Introduction
MicroRNAs (miRNAs/miRs) are small, endogenous,
non-coding RNAs composed of 18–25 nucleotides that regulate gene
expression at the post-transcriptional level (1). Certain miRNAs act as oncogenes or tumor
suppressors and are considered to be among the key regulators of
cancer progression (2,3). Several previous studies have reported
that miRNA levels in tumor tissues may be potential biomarkers for
immunotherapy (4,5).
Active immunotherapy using tumor-associated
antigen-derived epitope peptides (6,7), which
aims to induce an in vivo response from tumor-specific
cytotoxic T lymphocytes (CTLs), requires the preservation of the
host immune system as indicated in the 2011 US Food and Drug
Administration guidance report for therapeutic cancer vaccines
(8). However, since advanced-stage
patients with poor immune status are usually allowed to enroll into
clinical studies in early-phase drug development, it is often
difficult to evaluate the clinical benefit of these treatments
(9). Therefore, it is necessary to
identify predictive biomarkers to select patients who are likely to
respond well and induce responses from specific CTLs to epitope
peptides (10,11).
The authors of the present study previously reported
a phase I study where 5 epitope peptides were administered to
patients with advanced-stage colon cancer (6). Of these, 3 peptides were derived from
oncoantigens: Ring finger protein 43 (RNF43) (12), 34 kDa translocase of the outer
mitochondrial membrane (TOMM34) (13)
and insulin growth factor-II mRNA binding protein 3 (KOC1, also
known as IMP3) (14), and the
remaining 2 peptides were derived from vascular endothelial growth
factor receptors (VEGFRs) (15,16). A
phase II study using the same vaccine regimen in combination with
oxaliplatin-based chemotherapy was performed to further verify the
safety and the potential of the vaccine to induce CTLs and to
improve overall survival (OS) (17).
In these studies, a high CTL response following vaccination and an
injection site skin reaction were revealed to be potential markers
for the outcome of vaccine treatment (6), and a low neutrophil/lymphocyte ratio was
also a potential marker for improved survival time of vaccinated
patients (17).
The purpose of the present study was to identify
novel biomarkers in order to predict the efficacy of
immunotherapies prior to treatment, and to use such information to
select patients who are likely to exhibit improved treatment
outcomes following vaccination. The results of a comprehensive
miRNA microarray analysis of cancer tissues are presented in the
present study, and a number of miRNAs were identified as novel
biomarkers for predicting the efficiency of active
immunotherapies.
Materials and methods
Summary of the phase I study
The detailed protocol of the phase I study (UMIN
clinical trial registration no. UMIN000004948) was described
previously (6). Enrollment criteria
were as follows: i) Histological confirmation of colorectal cancer
(CRC) without surgical resection; ii) failure to respond to
previous standard chemotherapy or intolerance to standard therapy
and iii) positive DNA typing result for human leukocyte antigen
(HLA)-A*2402.
The phase I study was primarily conducted to
evaluate the safety and to investigate the recommended dose (RD) of
these peptides. Good Manufacturing Practice grade RNF43-721
(NSQPVWLCL) (18), TOMM34-299
(KLRQEVKQNL) (13), KOC1 (IMP-3)-508
(KTVNELQNL) (19), VEGFR1-1084
(SYGVLLWEI) (20) and VEGFR2-169
(RFVPDGNRI) (21) peptides restricted
with HLA-A*2402 were used. Dose escalation was performed in 3
patients with doses of 0.5, 1, and 3 mg for each peptide. Each
peptide was mixed with 0.5 ml of incomplete Freund's adjuvant (IFA)
and administered to patients subcutaneously into the thigh or
axilla regions on days 1, 8, 15, and 22 of a 28-day treatment
course. According to the results of this previous study, it was
decided that the RD of each peptide was 3.0 mg, and a dose of 3.0
mg was administered to an additional 3 patients to confirm the
safety of the peptides.
Next, a single injection of a cocktail of 5 peptides
was performed, and this was expected to induce immune responses at
the same level as separate injections of each of the 5 peptides.
The cocktail of the 5 peptides (at the dose of 3 mg) was
administered to 6 patients. A total of 8 out of 18
HLA-A*2402-positive CRC patients enrolled in the phase I clinical
trial were available for miRNA analysis in the present study.
Summary of the phase II study
To evaluate the clinical benefits of cancer
vaccination treatment, a phase II trial (UMIN clinical trial
registration no. UMIN000001791) was conducted using the previously
described 5 peptides. The phase II trial was a non-randomized,
HLA-A status double-blind study. The detailed protocol of this
study was described previously (17).
Briefly, the therapy consisted of a cocktail of 5 therapeutic
epitope peptides (the same as those used in the phase I study), in
addition to oxaliplatin-based chemotherapy. Although the peptides
used in the phase II study were HLA-A*2402 restricted peptides, the
same regime of peptide cocktail and oxaliplatin-based chemotherapy
was administered to all enrolled patients, whose HLA-A status was
double-blinded. The cocktail of (3 mg of each peptide) was mixed
with 1.5 ml IFA and administered subcutaneously into the thigh or
axilla regions once per week for 13 weeks. The vaccination schedule
was subsequently reduced to once every 2 weeks. Vaccination was
continued even if the disease progressed when the patient wished
and a primary doctor who provided additional chemotherapies agreed.
Enrollment criteria for the phase II study were as follows: i) ≥20
years old with histologically confirmed advanced CRC; ii)
chemotherapy-naïve; iii) adequate function of organs and iv) a life
expectancy of ≥3 months. Between February 2009 and November 2012,
96 chemotherapy-naïve CRC patients were enrolled and their HLA-A
status was masked. Among the 96 patients who were enrolled in the
phase II study, 26 cases were available for miRNA analysis in the
present study.
Sample collection
Among the 18 patients who participated in the phase
I trial (6), tissues from 8 cases of
CRC were obtained during surgery between October 2003 and June
2008. Of the patients, 4 were male and 4 were female, with an age
range of 56–75 years and a mean age of 64 years. From these 8
patients, CRC tissues were obtained, and normal colorectal tissues
were also obtained from surgical specimens of 5 patients between
January 2004 and July 2006. Of the normal patients, 2 were male and
3 were female, with an age range of 59–75 years and a mean age of
66 years. Tissues were snap-frozen in liquid nitrogen and stored at
−80°C. Similarly, CRC tissues were obtained from 26 patients who
participated in the phase II trial of vaccine treatment in
combination with oxaliplatin-based chemotherapy between October
2008 and September 2011 (17). Of
these patients, 11 were male and 15 were female, with an age range
of 47–82 years and a mean age of 67 years. CRC tissues were
obtained from 16 cases from the HLA-A*2402-matched group and 10
cases from the HLA-A*2402-unmatched group.
All patients underwent resection of the cancer prior
to phase I and II vaccine trials at Yamaguchi University Hospital
(Yamaguchi, Japan). Written informed consent was obtained from all
patients, and the present study was approved by the Institutional
Ethics Review Boards of Yamaguchi University and was conducted in
accordance with the Declaration of Helsinki.
miRNA microarray
The median miRNA expression values in 34 cancer
tissues were used as cut off values to classify patients into high
or low expression group. Total RNA from CRC tissues and normal
colorectal samples was analyzed by miRNA microarray. Total RNA was
extracted from tissues using the mirVana miRNA Isolation kit
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. The concentration and purity of RNAs was
assessed with a spectrophotometer and RNA integrity was verified
using an Agilent RNA 6000 Nano kit (Agilent Technologies, Inc.,
Santa Clara, CA, USA). The optical density (OD) 260/OD280 ratios of
each sample were within 2.00–2.10, which were accepted to be
adequate for microarray analysis. The miRNA array was performed
using 1 µg total RNA, the miRCURY LNA microRNA Array 6th generation
(EXIQON; Qiagen, Inc., Valencia, CA, USA) and the GenePix 4000B
(Molecular Devices, LLC; Sunnyvale, CA, USA). The relative
intensity of each hybridization signal was evaluated using the
Microarray Data Analysis tool (version 3.2; Filgen, Inc., Nagoya,
Japan).
Statistical analysis
Following the calculation of expression signals by
log2 transformation of the normalized data,
differentially expressed miRNAs were detected by using the
fold-change value and Fisher index using Microsoft Excel 2010
(Microsoft Corporation, Redmond, WA, USA) according to the
previously reported formula (22).
Differences between groups were estimated using the Mann-Whitney
U-test. Categorical variables were compared using χ2 and
Fisher's exact tests. A Cox's proportional hazards model was used
to estimate the hazard ratios for the treatment effect in relation
to OS and miRNA expression, and OS was measured in days from the
first vaccination until patient mortality (due to any cause).
Survival curves were analyzed using the Kaplan-Meier method and the
log-rank test. Statistical analyses were performed with JMP
software (version 11; SAS Institute Inc., Cary, NC, USA) and
GraphPad Prism software (version 5.0; GraphPad Software, Inc., San
Diego, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Selection of candidate miRNAs to
predict the efficacy of vaccination
The expression profiles of 1,425 miRNAs in CRC
tissues from 8 patients enrolled in the phase I study were
analyzed. The characteristics of the 8 patients are listed in
Table I. The patients were divided
into 2 groups (responder and non-responder group) with 4 patients
in each group. A responder is defined using the following criteria:
i) Progression free survival (PFS) >150 days or ii) OS >300
days without additional chemotherapy. Criteria for non-responders
were as follows: i) PFS <90 days and ii) OS <200 days.
 | Table I.Patient characteristics and outcomes
of the phase I study. |
Table I.
Patient characteristics and outcomes
of the phase I study.
|
|
|
|
|
|
|
| Prognosis |
|
|
|---|
|
|
|
|
|
|
|
|
|
|
|
|---|
| Patient no. | Dose of peptide
(mg) | Age | Sex | Sites of
disease | Previous
treatmentsa | Response | PFS (days) | OS (days) | Status | Treatments
following vaccination | Responder or
non-responderb |
|---|
| 1 | 0.5 | 56 | M | LN | IRI, OX, FU | CR | 2,150 | 2,150 | A | None | Responder |
| 2 | 0.5 | 72 | F | Lungs, bone | IRI, OX, FU | PD | 60 | 191 | D | None | Non-responder |
| 3 | 0.5 | 75 | M | Liver, lung | IRI, OX, FU | SD | 158 | 406 | D | OX, FU, BEV | Responder |
| 4 | 1.0 | 59 | F | Dissemi. | IRI, OX, FU | PD | 36 | 80 | D | None | Non-responder |
| 6 | 1.0 | 69 | M | Lungs, LN | IRI, OX, FU | PD | 62 | 110 | D | None | Non-responder |
| 9 | 3.0 | 59 | M | Lungs | IRI, OX, FU | SD | 221 | 885 | D | None | Responder |
| 14 | 3.0 (mix) | 65 | F | Dissemi. | IRI, OX, FU | PD | 50 | 342 | D | None | Responder |
| 19 | 3.0 (mix) | 58 | F | Liver,
dissemi. | OX, FU, BEV | PD | 37 | 52 | D | None | Non-responder |
On the basis of the relative comparison of
hybridization signals, miRNAs with significant differences in
expression between the responder and non-responder groups were
selected. The 10 miRNAs with the most significant differences in
expression between the two groups are listed in Table II.
 | Table II.miRNA expression of cancer tissues of
patients in the phase I study. |
Table II.
miRNA expression of cancer tissues of
patients in the phase I study.
|
|
| Signal intensity in
cancer tissues |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
|
|
| Responder
(n=4) | Non-responder
(n=4) |
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Rank | Name of miRNA | Mean | SD | Mean | SD | Log2
ratio >0.8 | Fisher's ratio |
P-valuea | miRNA expression in
the responder group |
|---|
| 1 | hsa-miR-142-5p | 1,606.3 | 275.3 | 3,697.8 | 1,734.8 | −1.2 | 4.8 | 0.055 | Low |
| 2 |
hsa-miR-196b-3p | 295.7 | 83.2 | 164.2 | 26.1 | 0.8 | 4.5 | 0.024 | High |
| 3 |
hsa-miR-196b-5p | 678.0 | 381.6 | 172.0 | 56.5 | 2.0 | 3.6 | 0.039 | High |
| 4 | hsa-miR-147b | 332.9 | 86.0 | 778.3 | 366.0 | −1.2 | 3.4 | 0.090 | Low |
| 5 | hsa-miR-320b | 4480.4 | 2085.6 | 8208.7 | 1799.7 | −0.9 | 3.4 | 0.035 | Low |
| 6 |
hsa-miR-378a-3p | 5711.6 | 2043.9 | 9709.1 | 2499.3 | −0.8 | 3.0 | 0.048 | Low |
| 7 | hsa-miR-320e | 3293.1 | 1831.7 | 6073.7 | 1557.0 | −0.9 | 2.9 | 0.060 | Low |
| 8 | hsa-miR-486-5p | 289.0 | 53.4 | 516.7 | 244.3 | −0.8 | 2.7 | 0.119 | Low |
| 9 | hsa-miR-378c | 950.8 | 522.4 | 1978.7 | 732.5 | −1.1 | 2.6 | 0.062 | Low |
| 10 | hsa-miR-320a | 4823.1 | 2574.0 | 9183.3 | 2923.4 | −0.9 | 2.4 | 0.066 | Low |
Comparison of miRNA expression levels
between cancer tissues and normal colorectal tissues
miRNA microarray profiles of cancer tissues from the
8 patients enrolled in the phase I study and 26 patients enrolled
in the phase II study were analyzed. Subsequently, the miRNA
microarray profiles of the cancer tissues were compared with the
microarray profiles of 5 normal colorectal tissues using the
Mann-Whitney U-test. miR-196b-3p (P=0.011) and miR-196b-5p
(P<0.001) were demonstrated to be upregulated compared with the
normal tissues (Table III). In
addition, miR-147b (P<0.001), miR-486-5p (P=0.003) and miR-378c
(P=0.056; not significant) were downregulated in CRC tissues
compared with normal tissues. A cut-off value for each miRNA
(miR-142-5p, −196b-3p, −196b-5p, −147b, −320b, −378a-3p, −320e,
−486-5p, −378c and −320a) was set at 1,400, 180, 450, 440, 6,000,
6,500, 5,800, 350, 1,200 and 6,500, respectively according to the
median expression value (1,419, 184, 446, 435, 5,947, 6,496, 5,764,
350, 1,210, and 6,426, respectively) in 34 cancer tissues. The
patients were classified into high or low expression groups based
on a cut-off value of miRNA expression for subsequent analysis
(Table III).
 | Table III.miRNA expression of cancer tissues
and normal colorectal tissues. |
Table III.
miRNA expression of cancer tissues
and normal colorectal tissues.
|
|
| Signal intensity in
cancer tissues (n=34) | Signal intensity in
normal colorectal tissues (n=5) |
|
|---|
|
|
|
|
|
|
|---|
| miR rank in the
phase I study | Name of miRNA | Mean ± SD
(n=34) | Median | Cut off value | Mean ± SD
(n=5) | Median |
P-valuea |
|---|
| 1 | miR-142-5p | 1,874.4±1406.5 | 1,419 | <1400 | 2,782.8±3284.3 | 1,129 | 0.850 |
| 2 | miR-196b-3p | 212.9±96.3 | 184 | >180 | 119.2±15.2 | 119 | 0.011 |
| 3 | miR-196b-5p | 627.0±615.2 | 446 | >450 | 105.4±10.5 | 106 | <0.001 |
| 4 | miR-147b | 500.5±227.6 | 435 | <440 | 1243.8±188.7 | 1213 | <0.001 |
| 5 | miR-320b | 6079.2±2127.6 | 5947 | <6000 | 6792.0±2861.0 | 5676 | 0.850 |
| 6 | miR-378a-3p | 6575.0±2459.3 | 6496 | <6500 | 8035.4±2335.6 | 9575 | 0.200 |
| 7 | miR-320e | 5981.1±2301.0 | 5764 | <5800 | 4623.2±1081.0 | 4390 | 0.159 |
| 8 | miR-486-5p | 358.8±115.1 | 350 | <350 | 528.6±124.0 | 483 | 0.003 |
| 9 | miR-378c | 1341.7±702.0 | 1210 | <1200 | 2062.2±707.0 | 2056 | 0.056 |
| 10 | miR-320a | 6647.1±2666.4 | 6426 | <6500 | 7695.8±2724.2 | 6413 | 0.437 |
Validation of candidate miRNA
expression in patients enrolled in the phase II study
A total of 24 CRC tissues from the phase II study
were analyzed using miRNA microarray. The profiles of the 26
patients are summarized in Table IV.
A total of 10 candidate miRNAs were selected by analyzing cancer
tissues in the phase I study (Table
II) and the clinical outcome of 16 patients in the
HLA-A*2402-matched group in the phase II study, where the same
peptide vaccines were used in combination with oxaliplatin-based
chemotherapy (Table V).
 | Table IV.Characteristics of patients in the
phase II study. |
Table IV.
Characteristics of patients in the
phase II study.
|
| HLA-A*2402 |
|
|---|
|
|
|
|
|---|
|
Characteristics | Matched (n=16) | Unmatched
(n=10) | P-value |
|---|
| Sex |
|
|
|
|
Male | 5 | 6 | >0.05 |
|
Female | 11 | 4 |
|
| Age |
|
|
|
|
Mean | 69.8 | 63.5 | >0.05 |
|
Range | 58–82 | 47–76 |
|
| Unresectable
site |
|
|
|
|
Liver | 8 | 7 | >0.05 |
|
Lung | 5 | 5 |
|
|
Dissemination | 1 | 2 |
|
|
Bone | 0 | 1 |
|
| Lymph
node | 3 | 3 |
|
|
Other | 1 | 1 |
|
| Resection of
primary lesion |
|
|
|
|
Yes | 16 | 10 |
|
| No | 0 | 0 |
|
| Chemotherapy |
|
|
|
|
FOLFOX | 15 | 10 | >0.05 |
|
XELOX | 1 | 0 |
|
|
Bevacizumab | 0 | 1 |
|
| Location of
cancer |
|
|
|
|
Colon | 12 | 5 | >0.05 |
|
Rectum | 4 | 5 |
 | Table V.Overall survival according to miRNA
expression (n=16, patients with HLA-A*2402). |
Table V.
Overall survival according to miRNA
expression (n=16, patients with HLA-A*2402).
|
|
|
| Number of
patients |
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| miR rank in the
phase I study | Name of
microRNA | Cut off
valuea | Low expression | High
expression | Hazard ratio | 95% CI | P-value |
|---|
| 1 | miR-142-5p | <1,400 | 8 | 8 | 0.785 | 0.243–2.534 | 0.678 |
| 2 | miR-196b-3p | >180 | 7 | 9 | 0.447 | 0.136–1.463 | 0.178 |
| 3 | miR-196b-5p | >450 | 7 | 9 | 0.099 | 0.014–0.441 | 0.002 |
| 4 | miR-147b | <440 | 8 | 8 | 0.320 | 0.084–1.041 | 0.059 |
| 5 | miR-320b | <6,000 | 10 | 6 | 0.933 | 0.292–3.219 | 0.908 |
| 6 | miR-378a-3p | <6,500 | 9 | 7 | 0.223 | 0.046–0.832 | 0.025 |
| 7 | miR-320e | <5,800 | 7 | 9 | 0.624 | 0.165–2.000 | 0.435 |
| 8 | miR-486-5p | <350 | 9 | 7 | 0.248 | 0.070–0.820 | 0.023 |
| 9 | miR-378c | <1,200 | 5 | 11 | 0.630 | 0.139–2.125 | 0.475 |
| 10 | miR-320a | <6,500 | 9 | 7 | 0.706 | 0.217–2.308 | 0.556 |
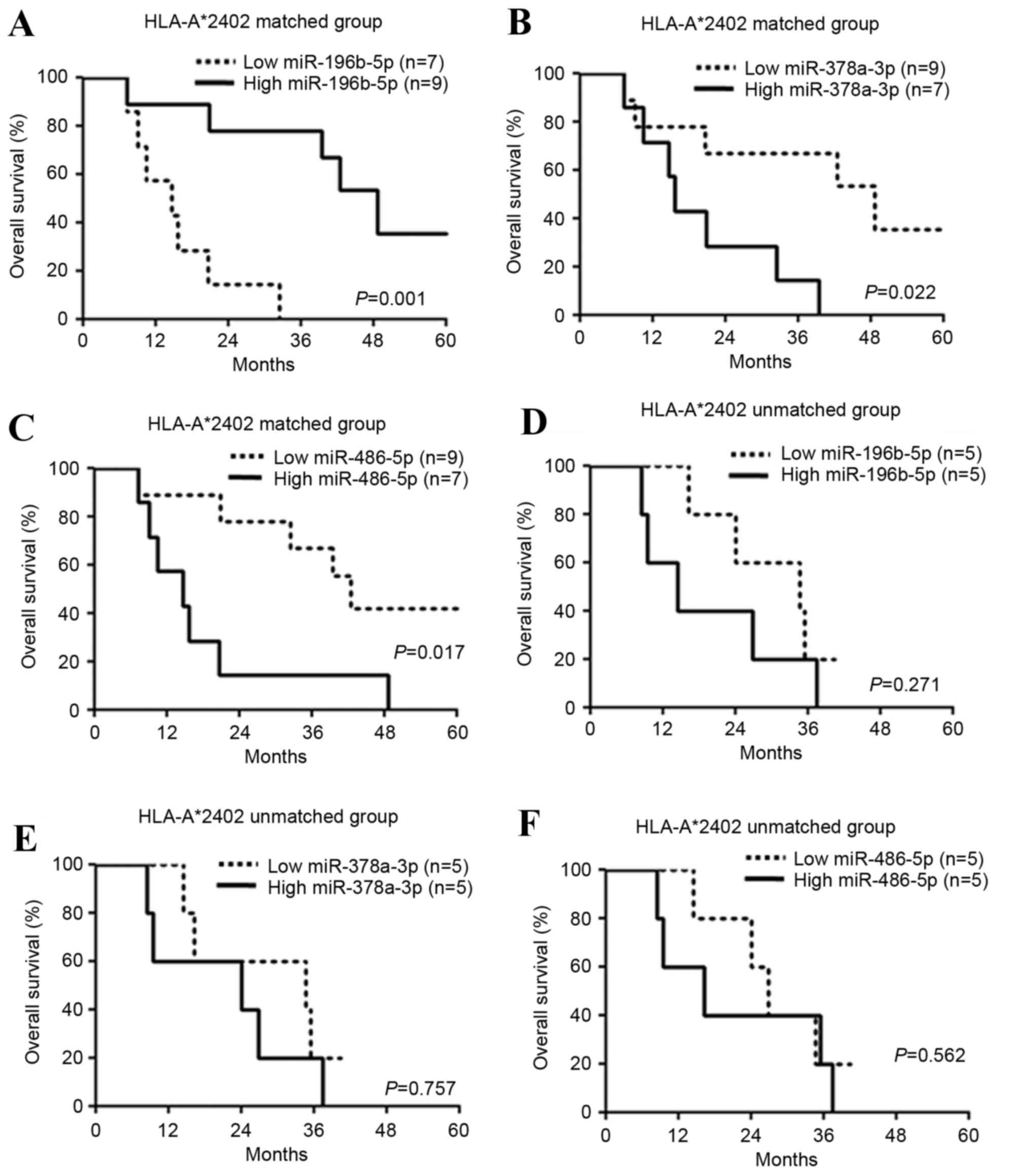
The expression levels of 10 candidate miRNAs were
subsequently examined in 16 patients in the HLA-A*2402-matched
group (Fig. 1). Significantly
improved prognosis was observed in the patients with increased
(>450) miR-196b-5p expression (hazard ratio=0.099, P=0.002,
Table V; P=0.001, Fig. 1A), decreased (<6,500) miR-378a-3p
expression (hazard ratio=0.223, P=0.025, Table V; P=0.022, Fig. 1B) and decreased (<350) miR-486-5p
expression (hazard ratio=0.248, P=0.023, Table V; P=0.017; Fig. 1C) compared with the other patients
with high or low expression of each miRNA. On the other hand, there
was no difference in the prognoses in 10 patients in the
HLA-A*2402-unmatched group according to expression levels of
miR-196b-5p (P=0.271; Fig. 1D),
miR-378a-3p (P=0.757; Fig. 1E) and
miR-486-5p (P=0.562; Fig. 1F).
Discussion
Due to the rapid and impressive progress made in the
field of cancer immunology (23), a
large number of novel vaccine approaches for the treatment of
cancer are being developed (24,25).
However, useful biomarkers that predict the clinical outcome of
immunotherapy remain to be identified (10). In addition, only a small number of
immunological or other biomarkers are available in clinical trials
for immunotherapy (26). The
identification of biomarkers would be useful for selecting
appropriate patient populations for evaluation at earlier stages of
clinical trials and for developing vaccines for cancer treatment
(10,11). In the present study, the efficacy of
novel biomarkers for immunotherapy was investigated by performing
miRNA microarray analysis of cancer tissues and by comparing the
clinical outcomes of phase I and II studies (6,17) testing
a vaccine treatment for metastatic CRC using a cocktail of 5
HLA-A*2402 restricted peptides.
As the first step, miRNA expression patterns in 8
CRC tissues obtained from the patients in the phase I study were
examined, and 10 candidate miRNAs were selected as potential
predictive markers for the efficacy of immunotherapy by comparing
miRNA expression of the responder and non-responder groups, one
with improved prognosis and the other with poorer prognosis, using
Fisher's ratio (Table II).
Subsequently, the expression levels of the 10
candidate miRNAs were examined in 34 cancer tissues (8 cases from
the phase I study and 26 cases from the phase II study) and
compared with the expression levels in 5 normal colorectal tissues.
Upregulation of miR-196b-3p (P=0.011) and miR-196b-5p (P<0.001),
and downregulation of miR-147b (P<0.001), miR-486-5p (P=0.003)
and miR-378c (P=0.056; not significant) were observed in the cancer
tissues (Table III). Cut-off values
for each miRNA were set according to the median expression value in
34 cancer tissues (Table III), and
patients were classified into 2 groups (high and low expression)
for subsequent analysis (Table
V).
The 10 candidate miRNAs were analyzed for
association with prognoses of patients (n=26) enrolled in the phase
II study. HLA-A*2402-restricted peptides were used in the phase II
study. Therefore, the HLA-A*2402-matched group (n=16) was
considered to be the immunological treatment group and the
HLA-A*2402-unmatched group (n=10) the control group. Significantly
improved prognoses were observed in the HLA-A*2402-matched group
with increased (>450) miR-196b-5p expression (hazard
ratio=0.099, P=0.002, Table V;
log-rank test, P=0.001, Fig. 1A),
decreased (<6,500) miR-378a-3p expression (hazard ratio=0.223,
P=0.025, Table V; log-rank test,
P=0.022, Fig. 1B) and decreased
(<350) miR-486-5p expression (hazard ratio=0.248, P=0.023,
Table V; log-rank test, P=0.017,
Fig. 1C) compared with the other
patients with high or low expression levels of each miRNA. However,
there was no difference in prognosis in the HLA-A*2402-unmatched
group (n=10) according to the expression of miR-196b-5p (P=0.271;
Fig. 1D), miR-378a-3p (P=0.757;
Fig. 1E) and miR-486-5p (P=0.562;
Fig. 1F). These results indicated
that these miRNAs may be used as biomarkers for immunotherapy.
The upregulation of miR-196b has previously been
reported in lung adenocarcinoma, oral cancer, CRC, leukemia and
lymphoma, and has been associated with the malignant phenotype of
these types of cancer (27–30). By contrast, the results of the present
study indicated that high miR-196b expression appeared to be
associated with improved prognosis for patients treated with
immunotherapy (Tables II and
V; Fig.
1A). High miR-196b expression levels may be associated with the
augmentation of the immune response to cancer, which may explain
these paradoxical results. miR-196b has previously been reported to
be a potential target of inflammatory signals, as it is known to be
overexpressed in the inflammatory epithelial lesions of patients
with Crohn's disease (31).
The cancer tissues used in the present study may
contain not only cancer cells but also tumor infiltrating immune
cells, as this has previously been suggested to be a biomarker for
improved prognosis of CRC (32). The
results of the present study indicated that high miR-196b
expression may be used as a biomarker for predicting whether
patients respond to immunotherapy, although the function of
miR-196b requires clarification through further investigations.
miR-378a has been reported as a tumor suppressor and
to be downregulated in CRC (33).
However, the results of the present study indicated that reduced
expression levels of miR-378a-3p were significantly associated with
improved prognosis (Table V; Fig. 1C). miR-378a was also reported to
promote cell survival, tumor growth and angiogenesis through
repression of the expression of two tumor suppressors, SUFU
negative regulator of hedgehog signaling and FUS RNA binding
protein (34). As miR-378a is able to
act as oncogene or a tumor suppressor, its function is complicated
(33,34). Therefore, it is essential to further
investigate the function of miR-378a-3p in the future.
miR-486 has been reported to be a cancer-suppressor
miRNA, and it is often downregulated in a number of cancer types in
multiple organs (35,36). Downregulation of miR-486 has been
reported to contribute to tumor progression and metastasis
(35). However, the results of the
present study indicated that low expression of miR-486-5p was
significantly associated with improved prognosis (Table V; Fig.
1E). Since miR-486 has been reported to sustain the activity of
nuclear factor-κB (NF-κB) by disrupting multiple NF-κB-negative
feedback loops (37) and suppressing
the antitumor immunity of the host (38,39).
Downregulation of miR-486 may reduce NF-κB expression and suppress
immune-suppressive cells in the microenvironment of the tumor.
In conclusion, high miR-196b-5p, low miR-378a-3p and
low miR-486-5p expression levels may be considered as novel
biomarkers to predict the efficacy of immunotherapy. However, the
results of the present study are preliminary, and the function of
miRNAs in immune responses remain unclear. In a planned phase III
study, it may be applicable to use these miRNAs as biomarkers to
assess the selection of patients who may expect an improved outcome
with the vaccine treatment.
Acknowledgements
The present study was part of the research program
of the Project for Development of Innovative Research on Cancer
Therapeutics, the Japan Agency for Medical Research and
Development. The authors would like to thank Dr. Takuya Tsunoda and
Dr. Koji Yoshida (Laboratory of Molecular Medicine, Human Genome
Center, Institute of Medical Science, The University of Tokyo,
Tokyo, Japan) for their excellent advice and cooperation and
providing all the peptides.
Glossary
Abbreviations
Abbreviations:
|
miRNA/miR
|
microRNA
|
|
HLA
|
human leukocyte antigen
|
|
CRC
|
colorectal cancer
|
|
CTL
|
cytotoxic T lymphocytes
|
|
RNF43
|
ring finger protein 43
|
|
TOMM34
|
34 kDa-translocase of the outer
mitochondrial membrane
|
|
KOC1
|
insulin-like growth factor-II mRNA
binding protein 3
|
|
VEGFR
|
vascular endothelial growth factor
receptor
|
|
OS
|
overall survival
|
|
RD
|
recommended dose
|
|
IFA
|
incomplete Freund's adjuvant
|
|
NF-κB
|
nuclear factor-κB
|
References
|
1
|
Zaharie F, Muresan MS, Petrushev B, Berce
C, Gafencu GA, Selicean S, Jurj A, Cojocneanu-Petric R, Lisencu CI,
Pop LA, et al: Exosome-carried microRNA-375 inhibits cell
progression and dissemination via Bcl-2 blocking in colon cancer. J
Gastrointestin Liver Dis. 24:435–443. 2015.PubMed/NCBI
|
|
2
|
Takeyama H, Yamamoto H, Yamashita S, Wu X,
Takahashi H, Nishimura J, Haraguchi N, Miyake Y, Suzuki R, Murata
K, et al: Decreased miR-340 expression in bone marrow is associated
with liver metastasis of colorectal cancer. Mol Cancer Ther.
13:976–985. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Konno Y, Dong P, Xiong Y, Suzuki F, Lu J,
Cai M, Watari H, Mitamura T, Hosaka M, Hanley SJ, et al:
MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation,
invasion and stem cell-like phenotype of aggressive endometrial
cancer cells. Oncotarget. 5:6049–6062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Achberger S, Aldrich W, Tubbs R, Crabb JW,
Singh AD and Triozzi PL: Circulating immune cell and microRNA in
patients with uveal melanoma developing metastatic disease. Mol
Immunol. 58:182–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okada H, Kohanbash G and Lotze MT:
MicroRNAs in immune regulation-opportunities for cancer
immunotherapy. Int J Biochem Cell Biol. 42:1256–1261. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hazama S, Nakamura Y, Takenouchi H, Suzuki
N, Tsunedomi R, Inoue Y, Tokuhisa Y, Iizuka N, Yoshino S, Takeda K,
et al: A phase I study of combination vaccine treatment of five
therapeutic epitope-peptides for metastatic colorectal cancer;
safety, immunological response, and clinical outcome. J Transl Med.
12:632014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suzuki N, Hazama S, Ueno T, Matsui H,
Shindo Y, Iida M, Yoshimura K, Yoshino S, Takeda K and Oka M: A
phase I clinical trial of vaccination with KIF20A-derived peptide
in combination with gemcitabine for patients with advanced
pancreatic cancer. J Immunother. 37:36–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
U.S Department of Health and Human
Services FaDA, . October;2011.U.S. Department of Health and Human
Services, Food and Drug Administration, October 2011: Guidance for
Industry. Clinical Considerations for Therapeutic Cancer Vaccines.
2011.
|
|
9
|
Nagorsen D and Thiel E: Clinical and
immunologic responses to active specific cancer vaccines in human
colorectal cancer. Clin Cancer Res. 12:3064–3069. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Copier J, Whelan M and Dalgleish A:
Biomarkers for the development of cancer vaccines: Current status.
Mol Diagn Ther. 10:337–343. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Whiteside TL: Immune responses to cancer:
Are they potential biomarkers of prognosis? Front Oncol. 3:1072013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yagyu R, Furukawa Y, Lin YM, Shimokawa T,
Yamamura T and Nakamura Y: A novel oncoprotein RNF43 functions in
an autocrine manner in colorectal cancer. Int J Oncol.
25:1343–1348. 2004.PubMed/NCBI
|
|
13
|
Shimokawa T, Matsushima S, Tsunoda T,
Tahara H, Nakamura Y and Furukawa Y: Identification of TOMM34,
which shows elevated expression in the majority of human colon
cancers, as a novel drug target. Int J Oncol. 29:381–386.
2006.PubMed/NCBI
|
|
14
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: Identification of genes for prediction of
lymph-node metastasis and sensitivity to anti-cancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Olofsson B, Korpelainen E, Pepper MS,
Mandriota SJ, Aase K, Kumar V, Gunji Y, Jeltsch MM, Shibuya M,
Alitalo K and Eriksson U: Vascular endothelial growth factor B
(VEGF-B) binds to VEGF receptor-1 and regulates plasminogen
activator activity in endothelial cells. Proc Natl Acad Sci USA.
95:pp. 11709–11714. 1998; View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Millauer B, Wizigmann-Voos S, Schnurch H,
Martinez R, Møller NP, Risau W and Ullrich A: High affinity VEGF
binding and developmental expression suggest Flk-1 as a major
regulator of vasculogenesis and angiogenesis. Cell. 72:835–846.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hazama S, Nakamura Y, Tanaka H, Hirakawa
K, Tahara K, Shimizu R, Ozasa H, Etoh R, Sugiura F, Okuno K, et al:
A phase II study of five peptides combination with
oxaliplatin-based chemotherapy as a first-line therapy for advanced
colorectal cancer (FXV study). J Transl Med. 12:1082014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Uchida N, Tsunoda T, Wada S, Furukawa Y,
Nakamura Y and Tahara H: Ring finger protein 43 as a new target for
cancer immunotherapy. Clin Cancer Res. 10:8577–8586. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suda T, Tsunoda T, Daigo Y, Nakamura Y and
Tahara H: Identification of human leukocyte antigen-A24-restricted
epitope peptides derived from gene products upregulated in lung and
esophageal cancers as novel targets for immunotherapy. Cancer Sci.
98:1803–1808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishizaki H, Tsunoda T, Wada S, Yamauchi M,
Shibuya M and Tahara H: Inhibition of tumor growth with
antiangiogenic cancer vaccine using epitope peptides derived from
human vascular endothelial growth factor receptor 1. Clin Cancer
Res. 12:5841–5849. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wada S, Tsunoda T, Baba T, Primus FJ,
Kuwano H, Shibuya M and Tahara H: Rationale for antiangiogenic
cancer therapy with vaccination using epitope peptides derived from
human vascular endothelial growth factor receptor 2. Cancer Res.
65:4939–4946. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iizuka N, Oka M, Yamada-Okabe H, Nishida
M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al:
Oligonucleotide microarray for prediction of early intrahepatic
recurrence of hepatocellular carcinoma after curative resection.
Lancet. 361:923–929. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boon T and van der Bruggen P: Human tumor
antigens recognized by T lymphocytes. J Exp Med. 183:725–729. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Okabe H, Satoh S, Kato T, Kitahara O,
Yanagawa R, Yamaoka Y, Tsunoda T, Furukawa Y and Nakamura Y:
Genome-wide analysis of gene expression in human hepatocellular
carcinomas using cDNA microarray: Identification of genes involved
in viral carcinogenesis and tumor progression. Cancer Res.
61:2129–2137. 2001.PubMed/NCBI
|
|
25
|
Okuno K, Sugiura F, Itoh K, Yoshida K,
Tsunoda T and Nakamura Y: Recent advances in active specific cancer
vaccine treatment for colorectal cancer. Curr Pharm Biotechnol.
13:1439–1445. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kijima T, Hazama S, Tsunedomi R, Tanaka H,
Takenouchi H, Kanekiyo S, Inoue Y, Nakashima M, Iida M, Sakamoto K,
et al: Micro RNA-6826 and −6875 in plasma are valuable non-invasive
biomarkers that predict the efficacy of vaccine treatment against
metastatic colorectal cancer. Oncol Rep. 37:23–30. 2017.PubMed/NCBI
|
|
27
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu YC, Chang JT, Liao CT, Kang CJ, Huang
SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al:
OncomiR-196 promotes an invasive phenotype in oral cancer through
the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 13:2182014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ge J, Chen Z, Li R, Lu T and Xiao G:
Upregulation of microRNA-196a and microRNA-196b cooperatively
correlate with aggressive progression and unfavorable prognosis in
patients with colorectal cancer. Cancer Cell Int. 14:1282014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan Y, Meng M, Zhang G, Han H and Zhou Q:
Oncogenic microRNAs in the genesis of leukemia and lymphoma. Curr
Pharm Des. 20:5260–5267. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen WX, Ren LH and Shi RH: Implication of
miRNAs for inflammatory bowel disease treatment: Systematic review.
World J Gastrointest Pathophysiol. 5:63–70. 2014.PubMed/NCBI
|
|
32
|
Galon J, Mlecnik B, Bindea G, Angell HK,
Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, et
al: Towards the introduction of the ‘Immunoscore’ in the
classification of malignant tumours. J Pathol. 232:199–209. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang GJ, Zhou H, Xiao HX, Li Y and Zhou
T: MiR-378 is an independent prognostic factor and inhibits cell
growth and invasion in colorectal cancer. BMC Cancer. 14:1092014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee DY, Deng Z, Wang CH and Yang BB:
MicroRNA-378 promotes cell survival, tumor growth and angiogenesis
by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA.
104:pp. 20350–20355. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen H, Ren C, Han C, Wang D, Chen Y and
Fu D: Expression and prognostic value of miR-486-5p in patients
with gastric adenocarcinoma. PLoS One. 10:e01193842015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang J, Tian X, Han R, Zhang X, Wang X,
Shen H, Xue L, Liu Y, Yan X, Shen J, et al: Downregulation of
miR-486-5p contributes to tumor progression and metastasis by
targeting protumorigenic ARHGAP5 in lung cancer. Oncogene.
33:1181–1189. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song L, Lin C, Gong H, Wang C, Liu L, Wu
J, Tao S, Hu B, Cheng SY, Li M and Li J: miR-486 sustains NF-κB
activity by disrupting multiple NF-κB-negative feedback loops. Cell
Res. 23:274–289. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fisher DT, Appenheimer MM and Evans SS:
The two faces of IL-6 in the tumor microenvironment. Semin Immunol.
26:38–47. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gajewski TF, Schreiber H and Fu YX: Innate
and adaptive immune cells in the tumor microenvironment. Nat
Immunol. 14:1014–1022. 2013. View Article : Google Scholar : PubMed/NCBI
|