Introduction
Hepatocellular carcinoma (HCC) is one of the most
common cancer types and a major cause of cancer-associated
mortality worldwide (1). The majority
of HCC cases are caused by chronic hepatitis B or hepatitis C
infections (2). HCC, characterized by
rapid recurrence and poor survival, remains a challenging disease
to treat (3). As HCC is not sensitive
to radiotherapy or chemotherapy, surgery is the only effective
treatment (4). However, the rate of
recurrence is high and metastasis is common following surgery,
which leads to the poor prognosis for HCC (3). Therefore, it is necessary to understand
the molecular mechanisms underlying the growth and metastasis of
HCC, which may help to identify effective diagnosis and therapeutic
targets to improve the survival. However, the associated molecular
mechanisms of HCC progression are not well understood.
The R-spondin (RSPO) protein family consists of four
homologous members, which are evolutionarily conserved in
vertebrates and are involved in a broad range of developmental and
physiological processes (5,6): RSPO1 is important for sex determination
(7); RSPO2 is required for limb,
laryngeal-tracheal and lung development (8); RSPO3 is critical for placental formation
(9); and the mutation of RSPO4
results in inherited anonychia (10).
The association between RSPO and cancer has not been extensively
studied. It has been reported that RSPO2 and RSPO3 insertional
activation is observed in the mouse mammary tumor virus model
system (11,12). Administration of RSPO1 protein to mice
induces rapid crypt cell proliferation, which causes a marked
increase in the size of the small intestine (13). Seshagiri et al (14) identified that RSPO2 and RSPO3
transcript fusion occurs in 10% of colon tumors. In addition, it
was found that RSPO fusions occur exclusively in tumors without
adenomatous polyposis coli mutations, indicating that RSPO genes
have a role in the activation of Wnt signaling and tumorigenesis
(14). Subsequently, RSPO gene
fusions were also observed in a subset of colon tumors in the
Japanese population, and forced expression of the RSPO gene was
revealed to increase the growth of colorectal cells (15).
A number of previous studies have indicated that
RSPOs may potentiate Wnt signaling via stimulation of the
leucine-rich repeat-containing G-protein coupled receptors (LGR) 4,
LGR5 and LGR6 (16–18). Wnt proteins, comprising a large family
of extracellular, lipid-modified glycoproteins, are crucial for
embryonic development and cell proliferation, regulation,
differentiation, survival and tissue homeostasis in adults
(19,20). It was demonstrated that RSPOs
cooperate with Wnts during development, particularly by promoting
the transcriptional activity of β-catenin, which is an important
mechanism of the Wnt signaling pathway (5,16). It has
been revealed that canonical Wnt/β-catenin signaling serves an
important role in numerous cancer types, including lung, breast,
brain, colorectal and liver tumors (21–23).
Interaction between RSPOs and LGR4, as well as the intracellular
signaling proteins, promotes phosphorylation of LRP5/6, stabilizes
β-catenin expression and promotes its transcriptional activity
(18). Activation of Wnt/β-catenin
results in the increased expression level of its target genes,
including cyclin D1 and c-Myc, which are important for driving
tumorigenesis in numerous types of cancer (24).
In the present study, high expression of RSPO2 was
observed in certain HCC cell lines. Tissue microarray also revealed
that the expression of RSPO2 is increased in primary tumors
compared with the adjacent normal tissues. Functional study
revealed that overexpression of RSPO2 enhances the cell
proliferation and anchorage-independent growth of the human liver
QGY7703 cancer cell line. In addition, RSPO2 overexpression may
promote cell motility, which was demonstrated using Transwell and
wound healing assays. Similar to a previous study (25), the present study also revealed that
overexpression of RSPO2 is involved in Wnt/β-catenin activation via
increasing the expression of β-catenin and the downstream gene
c-Myc. The present study revealed the functional role of RSPO2 in
HCC and indicated that RSPO2 may be a potential drug target for
patients with liver tumors.
Materials and methods
Patients and liver tissue samples
A total of 72 human liver tissues were obtained from
24 patients with HCC who had undergone surgical resection between
January 2013 and December 2014 at the First Affiliated Hospital of
Jiaxing College (Jiaxing, China) with written informed consent and
ethical approval from the Local Ethics Committee of Jiaxing
College. Patients did not receive radiotherapy or chemotherapy
prior to surgery.
Tissue microarray
Tissue samples (24 tumor tissues, 24 paired
non-tumor adjacent tissues and 24 normal tissues from 24 HCC
patients) were formalin-fixed and paraffin-embedded. The samples
were used to construct tissue microarrays (TMAs) using a Beecher
Instrument (Sun Prairie, WI, USA) as described previously (26). A total of 3 tissue cylinders of 0.6 mm
in diameter were punched from each sample. Subsequent to sectioning
(4 mm for each section), tissue slides were baked at 60°C for 2 h,
and kept at 4°C for subsequent analysis. TMAs were stained with
hematoxylin and eosin (H&E). A trained pathologist reevaluated
H&E-stained samples to determine the tumor stage and grade
according to the WHO criteria (27).
The slides with tissue sections were subjected to
immunohistochemical detection of RSPO2, using the primary antibody
against RSPO2 (dilution, 1:50; cat. no. ab73761; Abcam, Cambridge,
MA, USA). Following rinsing with 1X PBS, slides were incubated with
biotinylated anti-rabbit IgG for 30 min (dilution, 1:1,000) using
Vectastain Elite ABC kit (cat. no. PK-6100; Vector Laboratories,
Inc., Burlingame, CA, USA). The detection was achieved using the
avidin-biotin peroxidase method with a diaminobenzidine chromogen
kit (cat. no. SK-4100; Vector Laboratories, Inc.). The TMAs were
evaluated for RSPO2 expression by a trained pathologist and were
scored as strong (+++) with >80% positive cells, moderate (++)
with 30–80% positive cells, weak (+) with observed <30% stained
cells or absent (−). The subcellular localization of the staining
was noted for each score using CI microscopy by NIS-Elements F
under ×100 magnification.
Cell culture
The QSG-7701 cell line derived from human normal
liver tissue and the human hepatoma cell lines Bel7404, QGY7703,
HepG2, Huh7, 293T and Hep3B were purchased from the Cell Bank of
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). QSG-7701 and QGY7703 cells were cultured in
RPMI-1640 (cat. no. SH30809.01; HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) medium containing 10% fetal bovine serum
(FBS) (cat. no. 10099141; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Bel7404, HepG2, Huh7 and Hep3B cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; cat. no.
SH30243.01B; HyClone; GE Healthcare Life Sciences) with high
glucose, supplemented with 10% FBS. All cells were cultured in an
atmosphere of 95% air and 5% CO2 under humidified
conditions.
Western blot analysis
All cultured cells were lysed using
radioimmunoprecipitation assay lysis and extraction buffer (cat.
no. 89900; Thermo Fisher Scientific, Inc.). Total proteins were
subjected to concentration determination using a commercial
bicinchoninic acid quantification kit, according to the
manufacturer's protocol (cat. no. 23227; Thermo Fisher Scientific,
Inc.).
Lysates from QGY7703 cells or HCC tissue (15 µg)
were subjected to 12% SDS-PAGE for protein separation and then
electrophoretically transferred to nitrocellulose membranes (Axygen
Scientific, Union City, CA, USA). Subsequent to being blocked by
PBS containing 5% fat-free milk, the nitrocellulose membranes were
incubated with rabbit polyclonal antibody for RSPO2 (cat. no.
ab73761; dilution, 1:1,000; Abcam), β-catenin (cat. no. 8480;
dilution, 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA), c-Myc (cat. no. 5605; dilution, 1:1,000; Cell Signaling
Technology, Inc.), vascular endothelial growth factor (VEGF)-A
(cat. no. ab51745; dilution, 1:1,000; Abcam), phosphorylated signal
transducer and activator of transcription 3 (p-STAT3) (cat. no.
9130; dilution, 1:1,000; Cell Signaling Technology, Inc.), leptin
(cat. no. ab3583; dilution, 1:1,000; Abcam), p21 (cat. no. SC-397;
dilution, 1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
and rabbit polyclonal antibody for β-actin (cat. no. 10303001;
dilution, 1:5,000; Harmonious One Biotechnology, Shanghai, China)
overnight at 4°C and then incubated with horseradish
peroxidase-conjugated rabbit IgG (cat. no. 7074; dilution, 1:3,000;
Cell Signaling Technology, Inc.) for 1.5 h at room temperature. The
immunolabeled proteins were detected using a commercial enhanced
chemiluminescent detection kit (cat. no. 108070002; Harmonious One
Biotechnology, Shanghai, China). Results were quantified by a
luminescent digital image analyzer Bio-Spectrum600 (UVP; Upland,
CA, USA). Band intensity was assessed using a Gel-Pro analyzer
(V6.3; Media Cybernetics, Inc., Rockville, MD, USA).
Lentivirus vector construction
The total RNA from 5×106 HepG2 cells was
extracted using the phenol-chloroform method following TRIzol (cat.
no. 15596-026; Invitrogen; Thermo Fisher Scientific, Inc.) lysis,
according to the manufacturer's protocol. The cDNA was prepared by
reverse transcription using a random primer (D3801; Takara
Biotechnology Co., Ltd., Dalian, China) at 37°C for 1 h, following
the denaturation of total RNA by heating for 5 min at 37°C,
followed by immediate chilling on ice. The full coding region of
the RSPO2 gene was isolated following denaturation at 95°C for 5
mint, 36 cycles of 95°C for 30 sec, 56°C for 30 sec and 72°C for 45
sec, and then subjected to post-elongation for 10 min at 72°C. The
mixture for polymerase chain reaction (PCR) was composed of 5 µl of
10X PCR buffer, 0.2 mM dNTP, 0.2 µM RSPO2-EcoRI-F primer
(5′-CCGGAATTCATGCAGTTTCGCCTTTTCTC-3′), 0.2 µM RSPO2-BamHI-R primer
(5′-CGCGGATCCTTATTGGTTAGCTCTGTCTGTAGC-3′), 2 U PFU polymerase (cat.
no. 101060002; Harmonious One Biotechnology) and sterile distilled
water. The human RSPO2 gene was then subcloned into EcoRI
and BamHI sites of the lentiviral vector pLV-mCherry (2A)
puro (cat. no. VL3405; Inovogen Biotechnology Pvt. Ltd., Beijing,
China) with the selective marker gene puromycin by a classic
ligation and transformation method using T4 ligase (cat. no.
EL0011; Fermentas; Thermo Fisher Scientific, Inc.) and DH5α
chemical competent E. coli cells (cat. no. C502-03; Vazyme,
Piscataway, NJ, USA).
Lentivirus production and cells
transduction
Packaging of pseudotyped recombinant lentivirus was
performed by transfection of 293T cells. Briefly,
1.5×106 293T cells were plated in a 6-cm dish and
cultured for 20 h. The cells were then cotransfected with either
1.7 µg pLV-mCherry (2A) puro or pLV-mCherry (2A) puro-RSPO2, 1.13
µg pCMV Δ8.91 and 0.57 µg pMD.G (a gift from Institute of
Biochemistry and Cellular Biology, Shanghai, China) using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
following the manufacturer's protocol. The supernatant containing
the lentivirus was harvested at 72 h and filtered through a 0.45 µm
low protein binding polysulfonic filter (EMD Millipore, Billerica,
MA, USA). QGY7703 cells were inoculated in 6-well plates in advance
at a density of 2×105 cells per well and presented with
~40% confluence following incubation for 20 h at 37°C. The cells
were then infected with 1 ml lentivirus suspension in the presence
of 8 µg/ml polybrene (Chemicon; EMD Millipore). Following
transduction for 48 h at 37°C, QGY7703 cells were selected with 2.0
µg/ml puromycin for 10 days when all the blank control cells
without transfection were eradicated. The selected cells were used
for growth, anchorage-independent growth and migration assays.
Viability analysis
The human hepatoma QGY7703 cell line with stable
overexpression of RSPO2 and the control QGY7703 cells transfected
with empty vectors were seeded onto 96-well plates at the density
of 2.0×103 per well. Cells were analyzed using an MTT
assay at day 1, 2, 3, 4 and 5 subsequent to cell seeding. Briefly,
100 µg of MTT (cat. no. 0793–1G; Amresco, LLC, Solon, OH, USA) was
added to each well. Following incubation for 4 h at 37°C, the
purple formazan crystals generated from viable cells were dissolved
by adding 100 µl of dimethyl sulfoxide to each well. The absorbance
of each well was then read at 570 nm.
Anchorage-independent growth
analysis
QGY7703 cells transfected with pLV-mCherry (2A)
puro-RSPO2 or pLV-mCherry (2A) puro were trypsinized and suspended
in culture medium. The cells were seeded on each well of a 24-well
plate with complete medium at the density of 1.0×103
cells. Following growth for 10 days, the cells were subjected to
fixation by methanol for 10 min and then stained by 0.1% crystal
violet at room temperature for 30 min. Following removal of the
dye, colonies containing >50 cells were counted in five random
fields using TiS microscopy by NIS-Elements Viewer (version 4.2;
Nikon, Tokyo, Japan) under ×100 magnification.
Migration assay
A 24-well Transwell chamber with 8.0 µm pore size
(Costar; Corning Incorporated, Corning, NY, USA) was used for the
migration assay. The pre-balance of the Transwell chamber was
performed by adding DMEM without FBS into the upper and bottom
chamber overnight at 37°C. QGY7703 cells transfected with
pLV-mCherry (2A) puro-RSPO2 or pLV-mCherry (2A) puro were
trypsinized and suspended in 200 µl serum-free medium and were
seeded in the upper chamber at a density of 1.5×105
cells per well. The bottom chamber was filled with DMEM containing
10% FBS. Following incubation for 24 and 48 h at 37°C, the
non-migrating cells were removed by soft scratch with small cotton
swabs and rinsed with 1X PBS. Migrated cells were then dried, fixed
with methanol and stained with 0.1% crystal violet at room
temperature for 30 min. The transmembrane cells were counted using
a TiS microscopy by NIS-Elements Viewer (version 4.2, Nikon) under
×100 magnification.
Wound healing assay
RSPO2-overexpressed QGY7703 cells or control cells
were plated onto a 24-well plate with 4×106 per well and
incubated for 24 h at 37°C. Linear scratch wounds were then
produced using a 10 µl pipette tip on the confluent cell monolayer.
The medium was replaced with the serum-free medium. Images were
captured at 0, 12, 24, 36 and 72 h and the wounding size was
quantified and analyzed by NIS-Elements Viewer (version 4.2,
Nikon).
Statistical analysis
Data are presented as the mean ± standard deviation.
A two-tailed Student's t-test was employed to evaluate the
differences between groups. P<0.05 was considered to indicate a
statistically significant. The differences of indexes between tumor
tissues and paired non-tumor adjacent tissues were analyzed using
the Wilcoxon signed-rank test. Differences between groups were
analyzed by the Mann-Whitney U test. Data were processed with R
Studio (v1.0; Boston, MA, USA).
Results
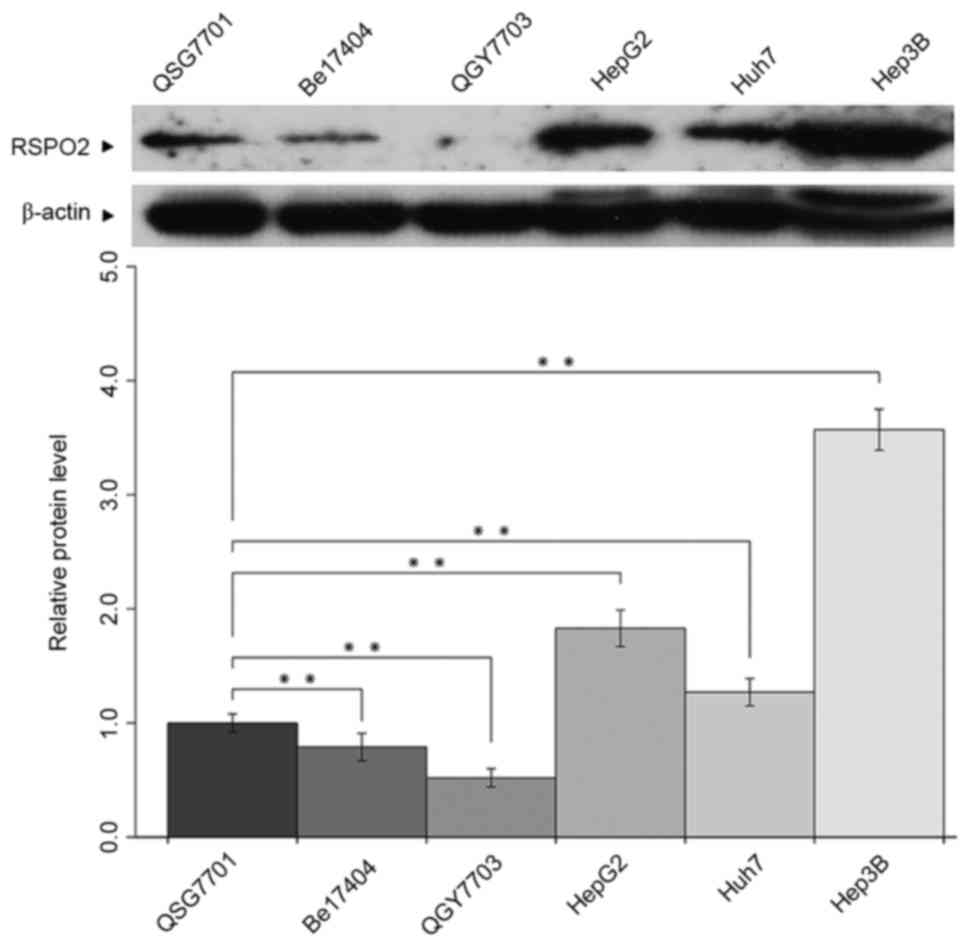
Expression of RSPO2 in various HCC
cells
The expression level of RSPO2 was detected in
various HCC cells by western blot analysis. As presented in
Fig. 1, increased expression levels
of RSPO2 were observed in HepG2, Huh7 and Hep3B cells compared with
the human normal liver QSG-7701 cell line. However, markedly
decreased expression of RSPO2 was found in the human hepatoma
Bel7404 and QGY7703 cell lines compared with QSG-7701 cells.
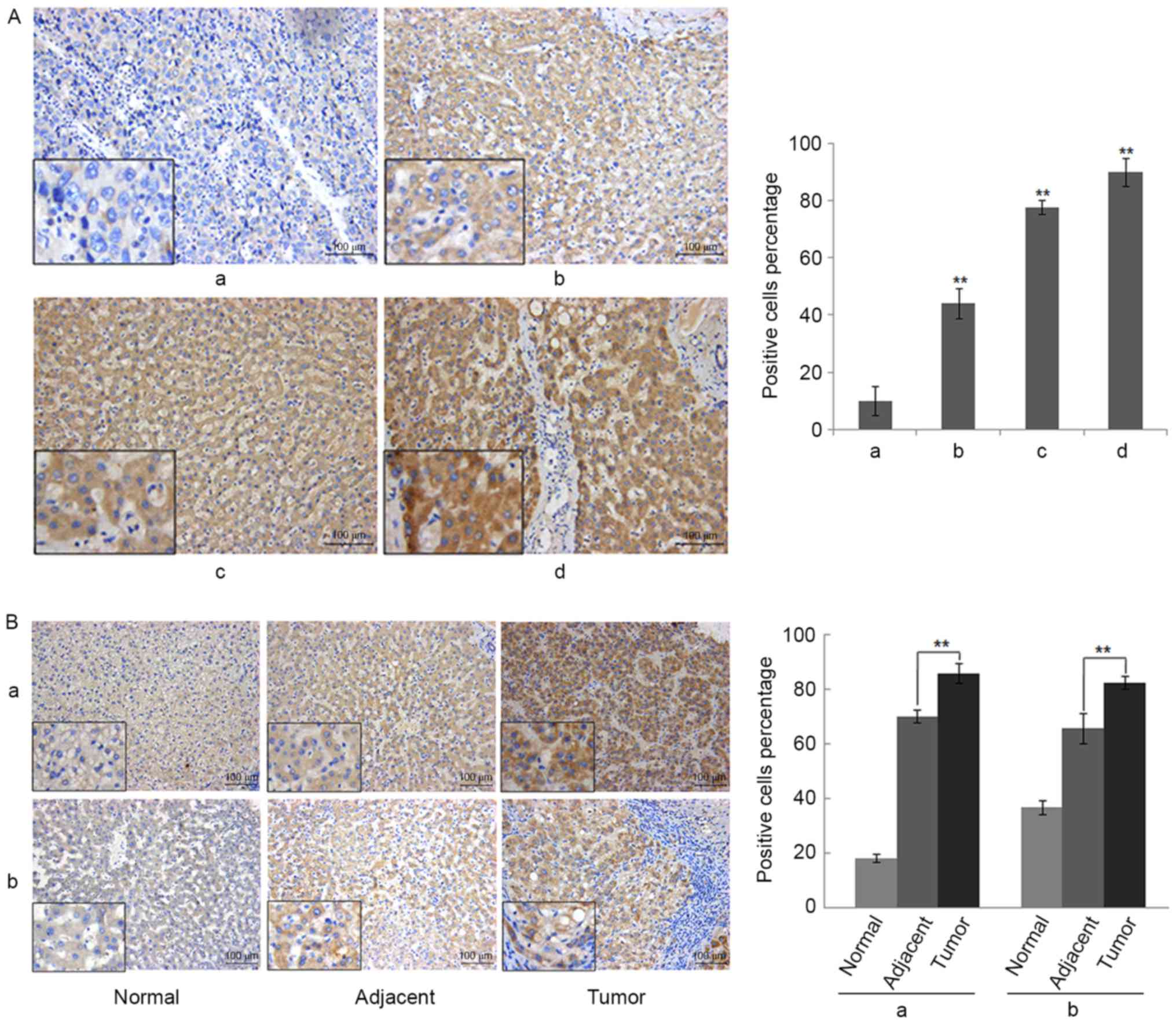
Overexpression of RSPO2 in liver
cancer tissues
The expression of RSPO2 in clinical samples of HCC
was analyzed. A total of 24 pairs of human hepatic carcinoma and
matched adjacent non-tumor tissues or normal tissues were examined
using immunohistochemical staining with an antibody against human
RSPO2. Samples were considered RSPO2-positive if either the cell
nucleus or cytoplasm stained positive. As presented in Fig. 2A and B, the staining of RSPO2 was
primarily observed in the cytoplasm of cancer cells. Fig. 2A presents representative examples from
the tissue microarray for each RSPO2 staining score, ranging from 0
to +++. Intense expression of RSPO2 in tumor tissue (score ++ or
+++) was identified in 16/24 patients (66.7%), whereas in other
patients (33.3%) a weak immunoreactivity (score +) was detected.
However, the expression of RSPO2 in non-tumor adjacent tissues was
significantly lower compared with tumor tissues (Wilcoxon
signed-rank test, P=0.007). Moderate expression of RSPO2 (score ++)
in non-tumor adjacent tissue was found in 6 of 24 patients (25%),
whereas the remaining 75% exhibited weak expression of RSPO2 (score
+). Similar expression patterns were found in the paired normal
tissues. The expression of RSPO2 in the paired normal tissues was
significantly lower compared with tumor tissues (Wilcoxon
signed-rank test, P=0.001). Representative images of RSPO2
expression in the paired normal, adjacent and tumor tissues are
shown in Fig. 2A.
Statistical analyses were performed to examine the
association between RSPO2 expression and the clinicopathological
characteristics of hepatic carcinoma. As shown in Table I, no association was observed between
the expression of RSPO2 and patient age or gender, tumor grading
and tumor staging in patients with HCC. A small sample size (n=24)
may be the reason that no statistically significant results were
identified.
 | Table I.Characteristics and RSO2 expression of
patients with hepatocellular carcinoma. |
Table I.
Characteristics and RSO2 expression of
patients with hepatocellular carcinoma.
|
|
| RSPO2 expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Patients, n (%) | Absent or weak,
n | Moderate to high,
n | P-value |
|---|
| Age |
|
|
| 0.155 |
| ≤50
years | 13 (54) | 6 | 7 |
|
| >50
years | 11 (46) | 2 | 9 |
|
| Gender |
|
|
| 0.766 |
|
Male | 18 (75) | 7 | 11 |
|
|
Female | 6 (25) | 1 | 5 |
|
| Tumor grading |
|
|
| 0.286 |
| I | 3 (3) | 0 | 3 |
|
| II | 19 (79) | 7 | 12 |
|
|
III | 2 (8) | 1 | 1 |
|
| Tumor staging |
|
|
| 0.996 |
| II | 4 (17) | 1 | 3 |
|
|
III | 20 (83) | 7 | 13 |
|
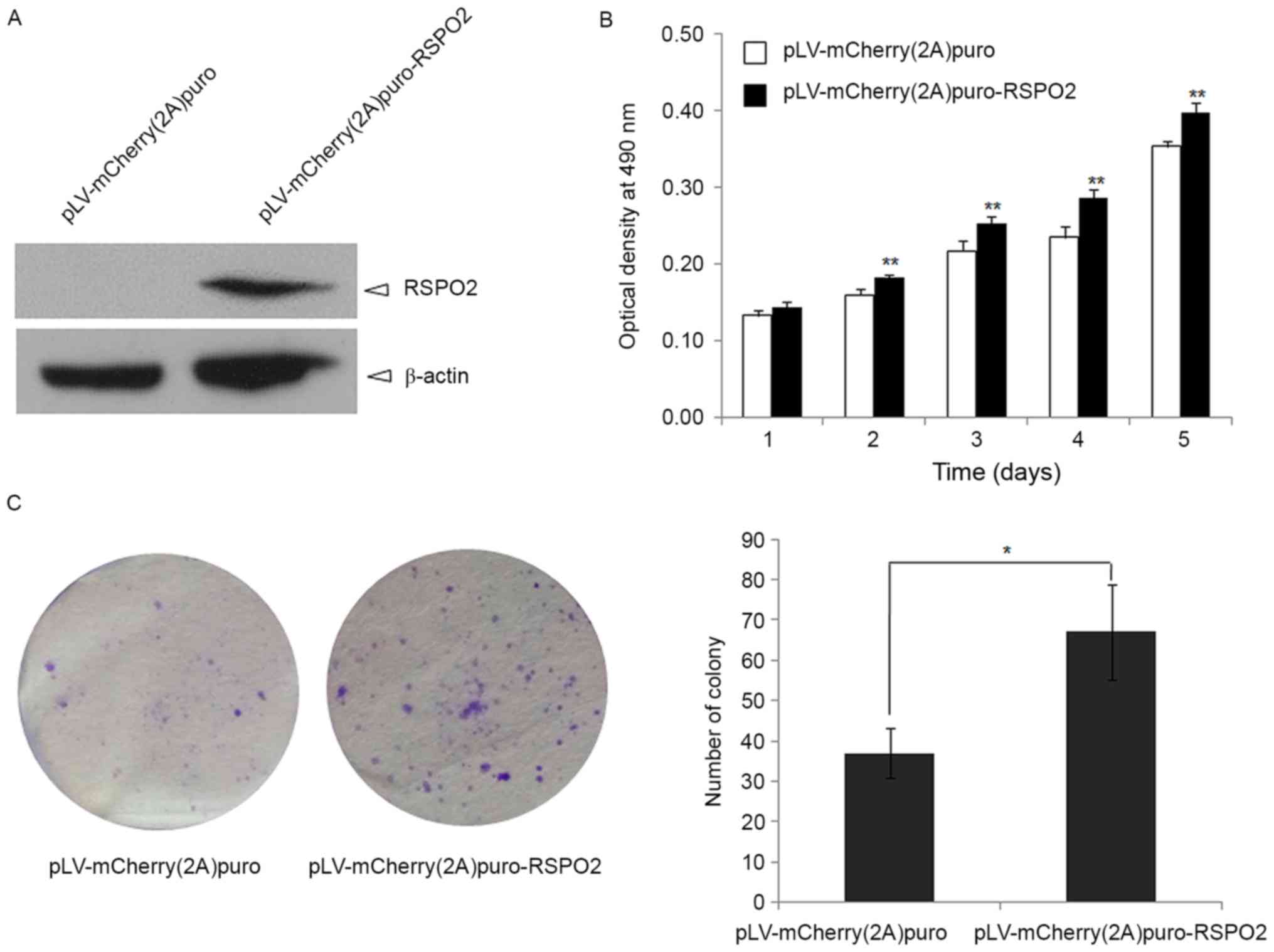
RSPO2 overexpression enhances the
proliferation and anchorage-independent growth of QGY7703
cells
To define the function of RSPO2 in HCC, RSPO2
overexpression in the QGY7703 cell line was achieved by lentivirus
delivery (Fig. 3A). The cell growth
of QGY7703 cells with stable overexpression of RSPO2 was firstly
evaluated by MTT assay. RSPO2 overexpression significantly promoted
QGY7703 cell growth at days 2, 3, 4 and 5 compared with the control
group, in which QGY7703 cells were transfected with empty vector
(P<0.01; Fig. 3B).
Anchorage-independent growth was also used to
examine whether the RSPO2 gene affects the tumorigenic growth of
QGY7703 cells. As shown in Fig. 3C,
RSPO2 overexpression significantly enhances soft agar growth of
QGY7703 cells, as evidenced by the decrease in colony number and
size compared with the control QGY7703 cells that were infected
with the pLV-mCherry (2A) puro empty vector (P<0.01).
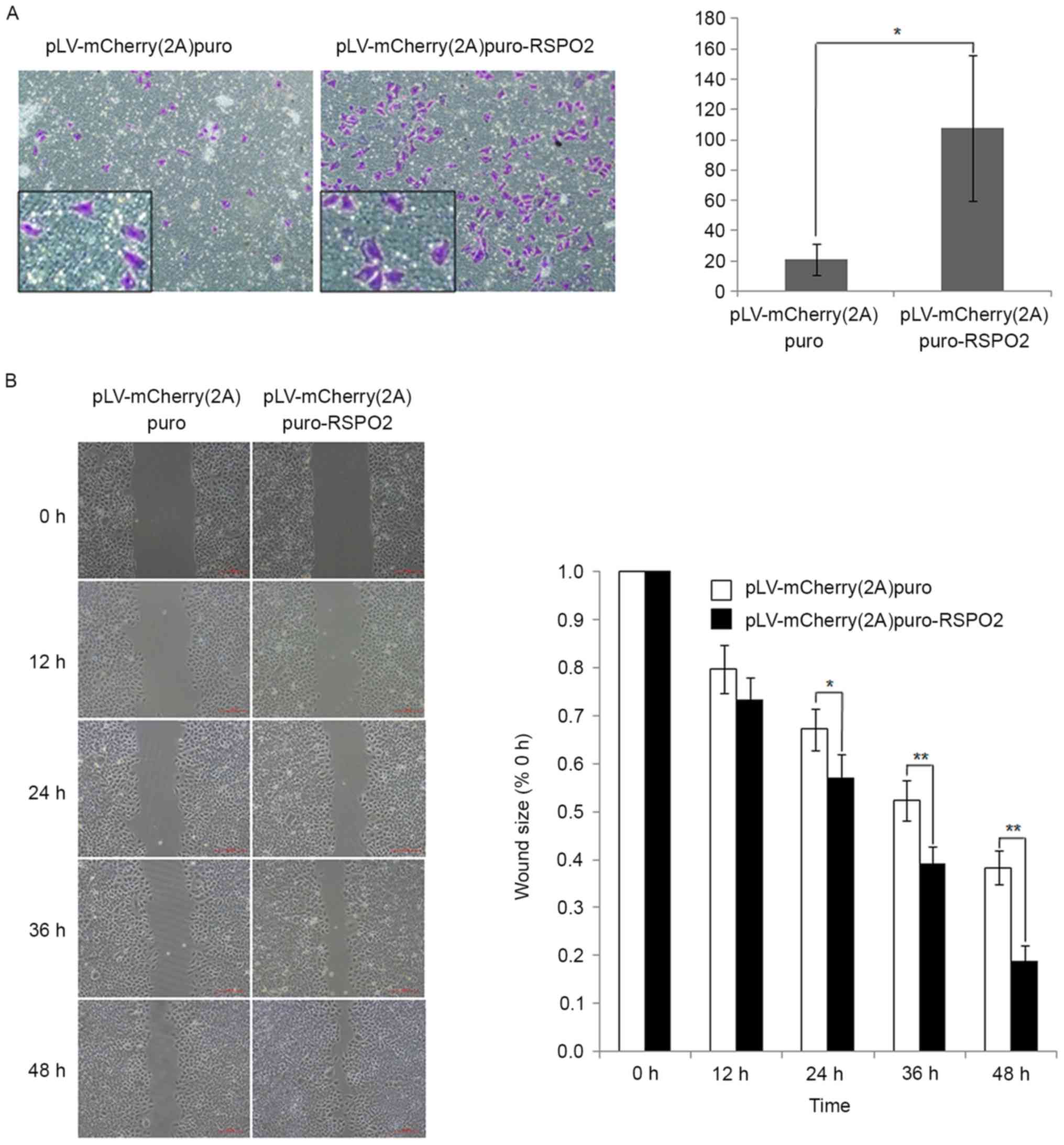
RSPO2 overexpression promotes the
migration of QGY7703 cells
To study the effect of RSPO2 on the motility of
QGY7703 cells, Transwell and wound healing assays were performed.
In the Transwell assay, the percentage of cells that migrated
through the membrane was significantly increased in cells with
RSPO2 overexpression compared with the control cells transfected
with empty vector (Fig. 4A). The
wound healing assay results also demonstrated that RSPO2
overexpression significantly enhanced the migration of QGY7703
cells compared with the control cells (Fig. 4B).
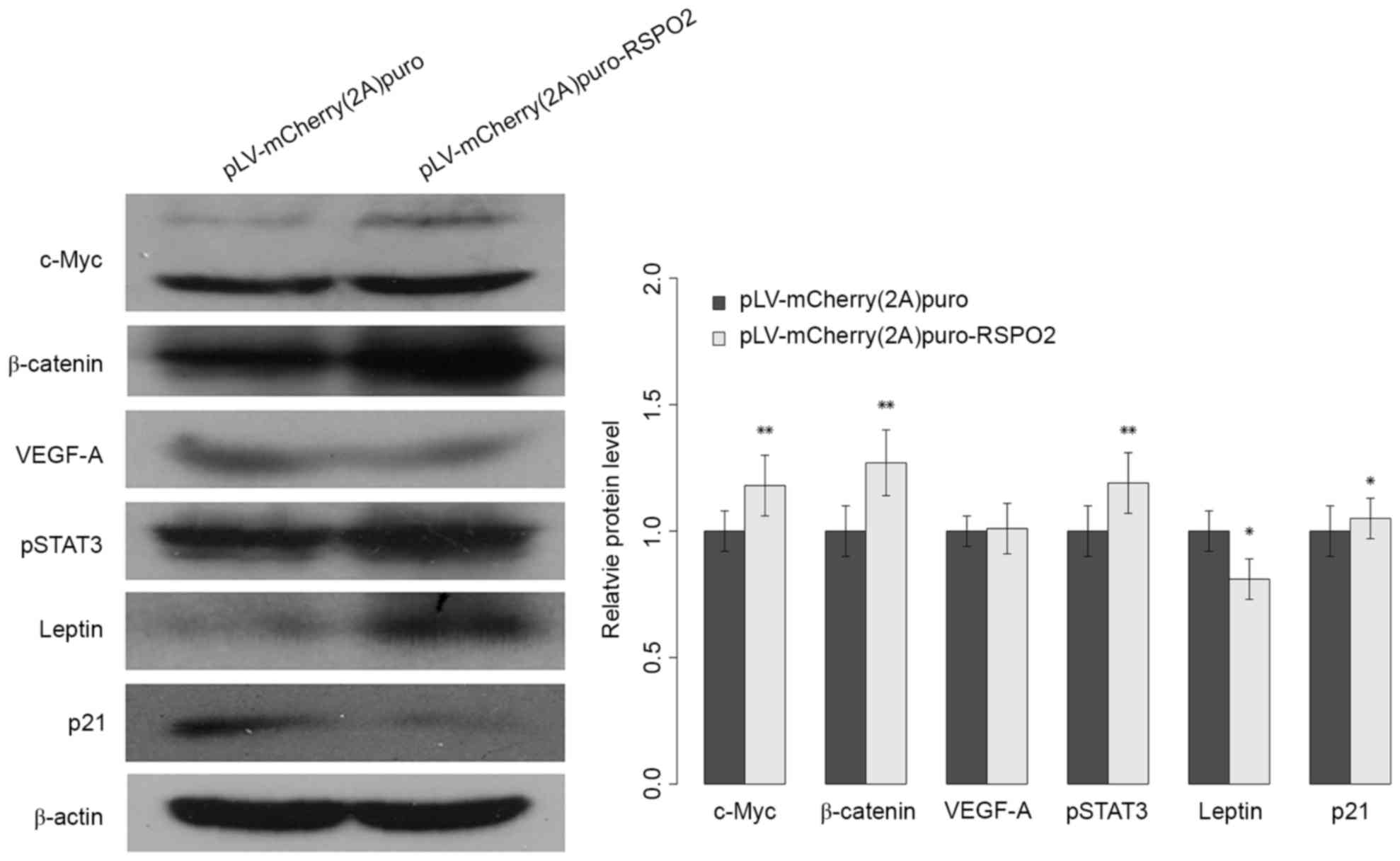
RSPO2 overexpression potentiates the
activation of Wnt/β-catenin
Previous studies have reported that the RSPO2 gene
is involved in the activation of the Wnt/β-catenin pathway. To
improve the understanding of the biological role of the RSPO2 gene
in HCC and the underlying mechanism of the aformentioned findings,
the expression level of nuclear β-catenin was analyzed in QGY7703
cells. As shown in Fig. 5, the
expression level of nuclear β-catenin was significantly increased
in QGY7703 cells with stable overexpression of RSPO2 gene compared
with the negative control group. The expression of c-Myc, one of
the target genes of Wnt/β-catenin signaling, was also analyzed.
Consistently, c-Myc gene expression was significantly increased in
QGY7703 cells with RSPO2 stable overexpression (Fig. 5). The present data indicated that
overexpression of RSPO2 is involved in Wnt/β-catenin activation via
increasing the expression of β-catenin and its downstream
genes.
RSPO2 regulates
proliferation-associated genes and signaling pathways
To further elucidate the molecular mechanism
underlying RSPO2-induced cell proliferation, the
proliferation-associated genes p21, leptin and VEGF-A, and the
STAT3 signaling pathway were investigated. The p21 and leptin genes
exhibited significantly reduced and increased expression,
respectively (Fig. 5). However, the
expression level of VEGF-A did not show a notable difference
between control and RSPO2-overexpressed QGY7703 cells. The elevated
expression of phosphorylated STAT3 indicated that the STAT3
signaling pathway may be involved in RSPO2-induced cell
proliferation.
Discussion
The carcinogenesis of HCC is a multi-factorial,
multi-step and complex process. Previous studies have documented
that the bidirectional interactions between tumors and hepatic
stellate cells (HSCs) compose an amplification loop to enhance
metastatic growth in the liver (28,29).
Previous studies revealed that the RSPO family may promote HSC
activation by enhancing the canonical Wnt pathway (30,31). In
the present study, the data provided evidence that RSPO2 expression
may contribute to malignant biological behavior in HCC. Additional
large-scale investigations are required to pinpoint the link
between the expression level of RSPO2 and the clinical
characteristics of human HCC. The present study demonstrated that
the expression of RSPO2 is upregulated in various HCC cell lines.
Paired HCC lesions and adjacent non-cancer tissues were found to
express RSPO2 differently. The tumor tissues exhibited
significantly increased expression of RSPO2 compared with adjacent
non-tumor tissues. Furthermore, the present data demonstrated that
RSPO2 overexpression enhances the cell proliferation and
anchorage-independent growth of human liver QGY7703 cell lines. In
addition, RSPO2 overexpression may also promote the motility of
QGY7703 cells. Study of the underlying molecular mechanism
indicated that overexpression of RSPO2 may be associated with
Wnt/β-catenin activation via increasing the expression of β-catenin
and its downstream gene c-Myc. Taken together, the present results
revealed the potential functional role of RSPO2 in HCC and
indicated that RSPO2 may be a potential drug target for patients
with liver tumors.
Previous studies have revealed that RSPOs are
involved in the activation of Wnt signaling, which is important for
tumorigenesis (14,32). However, the association between RSPO
and cancer has not been extensively studied. Studies on the role of
RSPO2 in cancer primarily focus on colon tumors (14,15). A
recent study reported that high-copy amplifications of RSPO2 gene
were observed in 231 HCC cases via whole exome sequencing (25). In the present study, significantly
increased expression levels of RSPO2 were detected in HCC tissues
compared with the adjacent non-tumor tissues or paired normal
tissue. Similarly, an increased expression level of RSPO2 was
observed in HepG2, Huh7 and Hep3B cells compared with human normal
liver QSG-7701 cell lines. High-copy amplifications of RSPO2 may
aid the explanation of the phenomenon of increased expression of
RSPO2 in tumor tissues.
In the present study, altered p21 and leptin
expression levels were observed, which may contribute to
RSPO2-induced proliferation on QGY7703 cells. Further work requires
an elucidation of the spatial-temporal association between RSPO2
and p21 or leptin. Furthermore, systematic investigation of pivotal
molecules located in the STAT3 signaling pathway may aid
understanding of the role of RSPO2 in liver cancer transformation.
Other signaling pathways may be examined to systematically
elucidate this molecular mechanism, potentially contributing to
identification of novel HCC therapeutics.
In conclusion, the present study revealed that
R-spondin 2 promotes proliferation and migration in various HCC
cell lines via the Wnt/β-catenin pathway. However, additional
studies are required to confirm the tumor-promoting effects of
R-spondin2 in mouse models.
Acknowledgements
The present study was funded by grants from the
Jiaxing Municipal Science and Technology Project (grant no.
2014AY21030-1), the Zhejiang Science and Technology Public Welfare
Project (grant no. 2015C33279), the Zhejiang Provincial Natural
Science Fund (grant nos. LY16H030016 and LY17H030012), the
Anesthesiology Center in North of Zhejiang Province and the Jiaxing
Key Laboratory of Neurology and Pain Medicine (2015-02-2).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liaw YF, Kao JH, Piratvisuth T, Chan HL,
Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH, et al:
Asian-Pacific consensus statement on the management of chronic
hepatitis B: A 2012 update. Hepatol Int. 6:531–561. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maluccio M and Covey A: Recent progress in
understanding, diagnosing, and treating hepatocellular carcinoma.
CA Cancer J Clin. 62:394–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu GQ, Shi KQ, Yu HJ, He SY, Braddock M,
Zhou MT, Chen YP and Zheng MH: Optimal adjuvant therapy for
resected hepatocellular carcinoma: A systematic review with network
meta-analysis. Oncotarget. 6:18151–18161. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Lau WB, Snel B and Clevers HC: The
R-spondin protein family. Genome Biol. 13:2422012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nam JS, Turcotte TJ and Yoon JK: Dynamic
expression of R-spondin family genes in mouse development. Gene
Expr Patterns. 7:306–312. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Parma P, Radi O, Vidal V, Chaboissier MC,
Dellambra E, Valentini S, Guerra L, Schedl A and Camerino G:
R-spondin1 is essential in sex determination, skin differentiation
and malignancy. Nat Genet. 38:1304–1309. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bell SM, Schreiner CM, Wert SE, Mucenski
ML, Scott WJ and Whitsett JA: R-spondin 2 is required for normal
laryngeal-tracheal, lung and limb morphogenesis. Development.
135:1049–1058. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aoki M, Mieda M, Ikeda T, Hamada Y,
Nakamura H and Okamoto H: R-spondin3 is required for mouse
placental development. Dev Biol. 301:218–226. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khan TN, Klar J, Nawaz S, Jameel M, Tariq
M, Malik NA, Baig SM and Dahl N: Novel missense mutation in the
RSPO4 gene in congenital hyponychia and evidence for a polymorphic
initiation codon (p.M1I). BMC Med Genet. 13:1202012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lowther W, Wiley K, Smith GH and Callahan
R: A new common integration site, Int7, for the mouse mammary tumor
virus in mouse mammary tumors identifies a gene whose product has
furin-like and thrombospondin-like sequences. J Virol.
79:10093–10096. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Theodorou V, Kimm MA, Boer M, Wessels L,
Theelen W, Jonkers J and Hilkens J: MMTV insertional mutagenesis
identifies genes, gene families and pathways involved in mammary
cancer. Nat Genet. 39:759–769. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim KA, Kakitani M, Zhao J, Oshima T, Tang
T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, et al: Mitogenic
influence of human R-spondin1 on the intestinal epithelium.
Science. 309:1256–1259. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seshagiri S, Stawiski EW, Durinck S,
Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman
V, Jaiswal BS, et al: Recurrent R-spondin fusions in colon cancer.
Nature. 488:660–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shinmura K, Kahyo T, Kato H, Igarashi H,
Matsuura S, Nakamura S, Kurachi K, Nakamura T, Ogawa H, Funai K, et
al: RSPO fusion transcripts in colorectal cancer in Japanese
population. Mol Biol Rep. 41:5375–5384. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim KA, Wagle M, Tran K, Zhan X, Dixon MA,
Liu S, Gros D, Korver W, Yonkovich S, Tomasevic N, et al:
R-Spondins family members regulate the Wnt pathway by a common
mechanism. Mol Biol Cell. 19:2588–2596. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huch M, Dorrell C, Boj SF, van Es JH, Li
VS, van de Wetering M, Sato T, Hamer K, Sasaki N, Finegold MJ, et
al: In vitro expansion of single Lgr5+ liver stem cells induced by
Wnt-driven regeneration. Nature. 494:247–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carmon KS, Gong X, Yi J, Thomas A and Liu
Q: RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc
Natl Acad Sci USA. 111:pp. E1221–E1229. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Logan CY and Nusse R: The Wnt signaling
pathway in development and disease. Annu Rev Cell Dev Biol.
20:781–810. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Clevers H: Wnt/beta-catenin signaling in
development and disease. Cell. 127:469–480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Monga SP: β-Catenin signaling and roles in
liver homeostasis, injury and tumorigenesis. Gastroenterology.
148:1294–1310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stewart DJ: Wnt signaling pathway in
non-small cell lung cancer. J Natl Cancer Inst. 106:djt3562014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gregorieff A, Liu Y, Inanlou MR, Khomchuk
Y and Wrana JL: Yap-dependent reprogramming of Lgr5(+) stem cells
drives intestinal regeneration and cancer. Nature. 526:715–718.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu T and Li C: Convergence between
Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer. 9:2362010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM,
Sung CO, Baek D, Haq F, Ansari AA, Lee SY, et al: Genomic portrait
of resectable hepatocellular carcinomas: Implications of RB1 and
FGF19 aberrations for patient stratification. Hepatology.
60:1972–1982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kononen J, Bubendorf L, Kallioniemi A,
Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G
and Kallioniemi OP: Tissue microarrays for high-throughput
molecular profiling of tumor specimens. Nat Med. 4:844–847. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boseman FT, Carneiro F, Hruban RH and
Theise ND: Tumours of the liver and intrahepatic bile ductsWorld
Health Organization Classification of Tumours of the Digestive
System. 4th. IARC; Lyon: pp. 195–262. 2010
|
|
28
|
Kang N, Gores GJ and Shah VH: Hepatic
stellate cells: Partners in crime for liver metastases? Hepatology.
54:707–713. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Coulouarn C and Clément B: Stellate cells
and the development of liver cancer: Therapeutic potential of
targeting the stroma. J Hepatol. 60:1306–1309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xinguang Y, Huixing Y, Xiaowei W, Xiaojun
W and Linghua Y: R-spondin1 arguments hepatic fibrogenesis in vivo
and in vitro. J Surg Res. 193:598–605. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin X, Yi H, Wu W, Shu J, Wu X and Yu L:
R-spondin2 activates hepatic stellate cells and promotes liver
fibrosis. Dig Dis Sci. 59:2452–3961. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chartier C, Raval J, Axelrod F, Bond C,
Cain J, Dee-Hoskins C, Ma S, Fischer MM, Shah J, Wei J, et al:
Therapeutic targeting of tumor-derived R-spondin attenuates
β-catenin signaling and tumorigenesis in multiple cancer types.
Cancer Res. 76:713–722. 2016. View Article : Google Scholar : PubMed/NCBI
|