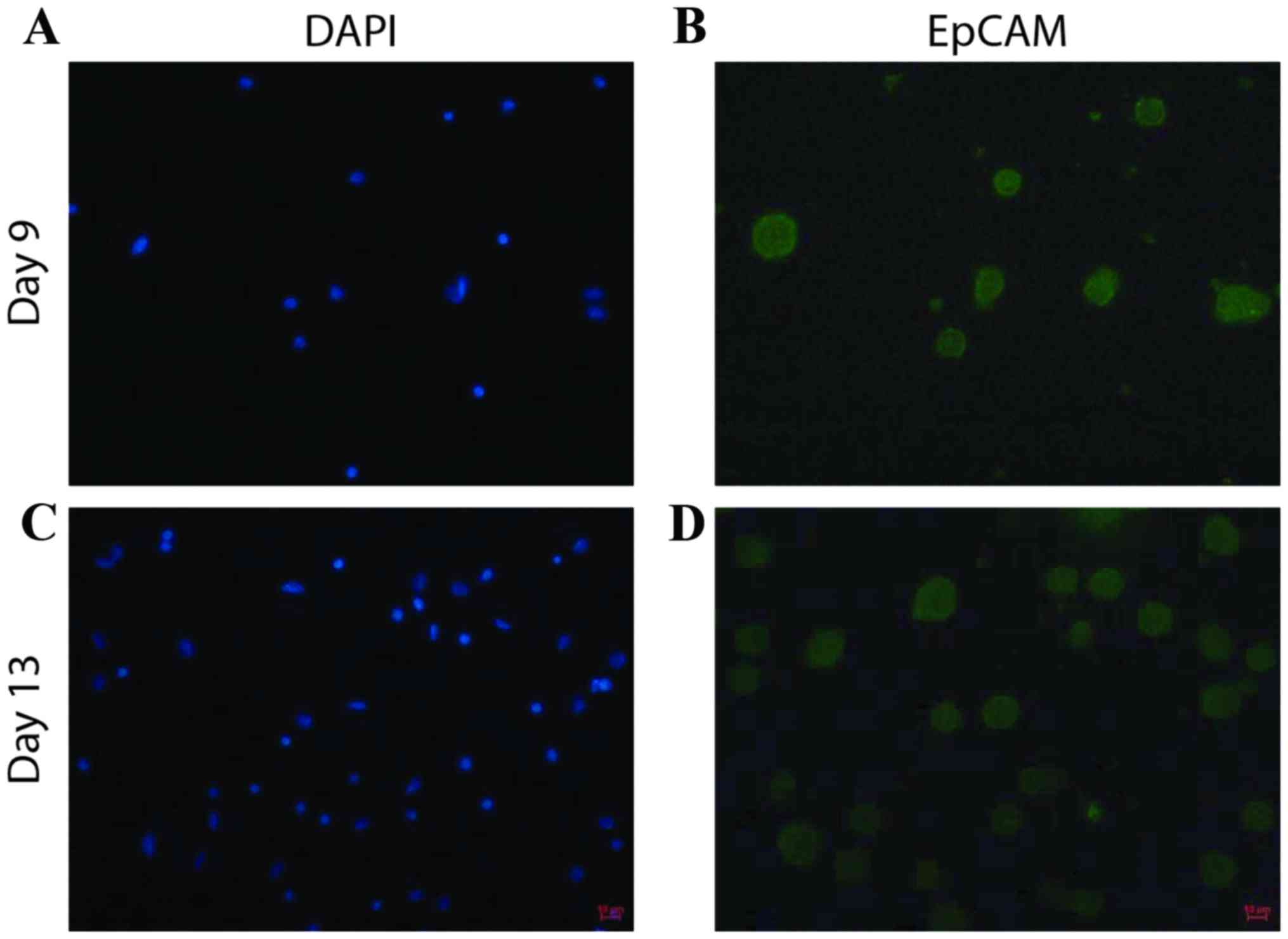
Following 16 to 18 days of cell culture,
immunofluorescence staining was performed for EpCAM, an epithelial
cell marker, to evaluate the proportion of cultured cells that were
CTCs (Fig. 2). The percentage of
EpCAM-positive cells in the samples from patients with breast
cancer ranged from 35–86% (Table I),
which suggested that CTCs may be the predominantly proliferating
cells.
Cancer gene panel analysis
COSMIC mutations in Platelet-Derived Growth Factor
Receptor Alpha (PDGFRA), MET (also known as Hepatocyte Growth
Factor Receptor), Phosphatase and Tensin Homolog (PTEN), Harvey Rat
Sarcoma Viral Oncogene Homolog (HRAS), SWI/SNF Related, Matrix
Associated, Actin Dependent Regulator of Chromatin Subfamily B
Member 1 (SMARCB1), Cyclin Dependent Kinase Inhibitor 2A (CDKN2A)
and MutL Homolog 1 genes (MLH1) genes were detected in 5/6 samples
of cultured CTCs (Table II). To
evaluate whether the cultured CTCs maintained genomic profiles that
were similar to those of the primary tumor tissues, mutations were
analyzed in cultured CTCs and compared with those detected in
primary tumor tissues. In the samples obtained from patient
AMC-15-02, an identical mutation in HRAS was detected in the
cultured CTCs and the primary tumor tissues (Table III). Similarly, 60% of the novel
mutations were identified in the cultured CTCs and the primary
tumor tissues (Table IV). Although
the cultured CTCs obtained from patient AMC-15-06 did not possess
mutations that have been identified in the COSMIC database that
were also detected in the primary tumor tissue, 80% of novel
mutations were identified in the cultured CTCs and the primary
tumor tissues (data not shown).
 | Table II.Ion AmpliSeq Cancer Panel V2 of
cultured CTCs from patients with breast cancer. |
Table II.
Ion AmpliSeq Cancer Panel V2 of
cultured CTCs from patients with breast cancer.
| Patient ID | Gene ID | Type of
mutation | AA mutation | COSMIC number |
|---|
| AMC-15-01 | PDGFRA | SNP | N659K | COSM22414 |
|
| MET | SNP | Unknown | COSM710 |
|
| PTEN | INS | N323fs*2 | COSM23626 |
|
| PTEN | INS | T321fs*3 | COSM4994 |
|
| PTEN | INS | N323fs*2 | COSM4990 |
| AMC-15-02 | PDGFRA | SNP | V824V | COSM22413 |
|
| HRAS | SNP | H27H | COSM249860 |
|
| SMARCB1 | SNP | Unknown | COSM1090 |
| AMC-15-03 | PDGFRA | SNP | V824V | COSM22413 |
|
| HRAS | SNP | H27H | COSM249860 |
|
| SMARCB1 | SNP | Unknown | COSM1090 |
| AMC-15-04 |
|
| N/A |
|
| AMC-15-05 | CDKN2A | SNP | H66R | COSM14253 |
| AMC-15-06 | MLH1 | SNP | V384D | COSM26085 |
|
| MET | SNP | Unknown | COSM710 |
|
| HRAS | SNP | H27H | COSM249860 |
 | Table III.Comparison of COSMIC mutations
detected in primary tumor tissue with those detected in cultured
CTCs from patient AMC-15002 with breast cancer. |
Table III.
Comparison of COSMIC mutations
detected in primary tumor tissue with those detected in cultured
CTCs from patient AMC-15002 with breast cancer.
| Tissue | Gene ID | Mutation type | AA mutation | COSMIC number |
|---|
| Primary tissue | NOTCH1 | DEL | V1578delV | COSM13047 |
|
| HRAS | SNP | H27H | COSM249860 |
|
| TP53 | SNP | H193Y | COSM10672 |
| CTCs | PDGFRA | SNP | V824V | COSM22413 |
|
| HRAS | SNP | H27H | COSM249860 |
|
| SMARCB1 | SNP | Unknown | COSM1090 |
 | Table IV.Comparison of Ion AmpliSeq Cancer
Panel V2 between primary tumor tissue and cultured CTCs from
patient AMC-15002 with breast cancer. |
Table IV.
Comparison of Ion AmpliSeq Cancer
Panel V2 between primary tumor tissue and cultured CTCs from
patient AMC-15002 with breast cancer.
| A, Primary tumor
tissue |
|---|
| Gene ID | Type of
mutation | Allele source | COSMIC number |
|---|
| ERBB4 | SNP | Novel | – |
| VHL | SNP | Novel | – |
| FGFR3 | SNP | Novel | – |
| PDGFRA | SNP | Novel | – |
| APC | SNP | Novel | – |
| CSF1R | MNP | Novel | – |
| NOTCH1 | DEL | Hotspot | COSM13047 |
| RET | SNP | Novel | – |
| HRAS | SNP | Hotspot | COSM249860 |
| ATM | INS | Novel | – |
| FLT3 | SNP | Novel | – |
| TP53 | SNP | Novel | – |
| TP53 | SNP | Hotspot | COSM10672 |
| TP53 | SNP | Novel | – |
|
| B, Cultured
CTCs |
|
| ALK | SNP | Novel | – |
| ERBB4 | SNP | Novel | – |
| FGFR3 | SNP | Novel | – |
| PDGFRA | SNP | Novel | – |
| PDGFRA | SNP | Hotspot | COSM22413 |
| APC | SNP | Novel | – |
| CSF1R | MNP | Novel | – |
| EGFR | SNP | Novel | – |
| NOTCH1 | SNP | Novel | – |
| RET | SNP | Novel | – |
| HRAS | SNP | Hotspot | COSM249860 |
| FLT3 | SNP | Novel | – |
| TP53 | SNP | Novel | – |
| STK11 | SNP | Novel | – |
| SMARCB1 | SNP | Hotspot | COSM1090 |
Discussion
It has been previously reported that CTCs retain the
genomic characteristics of the primary tumor. Therefore, CTCs may
be used as a substitute for tissue biopsy to evaluate drug
responsiveness and predict an optimal therapy (7,8). The
authors of the current study performed cancer gene panel analyses
using uncultured CTCs (Lee et al, unpublished), which
indicated that CTCs are rare, but may be optimal in number for
molecular analysis without culturing. However, the expansion of the
CTC sample is required for chemosensitivity assays and
patient-derived xenograft (PDX) models.
This novel methodology is able to provide sufficient
cell numbers for the isolation and culture of CTCs. The number of
EpCAM-positive cells ranged from 35 to 86% of the total cells that
were obtained using the culture method and the final number of
cultured cells was between 4×105 and 8×105
(Table I). The cells were cultured
until there were >1×105 CTCs and these cells were
used for cancer gene panel analysis. Furthermore, the cultured CTCs
may be used in a chemosensitivity assay and in the PDX model of
breast cancer. The number of CTCs may have been underestimated in
the current study, as the described method was unable to detect
mesenchymal CTCs that may have undergone the epithelial to
mesenchymal transition (23).
Mutations in PDGFRA, MET, PTEN, HRAS, SMARCB1,
CDKN2A and MLH1 were identified from the genomic
analysis of cultured CTCs in the current study. Mutations in these
genes have previously been identified in breast tumor tissues
(24–26), and this may demonstrate that cultured
CTCs maintain genetic characteristics that are similar to those
detected in the primary tumor tissues. PDGFRA and
HRAS, which were mutated in 3/6 of the cultured CTC samples,
are established to be associated with breast cancer progression
(25–27).
Furthermore, the analyses of the genomic profiles of
primary tumor tissues and those of the corresponding cultured CTCs
identified that a large portion of mutations that were detected in
CTCs was also detected in the primary tumor tissues. Although the
cultured CTCs obtained from patient AMC-15-06 did not have any of
the COSMIC database identified mutations also identified in the
primary tumor tissue, a mutation of HRAS was detected in
CTCs of this patient that has previously been reported to be
associated with breast cancer recurrence and metastasis (27,28).
In conclusions, the evaluation of whether cultured
CTCs maintain the genomic characteristics of the primary tumor may
be the first step in the application of cultured CTCs to predict an
effective treatment for a patient with breast cancer. In the
present study, CTCs were isolated and cultured effectively, and
genomic analysis was performed on them. It was also demonstrated
that cultured CTCs may maintain a similar genomic profile compared
with primary tumor tissues and this suggests that the use of
cultured CTCs may provide a novel approach for breast cancer
diagnosis and treatment.
Acknowledgements
The current study was supported by a grant from the
National Research and Development Program, Ministry of Trade,
Industry and Energy, Republic of Korea (grant no. 10045947). The
authors would like to thank Enago (www.enago.kr.com) for the English language review.
References
|
1
|
Ashworth TR: A case of cancer in which
cells similar to those in the tumors were seen the blood after
death. Aust Med J. 14:146–149. 1869.
|
|
2
|
Aceto N, Bardia A, Miyamoto DT, Donaldson
MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al:
Circulating tumor cell clusters are oligoclonal precursors of
breast cancer metastasis. Cell. 158:1110–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cristofanilli M, Hayes DF, Budd GT, Ellis
MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC,
et al: Circulating tumor cells: A novel prognostic factor for newly
diagnosed metastatic breast cancer. J Clin Oncol. 23:1420–1430.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Markiewicz A, Książkiewicz M,
Wełnicka-Jaśkiewicz M, Seroczyńska B, Skokowski J, Szade J and
Żaczek AJ: Mesenchymal phenotype of CTC-enriched blood fraction and
lymph node metastasis formation potential. PLoS One. 9:e939012014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marchetti A, Del Grammastro M, Felicioni
L, Malatesta S, Filice G, Centi I, de Pas T, Santoro A, Chella A,
Brandes AA, et al: Assessment of EGFR mutations in circulating
tumor cell preparations from NSCLC patients by next generation
sequencing: Toward a real-time liquid biopsy for treatment. PLoS
One. 9:e1038832014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marrinucci D, Bethel K, Luttgen M, Bruce
RH, Nieva J and Kuhn P: Circulating tumor cells from
well-differentiated lung adenocarcinoma retain cytomorphologic
features of primary tumor type. Arch Pathol Lab Med. 133:1468–1471.
2009.PubMed/NCBI
|
|
7
|
van de Stolpe A, Pantel K, Sleijfer S,
Terstappen LW and den Toonder JM: Circulating tumor cell isolation
and diagnostics: Toward routine clinical use. Cancer Res.
71:5955–5960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Giuliano M, Giordano A, Jackson S, de
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Brest Cancer Res. 16:4402014. View Article : Google Scholar
|
|
9
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lorusso G and Rüegg C: New insights into
the mechanisms of organ-specific breast cancer metastasis. Semin
Cancer Biol. 22:226–233. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cardoso F, Harbeck N, Fallowfield L,
Kyriakides S and Senkus E; ESMO Guidelines Working Group, : Locally
recurrent or metastatic breast cancer: ESMO clinical practice
guidelines for diagnosis, treatment and follow-up. Ann Oncol. 23
Suppl 7:vii11–vii19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suzuki M and Tarin D: Gene expression
profiling of human lymph node metastases and matched primary breast
carcinomas: Clinical implications. Mol Oncol. 1:172–180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giuliano M, Giordano A, Jackson S, de
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Breast Cancer Res. 16:4402014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rack B, Schindlbeck C, Jückstock J,
Andergassen U, Hepp P, Zwingers T, Friedl TW, Lorenz R, Tesch H, et
al: Circulating tumor cells predict survival in early
average-to-high risk breast cancer patients. J Natl Cancer Inst.
106:pii: dju066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bidard FC, Fehm T, Ignatiadis M, Smerage
JB, Alix-Panabières C, Janni W, Messina C, Paoletti C, Müller V,
Hayes DF, et al: Clinical application of circulating tumor cells in
breast cancer: Overview of the current interventional trials.
Cancer Metastasis Rev. 32:179–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nadal R, Lorente JA, Rosell R and Serrano
MJ: Relevance of molecular characterization of circulating tumor
cells in breast cancer in the era of targeted therapies. Expert Rev
Mol Diagn. 13:295–307. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franken B, de Groot MR, Mastboom WJ,
Vermes I, van der Palen J, Tibbe AG and Terstappen LW: Circulating
tumor cells, disease recurrence and survival in newly diagnosed
breast cancer. Breast Cancer Res. 14:R1332012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cristofanilli M, Budd GT, Ellis MJ,
Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ,
Terstappen LW and Hayes DF: Circulating tumor cells, disease
progression, and survival in metastatic breast cancer. N Engl J
Med. 351:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fernandez SV, Bingham C, Fittipaldi P,
Austin L, Palazzo J, Palmer G, Alpaugh K and Cristofanilli M: TP53
mutations detected in circulating tumor cells present in the blood
of metastatic triple negative breast cancer patients. Breast Cancer
Res. 16:4452014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu M, Stott S, Toner M, Maheswaran S and
Haber DA: Circulating tumor cells: Approaches to isolation and
characterization. J Cell Biol. 192:373–382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th Edition of the AJCC Cancer Staging
Manual and the Future of TNM. Ann Surg Oncol. 17:14712010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim EH, Lee JK, Kim BC, Rhim SH, Kim JW,
Kim KH, Jung SM, Park PS, Park HC, Lee J and Jeon BH: Enrichment of
cancer cells from whole blood using a microfabricated porous
filter. Anal Biochem. 440:114–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gorges TM, Tinhofer I, Drosch M, Röse L,
Zollner TM, Krahn T and von Ahsen O: Circulating tumour cells
escape from EpCAM based detection due to epithelial-to mesenchymal
transition. BMC Cancer. 12:1782012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tung N, Battelli C, Allen B, Kaldate R,
Bhatnagar S, Bowles K, Timms K, Garber JE, Herold C, Ellisen L, et
al: Frequency of mutations in individuals with breast cancer
referred for BRCA1 and BRCA2 testing using next-generation
sequencing with a 25-gene panel. Cancer. 121:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Inês C, Fernanda M, Albino M, Rui MR and
Fernando S: Overexpression of platelet-derived growth factor
receptor alpha in breast cancer is associated with tumour
progression. Breast Cancer Res. 7:R788–R795. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fernández-Medarde A and Santos E: Ras in
cancer and developmental diseases. Genes Cancer. 2:344–358. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yong HY, Hwang JS, Son H, Park HI, Oh ES,
Kim HH, Kim DK, Choi WS, Lee BJ, Kim HR and Moon A: Identification
of H-Ras-specific motif for the activation of invasive signaling
program in human breast epithelial cells. Neoplasia. 13:98–107.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Watson DM, Elton RA, Jack WJ, Dixon JM,
Chetty U and Miller WR: The H-ras oncogene product p21 and
prognosis in human breast cancer. Breast Cancer Res Treat.
17:161–169. 1991. View Article : Google Scholar : PubMed/NCBI
|