Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most
common type of lymphoma worldwide, accounting for between 40 and
45% of non-Hodgkin's lymphoma, and its morbidity increases at an
annual rate of 3% (1). Forkhead box
P3 (FOXP3) is a transcription factor that is specifically expressed
by T cells. FOXP3 is a marker of regulatory T cells (Tregs) and
serves an important role in maintaining the immunosuppressive
function of Tregs (2). In the immune
system, T cells are responsible for negative regulation and inhibit
the proliferation and immunocompetence of immunological effector
cells to assist with the formation of autoimmune tolerance
(3). CD4+CD25+
T cells are a principal type of Treg and exhibit an
immunosuppressive function (4).
Previously, it was identified that CD4+CD25+
T cells exhibit a marked association with certain physiological
processes, including autoimmune disease, tumorigenesis, tolerance
and anaphylaxis (5). FOXP3 is the
basic transcription factor that is essential for differentiation of
Tregs, and is therefore a selection marker for Tregs (6) and their defining feature (7). A previous study has identified that the
immunosuppressive function of the T cells may be regulated, and the
antineoplastic immune response ability of the body may be enhanced,
by decreasing the expression of FOXP3 (8). The increased expression of FOXP3 in the
majority of tumors is a marker of poor prognosis of patients with
cancer (9–11). However, the association between FOXP3
and the prognosis of patients with DLBCL remains unclear.
MicroRNA (miRNA) is a type of non-coding RNA,
typically 22 nucleotides in length. A mature miRNA is able to
degrade a target signal to inhibit the transcription of RNA and
lead to the regulation of target gene expression (12). Previous studies have identified the
influence of miRNA on T cells: Studies in animals have suggested
that the lack of miRNA results in a deceased number of T cells in
the thymus, spleen and lymph gland (13,14). It
has also been identified that FOXP3 may influence the expression of
certain miRNAs in T cells; a FOXP3-binding site was identified in
specific parts of the miRNA coding area, and FOXP3 may directly or
indirectly regulate the expression of certain miRNAs in T cells
(15).
FOXP3 is the basic transcription factor of T cells.
The immunosuppressive function of T cells weakens the
antineoplastic immune response ability of the body. However, to the
best of our knowledge, previous studies have rarely investigated
the regulation of FOXP3 expression. miR-155 is a modulator of
FOXO3a protein has been identified as a potential regulator of
FOXP3 (16). It was hypothesized that
miR-155 may influence the expression of FOXP3 and exhibit an
interference effect on the progression of DLBCL. Therefore, to
determine the role of FOXP3 in DLBCL, the expression of FOXP3 was
investigated in the human DLBCL cell lines Ly1, Ly8 and Ly10, the
mouse DLBCL cell line A20, and normal B cells, and the expression
of FOXP3 in DLBCL tumor tumor-adjacent tissues was compared in
order to analyze the association between the expression of FOXP3 in
the tumor tissues and prognosis. The lentiviral transfection
technique was used to silence the miR-155 gene in the A20 cells in
order to analyze the effect of the miR-155 gene on FOXP3 in DLBCL.
The A20 cell line, following stable transfection, was used in a
tumorigenicity experiment in BALB/c mice to compare the
tumorigenicity and tumor growth rates, and provide evidence to
support the hypothesis that decreasing the expression of FOXP3 by
miR-155 inhibits DLBCL.
Materials and methods
Materials
A total of 60 male BALB/c mice (6-week-old; weight,
25±5 g) were purchased from the Central Laboratory of Animal
Science of the Zhejiang University Medical College (Jinhua, China).
All animals received humane care according to the criteria outlined
in the ‘Guide for Care and Use of Laboratory Animals℉ (17). The animal protocol was approved by the
Animal Care and Use Committee of Zhejiang University Medical
College.
Normal B cells, the human DLBCL cell lines Ly1, Ly8
and Ly10, and the BALB/c mouse-derived DLBCL A20 cell line were
kindly provided by Professor Zhao Tong (Nanfang Hospital, Southern
Medical University, Guangzhou, China). A total of 60 surgical
resection samples of human DLBCL (including from 42 males and 18
females, with a mean age of 62 years) were obtained from the
Department of Pathology of Jinhua Hospital of Zhejiang University
(Jinhua, China) between March 2009 to December 2013. Written
informed consent was obtained from all patients for the use of
their samples.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from cells was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. cDNA synthesis was
performed on 10 ng total RNA using a RevertAid First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. RT-qPCR was performed on a 7500/7500 Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using a SYBR-Green Premix Ex Taq™ kit (Takara Bio, Inc.,
Otsu, Japan). Primer sequences used for the amplification are
listed in Table I. A total of 4
replicates were used for each sample. The thermocycling conditions
were DNA deformation and polymerase activation at 95°C for 3 min,
then 38 cycles of denaturation at 95°C for 15 sec, and
amplification and fluorescence measurement at 57°C for 60 sec.
Melting curve analysis was performed between 55 and 95°C with
increments of 0.5°C every 3 sec. The relative quantification was
determined from Cq values for each target gene and the
internal control gene (β-actin) in the cell lines, as previously
described using GeNorm software (version 3.4) (18).
 | Table I.Primer sequences used for the
amplification of FOXP3 and β-actin. |
Table I.
Primer sequences used for the
amplification of FOXP3 and β-actin.
| Gene | Forward primer | Reverse primer | Product size,
bp |
|---|
| FOXP3 |
5′-CTCATGATAGTGCCTGTGTCCTCAA-3′ |
5′-AGGGCCAGCATAGGTGCAAG-3′ | 167 |
| β-actin |
5′-CATCCGTAAAGACCTCTATGCCAAC-3′ |
5′-ATGGAGCCACCGATCCACA-3′ | 250 |
Immunohistochemistry
Whole sections of human DLBCL samples were sectioned
at 0.3 µm thickness, deparaffinized in xylene and rehydrated in
graduate series of alcohols. Slides were pretreated with 10 mmol/l
citrate buffer (pH 6.0) (Thermo Fisher Scientific, Inc.) and
stained with a rabbit polyclonal immunoglobulin G anti-FOXP3
antibody (dilution, 1:100; cat. no. ab54501; Dako; Agilent
Technologies, Inc., Santa Clara, USA). Briefly, paraffin sections
were dewaxed with xylene, soaked for 5 min in 100% ethanol and then
soaked for 10 min in 95% alcohol at room temperature. Antigen
retrieval was performed by heating the samples in Dako REAL Target
Retrieval solution (Agilent Technologies GmbH, Waldbronn, Germany)
for 40 min at 98°C. The sections were then treated with
H2O2 to block endogenous peroxidase activity.
The sections were then incubated with diaminobenzidine (DAB)
substrate (Dako; Agilent Technologies, Inc.) and counterstained
with hematoxylin. Image analysis with an optical microscope
(magnification, ×200; Olympus Corporation, Tokyo, Japan) was
performed to determine the average integral optical density of the
positive cells using ImagePro Plus software (Image-Pro Plus version
6.0; Media Cybernetics, Rockville, MD, USA) (19). In order to analyze the association
between the expression of FOXP3 and the clinicopathological
features of patients with DLBCL, Cox's regression model was used to
conduct multifactor analysis of the age, expression of FOXP3 within
and around tumors, extra-nodal involvement and International
Prognostic Index rating clinical stages of patients with DLBCL.
Cell transduction
Human embryonic kidney 293T cells (HEK293T) cells
were maintained in Dulbecco's modified Eagle's medium (DMEM;
Beijing Solarbio Science & Technology Co., Ltd., Beijing,
China) supplemented with 10% FBS. The murine leukemia cell line A20
(B lymphocytic) was cultured in RPMI-1640 medium (Beijing Solarbio
Science & Technology Co., Ltd.) supplemented with 10% FBS and
0.05 mM 2-mercaptoethanol. pLKO-TRC short hairpin RNA (shRNA)
clones (cat. nos. TRCN0000068102, TRCN0000068101, TRCN0000068100,
TRCN0000068099 and TRCN0000068098) targeting mouse miR-155 were
purchased from Open Biosystems (Huntsville, AL, USA). cDNA encoding
human miR-155 was synthesized and cloned into a pBABE retroviral
vector (Cell Biolabs, Inc., San Diego, CA, USA). For cell
transduction, retroviruses were prepared by transient
co-transfection with helper viral vector into HEK293T cells using
calcium phosphate precipitation (MACGENE Biotechnology Ltd.,
Beijing, China). HEK293T cells were transfected with plasmid DNA
and cultured at 37°C for 6 h, prior to replacement of the medium.
At between 36 and 48 h post-transfection, the supernatant was
collected and filtered through a 0.45 µm filter. A20 cells were
transfected at ~2×106 cells/10-cm cell culture dish in
DMEM/RMPI-1640 medium (1:1 ratio) supplemented with 8 µg/ml
polybrene. After 24 h, the medium was changed to RPMI-1640 medium
supplemented with 10% FBS and 0.05 mM 2-mercaptoethanol prior to
culture for further assay. Cell transduction with lentiviral vector
encoding shRNA targeting mouse miR-155 was performed according to
the manufacturer's protocol (Open Biosystems; GE Healthcare,
Chicago, IL, USA). Western blot analysis and RT-qPCR were performed
to confirm the knockdown, and A20-miR-155+ and A20-miR-155-
subclones were obtained. Total RNA from transfected cells was
extracted, and FOXP3 protein expression was determined in
A20-miR-155+ and A20-miR-155- cells.
Animals and in vivo tests
The 60 BALB/c mice were randomly divided into three
groups of 20: A20-miR-155-, A20-miR-155+ and the control group.
Tumor cells (2×107) in 0.2 ml growth medium were subcutaneously
injected into the mouse left axillary fossa. Procedures were
conducted under sterile conditions. Tumor growth was determined by
calculating the tumor volume. Mice were sacrificed when exhibiting
external signs of suffering (including decreased mobility and
altered behavior). All remaining animals were sacrificed at 30
days. Immediately following excision, the tumor tissues were stored
in 10% neutral-buffered formalin. After 48 h, the samples were
paraffin-embedded and then sliced into 4-µm sections for
hematoxylin and eosin staining. The sections were deparaffinized
and rehydrated, followed by antigen retrieval with citrate buffer
(pH 6.0). Endogenous peroxidase activity was inhibited with 3% H2O2
for 15 min and the sections were incubated with 10% normal goat
serum to block non-specific binding. Following incubation with
anti-FOXP3 antibody (dilution, 1:200; cat. no. HPA027342; Santa
Cruz Biotechnology, Dallas, TX, USA) at 4°C overnight, the sections
were washed, treated with biotinylated anti-immunoglobulin antibody
(dilution, 1:150; cat. no. LP0012145; Vector Laboratories, Inc.,
Burlingame, CA, USA) for 20 min at 25°C and reacted with
horseradish peroxidase-conjugated streptavidin. A liquid DAB
substrate/chromogen system (Fuzhou Maixin Biotech. Co., Ltd.,
Fuzhou, China) was used, according to the manufacturer's protocol,
followed by counterstaining with hematoxylin. Representative images
of tumor tissues were captured using a light microscope (Olympus
Corporation).
Western blot analysis
A 200 mg sample of tumor tissue was ground with
liquid nitrogen using a mortar. A 1 ml volume of protein extraction
buffer (50 mM Tris/HCl, 150 mM NaCl, 0.5 mM EDTA, 1 mM DTT, 1%
Triton X-100, 0.5% sodium deoxycholate and 0.1% SDS) with 1X Halt
protease inhibitors (Pierce; Thermo Fisher Scientific, Inc.) was
added. A 1 ml volume of the protein extraction buffer mixture was
transferred to a sterile Eppendorf tube and placed in an ice bath
at 4°C for 20 min prior to centrifugation at 2,500 × g. The
supernatant was divided into aliquots and stored at −80°C.
Subsequently, 50 µg total cellular protein from each sample was
separated by SDS-PAGE (10% gel) and electrotransferred onto a
polyvinylidene fluoride membrane using a semi-dry blotting
apparatus (Bio-Rad Laboratories, Inc.). Following blocking for 2 h
at 25°C with 5% skimmed milk, the membranes were hybridized with a
mouse monoclonal anti-FOXP3 antibody (dilution, 1:800; cat. no.
SAB1402390; Sigma-Aldrich; Merck KGaA) overnight at 4°C, followed
by incubation with secondary antibodies for 8 h at 4°C (dilution,
1:3,000; cat. no. 7072; Cell Signaling Technology, Inc., Danvers,
MA, USA). The protein bands were then visualized using Pro-Lighting
horseradish peroxidase agent (Cell Signaling Technology, Inc) and
their densities were determined using ImageJ software (version
2.1.4.7; imagej.nih.gov/ij). GAPDH (dilution,
1:300; cat. no. SRT1546780; Sigma-Aldrich; Merck KGaA) was used as
a loading control.
Cell cycle analysis
Splenic lymphocytes were obtained under sterile
conditions following sacrifice of the mice by severing their necks.
From each mouse, 100 µl lymphocyte suspension was obtained, to
which 4 µl anti-cluster of differentiation (CD)4-phycoerthyrin
(PE)-cyanine 5.5 (Cy5.5) (dilution, 1:1,500; cat. no. 3324; Cell
Signaling Technology, Inc.) and anti-CD25-allophycocyanin (APC)
(dilution, 1:1,500; cat. no. 3654; Cell Signaling Technology, Inc.)
were each added, followed by aeration and even mixing. The samples
were refrigerated at 4°C for 30 min. A 1.5 ml volume of precooled
flow cytometry staining buffer (cat. no. 00-4222; Cell Signaling
Technology, Inc.) was added prior to aeration and stirring to
evenly mix the washed cells. Samples were centrifuged at 800 × g at
4°C for 5 min. The supernatant was discarded and cells were
resuspended in 1 ml prepared lysis/fixing solution from a PerFix-nc
kit (Beckman Coulter, Inc., Brea, CA, USA). Following mixing by
vortex, a further 1 ml fixed/ruptured membrane working solution was
added and cells were vortex-mixed again. Subsequently, cells were
incubated at 4°C for 3 h in darkness. Following incubation, 2 ml
prepared ruptured membrane buffer working solution was added prior
to aeration and stirring for even mixing with the washed cells.
Cells were centrifuged for 800 × g at 8°C and the supernatant was
discarded. Cells were resuspended in 100 µl ruptured membrane
working solution and 0.5 µg Fc-receptor blocking solution (cat. no.
42232; Cell Signaling Technology, Inc.) was added prior to
incubation at 4°C for 14 min in the dark. Anti-FOXP3-PE (dilution,
1:500; cat. no. 1716; Cell Signaling Technology, Inc.) or
PE-Rat-lgG2a (dilution, 1:500; cat. no., 1285; Cell Signaling
Technology, Inc.) was used for homotypic comparison. Following
incubation for 14 min, 2 ml lysis/fixing solution was added to the
washed cells prior to aeration and stirring. Cells were centrifuged
at 2,000 × g at 8°C for 30 min. The supernatant was discarded, and
cells were resuspended in 500 µl 0.1% polyoxymethylene and
refrigerated at 4°C for 24 h. Samples were examined by flow
cytometry using a FACScan cytometer and the results analyzed by
ModFitLT software (version 2.0; both from BD Biosciences, Franklin
Lakes, NJ, USA).
Statistical analysis
SPSS software (version 20.0; IBM Corp., Armonk, NY,
USA) was used for statistical analyses. Data were expressed as the
mean ± standard deviation. Student's t-test for paired samples was
performed. P<0.05 was considered to indicate a statistically
significant difference.
Results
Foxp3 expression in distinct cell
lines
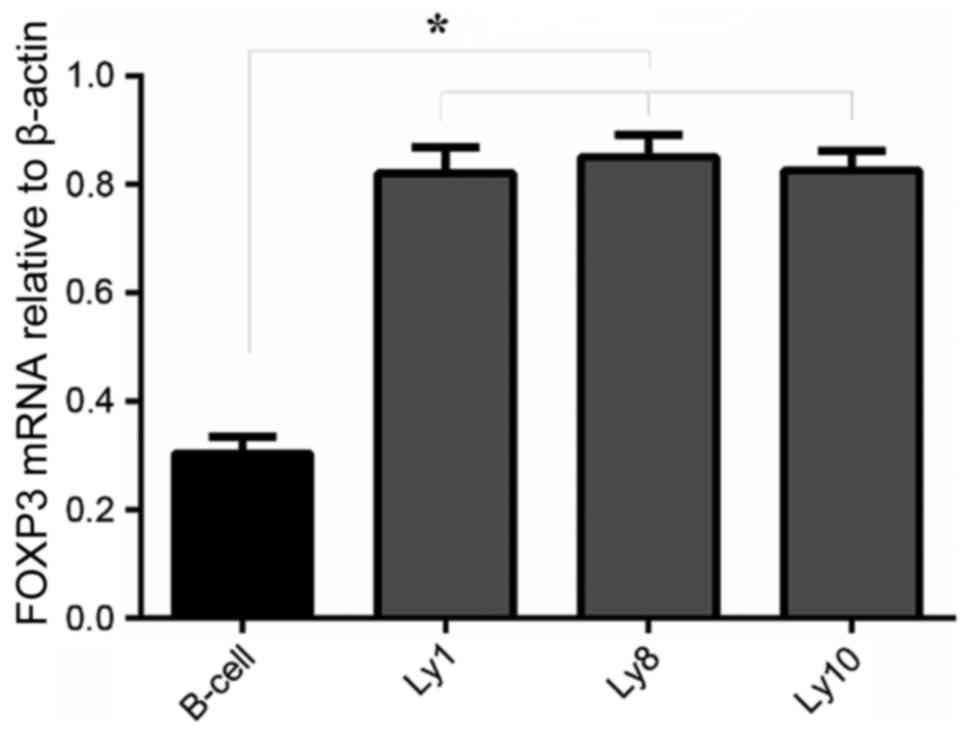
FOXP3 mRNA expression in normal B and DLBCL cell
lines was examined by RT-qPCR. All 4 cell lines, including normal B
and DLBCL cells (Ly1, Ly8 and Ly10), expressed FOXP3 mRNA. Compared
with normal B cells, FOXP3 mRNA expression in DLBCL cells (Ly1, Ly8
and Ly10) was significantly increased (P<0.05; Fig. 1).
Foxp3 protein expression in cancer
tissues and adjacent tissues
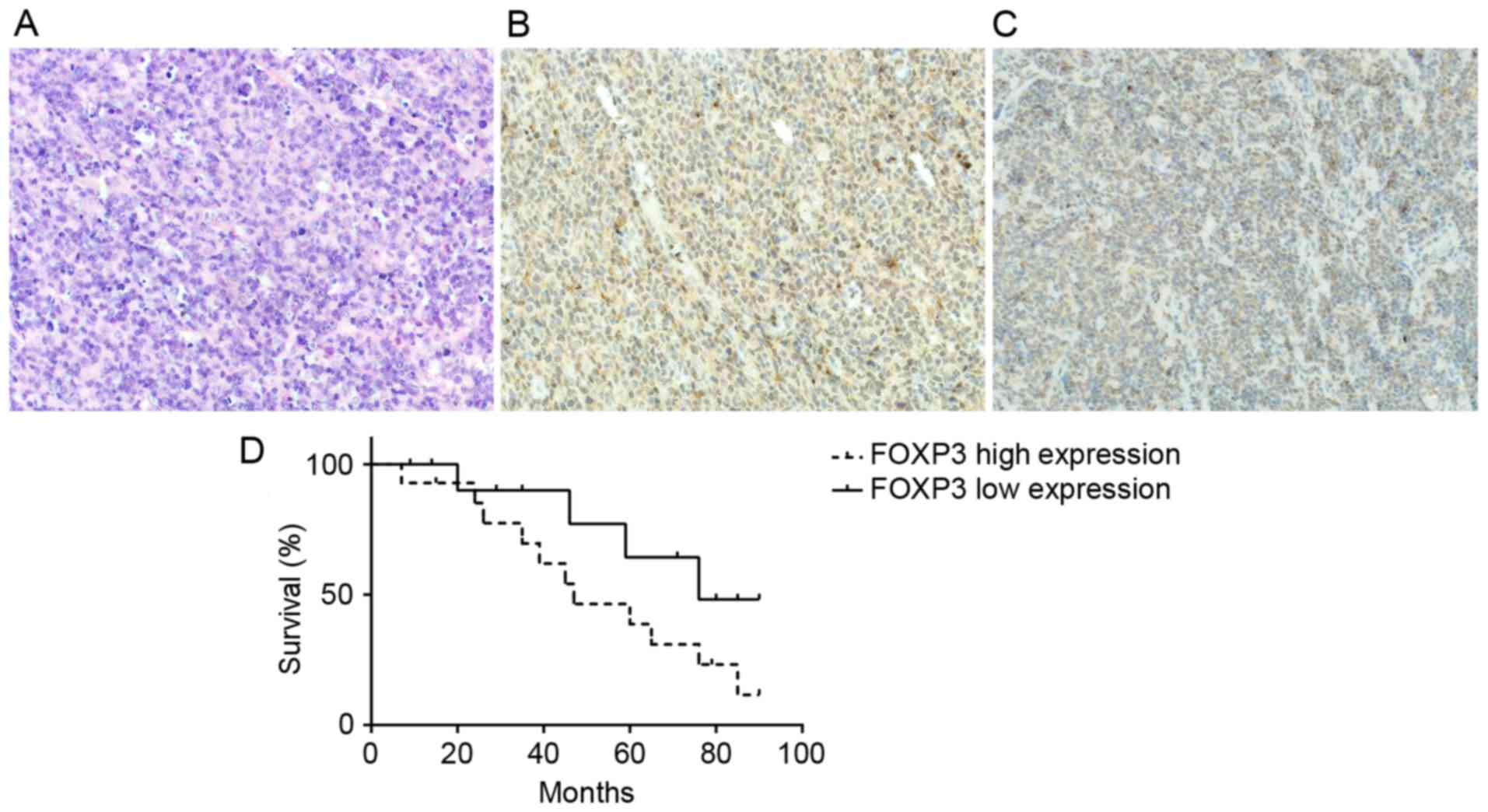
Immunohistochemistry was performed to determine the
expression of FOXP3 in DLBCL tumor tissues. The results indicated
that the protein expression levels of FOXP3 of cancer tissues were
decreased compared with that of adjacent tissues (Fig. 2A-C). By comparing the association
between the clinicopathological features of patients with DLBCL, it
was identified that increased expression of FOXP3 is a marker of
poor prognosis of patients with DLBCL (Table II). Increased expression of FOXP3 in
patients with DLBCL was identified to be associated with poor
prognosis (P<0.05; Fig. 2D).
 | Table II.Cox's multivariate analysis of
prognosis of diffuse large B-cell lymphoma. |
Table II.
Cox's multivariate analysis of
prognosis of diffuse large B-cell lymphoma.
|
| OS | PFS |
|---|
|
|
|
|
|---|
| Factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age | 1.28 | 0.58–1.34 | 0.451 | 1.09 | 0.38–2.47 | 0.623 |
| Metastasis | 0.91 | 0.43–1.92 | 0.820 | 0.59 | 0.41–1.23 | 0.701 |
| IPI score | 0.89 | 0.40–1.96 | 0.779 | 0.62 | 0.33–1.24 | 0.149 |
| Staging | 1.47 | 0.58–2.36 | 0.701 | 2.42 | 0.22–2.15 | 0.494 |
| FOXP3+
tumor | 2.13 | 1.99–3.42 | 0.026 | 1.08 | 0.52–1.64 | 0.348 |
Expression of FOXP3 in
miR-155-silenced A20 cells
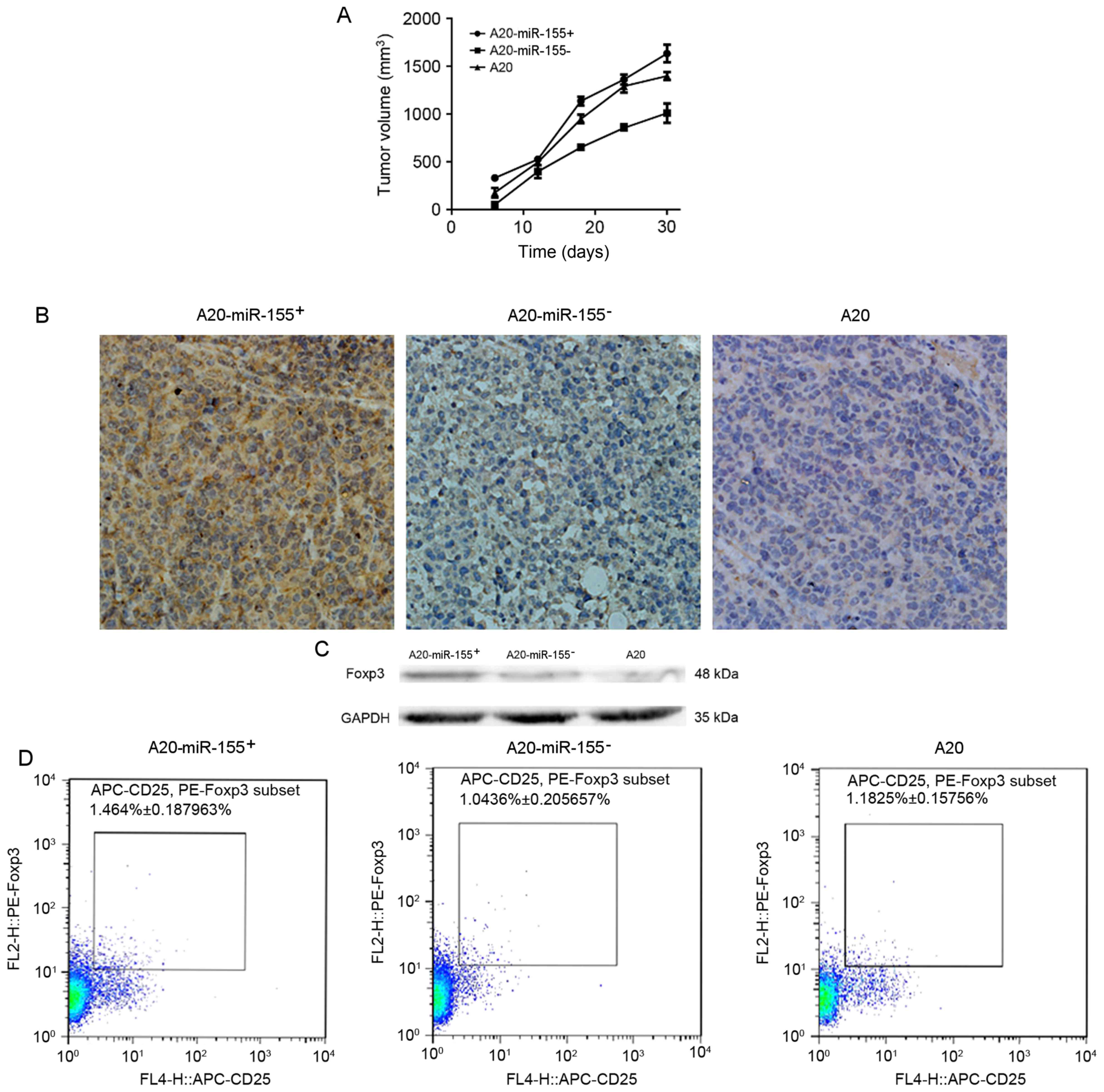
RT-qPCR was used to determine the expression levels
of FOXP3 protein within the A20-miR-155-, A20-miR-155+ and A20 cell
lines (Fig. 3A). Silencing miR-155 in
A20 cells led to a significant decrease in the expression of FOXP3
protein (P<0.05; Fig. 3B).
Histology of tumors in BALB/c
mice
Subcutaneous tumor models were successfully
established in BALB/c mice. Tumor growth of the mice injected with
A20-miR-155+ cells was increased compared with the A20-miR-155-
group (Table III). At 30 days after
injection of A20 cells, the mice with silenced miR-155 demonstrated
a decreased tumor volume (Fig. 4A).
Visible tumors were round or oval with nodules and were adherent to
the surface of the skin of the capillary-rich gray cut surface,
with no distant lymph node metastasis. Microscopic imaging of the
tumor tissue demonstrated that the tumor cells exhibited diffuse
homogeneous infiltrates consisting of large and cohesive tumor
cells with moderate cytoplasm and pleomorphic nuclei (Fig. 4B). Occasionally pathological mitotic
figures and a limited number of small lymphocytes were observed.
Western blot analysis was used to determine the protein expression
level of FOXP3, which was increased in A20-miR-155+ cells (Fig. 4C).
 | Table III.Tumor growth of mice following
injection of A20 cells. |
Table III.
Tumor growth of mice following
injection of A20 cells.
| Group | n (%) | Time, days | Weight, g | Volume,
mm3 |
|---|
| A20-miR-155 | 19/20 (95) | 7.0±0.9 | 2.11±0.42 |
1612±191.94a |
| A20-miR-155 | 12/20 (60.0) | 9.5±2.8 | 1.12±0.14 |
998.00±99.85a,b |
| A20 | 14/20 (70) | 9.0±1.8 | 1.67±0.22 |
1367±139.25b |
Flow cytometric analysis of
CD4+CD25+FOXP3+ T cells
The proportion of CD25+FOXP3+T
cells to total CD4+ T cells was determined using flow
cytometry in each group and was identified to be significantly
increased in A20-miR-155+-transducted mice compared with
the other groups (P<0.05; Fig.
4D).
Discussion
The results of the present study identified that:
Under the same conditions, the expression of FOXP3 in human DLBCL
cell lines (Ly1, Ly8 and Ly10) was increased compared with in
Bcells. By comparing the expression quantity of FOXP3 in the DLBCL
tumor tissue samples and tissue samples adjacent to tissues, it was
identified that the expression of FOXP3 in the former was increased
compared with that in the latter. The analysis of the clinical data
revealed that the increased expression of FOXP3 is one of the
factors indicating poor prognosis of DLBCL. This may because of an
immunosuppressive function of FOXP3+ T cells. T cells
participated in the immunoevasion of certain tumors and promoted
the generation and development of tumors by assisting in the
monitoring of the immunoevasion system of the tumor cells (18). CD4+CD25+ T cells
are an important CD4+ Treg lymphocyte subpopulation in
the immune microenvironment, which serve an important role in
maintaining the self-stability of the immune system and preventing
autoimmune diseases (20,21). As an indicator of the specificity of
the CD4+CD25+ T cells, FOXP3 is a major
regulatory factor that stimulates the development of
CD4+CD25+ T cells and the biological
function, and may decrease the immune response. Previous studies
have demonstrated that the proportion of T cells within the
lymphocytes that had infiltrated the tumor tissues of patients with
lung cancer, gastric cancer and ovarian cancer is high and is
associated with the poor prognosis of patients (22,23).
However, it has been identified that the increased level of FOXP3
protein expression within follicular lymphoma and skin T cell
lymphoma is a predictor of good prognosis (9,10).
Furthermore, a previous study examined the absolute value of
FOXP3+ T cells in 1,019 cases of distinct types of
lymphoma tissues and analyzed the association between its
expression level and the prognosis (24). This study demonstrated that patients
with DLBCL, follicular lymphoma and classic Hodgkin's lymphoma may
exhibit long survival times if the number of FOXP3+ T
cells increases. However, among the patients with DLBCL in the
middle and late stage, the increase in FOXP3+T cells is
associated with poor prognosis (24).
Previous studies have demonstrated that FOXP3+ Tregs
exist in tumor tissue mesenchyme and that FOXP3 is expressed in
various tumor cells (including pancreatic, breast, malignant
melanin and gastric cancer) (25–27). These
studies identified that FOXP3 is expressed in DLBCL tumor cells and
that the number of FOXP3+ T cells within the tumors is
significantly positively associated with overall survival and
provide a solid basis for the study of the effect of inhibiting the
expression of FOXP3 on DLBCL.
miR-155 is located within the third exon (21q21.3)
of the B-cell integration cluster (BIC) on human chromosome 21. The
gene does not include the open reading frame, so the expression
level is regulated and controlled by the BIC transcriptional level
and the processing of miRNA (28). It
promotes the abnormal proliferation of cells through its expression
and serves an important role in cancer. The copy number of miR-155
within tumor cells of DLBCL is between 10- and 30-fold that of
normal B cells, and its expression in the plasma of patients with
DLBCL is increased 5.24-fold compared with healthy individuals
(28). The expression level in the
activated B cell phenotype of DLBCL is increased between 2- and
3-fold compared with that of the germinal center phenotype of DLBCL
(29). Therefore, an association
between miR-155 and DLBCL has been identified. However, the results
of the present study suggested that, following silencing of the
expression of miR-155 in the DLBCL cell line A20, the expression
level of FOXP3 within the A20 cell line is decreased accordingly.
In the mouse experiment, the tumorigenicity rate and time of the
A20-miR-155−-transducted mice were significantly
decreased compared with those of the
A20-miR-155+-transducted mice. Using immunohistochemical
analysis of the expression of FOXP3 in the tumor tissues of mice,
it was identified that the expression of FOXP3 within the tumor
tissues of A20-miR-155+-transducted mice was decreased
markedly when compared with that of
A20-miR-155−-transducted mice. The subsequent flow
cytometry experiment also demonstrated that the number of T cells
within lymphocytes of A20-miR-155+-transducted mice
decreased compared with A20-miR-155−-transducted mice.
These results suggested that miR-155 may be a promoter of FOXP3,
but the underlying molecular mechanism remains unclear. A previous
study suggested that the increased expression of miR-155 in T cells
promotes the proliferation of T cells (30). In mice lacking miR-155, the number of
T cells within their spleen and thymus was significantly decreased;
however, it was indicated that the suppressive effect of Tregs that
lack miR-155 was the same as normal Tregs (31). It was also demonstrated that the
underlying molecular mechanism for how miR-155 adjusts the
proliferation of T cells may involve the interleukin (IL)-2
signaling pathway (32). A previous
study has demonstrated the vital role of IL-2 in maintaining the
dynamic balance of T cells (31).
However, suppressor of cytokine signaling 1 (SOCS1; the negative
regulatory factor in the IL-2 signaling pathway) is a direct target
gene of miR-155. The lack of miR-155 can result in the increased
expression of SOCS1, thus inhibiting the phosphorylation of signal
transducer and activator of transcription 5, resulting in damage to
the IL-2 signaling pathway and finally decreasing the proliferative
capacity of T cells (29). A previous
study has suggested that miR-155 promotes the proliferation of T
cells and Th17 cells through negative regulation of SOCS1 and
induces Th17cells to secrete IL-17, but is unable to stimulate T
cells to secrete IL-10 and transforming growth factor β (33), also demonstrating the influence of
miR-155 on T cells and their specific marker, FOXP3.
In the present study, RT-qPCR was used to determine
the FOXP3 mRNA expression level within human DLBCL cell lines and
normal B cells. Immunohistochemical analysis was performed to
determine the expression of FOXP3 in tumor tissues and surrounding
tumors. The association between the expression of FOXP3 in tumor
organization and the prognosis of patients with DLBCL was analyzed.
The results indicated that FOXP3 is an indicator of poor prognosis
of patients with DLBCL in the middle and late stage. It was also
identified that, following silencing of miR-155 in the A20 cell
line, the FOXP3 expression level also decreased. In the studies
using mice, the tumorigenicity of mice transfected with
A20-miR-155−was decreased compared with those
transfected with A20-miR-155+. The number of FOXP3 and T
cells exhibited the same trend. It may be that miR-155 is an
upstream factor of FOXP3. This potential underlying molecular
mechanism provides the experimental basis for the inhibition of the
expression of T cells and their specificity marker, FOXP3, and the
treatment of DLBCL via targeting FOXP3.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Zhejiang Province (grant no.
LY15H160049).
References
|
1
|
Cultrera JL and Dalia SM: Diffuse large
B-cell lymphoma: Current strategies and future directions. Cancer
Control. 19:204–213. 2012.PubMed/NCBI
|
|
2
|
Campbell DJ and Ziegler SF: FOXP3 modifies
the phenotypic and functional properties of regulatory T cells. Nat
Rev Immunol. 7:305–310. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu CT, Li WM and Yao YM: Regulating
mechanism of regulatory T cells in immunoregulatory responses. Int
J Pathol Clin Med. 28:0199–0106. 2008.
|
|
4
|
Schwartz RH: Natural regulatory T cells
and self-tolerance. Nat Immunol. 6:327–330. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Curiel TJ: Tregs and rethinking cancer
immunotherapy. J Clin Invest. 117:1167–1174. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sakaguchi S, Miyara M, Costantino CM and
Hafler DA: FOXP3+ regulatory T cells in the human immune system.
Nat Rev Immunol. 10:490–500. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kryczek I, Liu R, Wang G, Wu K, Shu X,
Szeliga W, Vatan L, Finlayson E, Huang E, Simeone D, et al: FOXP3
defines regulatory T cells in human tumor and autoimmune disease.
Cancer Res. 69:3995–4000. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fontenot JD, Rasmussen JP, Williams LM,
Dooley JL, Farr AG and Rudensky AY: Regulatory T cell lineage
specification by the forkhead transcription factor foxp3. Immunity.
22:329–341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karanikas V, Speletas M, Zamanakou M,
Kalala F, Loules G, Kerenidi T, Barda AK, Gourgoulianis KI and
Germenis AE: FOXP3 expression in human cancer cells. J Transl Med.
6:192008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Generali D, Bates G, Berruti A, Brizzi MP,
Campo L, Bonardi S, Bersiga A, Allevi G, Milani M, Aguggini S, et
al: Immunomodulation of FOXP3+ regulatory T cells by the aromatase
inhibitor letrozole in breast cancer patients. Clin Cancer Res.
15:1046–1051. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tzankov A, Meier C, Hirschmann P, Went P,
Pileri SA and Dirnhofer S: Correlation of high numbers of
intratumoral FOXP3+ regulatory T cells with improved survival in
germinal center-like diffuse large B-cell lymphoma, follicular
lymphoma and classical Hodgkin's lymphoma. Haematologica.
93:193–200. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cobb BS, Hertweck A, Smith J, O'Connor E,
Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG and
Merkenschlager M: A role for Dicer in immune regulation. J Exp Med.
203:2519–2527. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seo KH, Zhou L, Meng D, Xu J, Dong Z and
Mi QS: Loss of microRNAs in thymus perturbs invariant NKT cell
development and function. Cell Mol Immunol. 7:447–453. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sadlon TJ, Wilkinson BG, Pederson S, Brown
CY, Bresatz S, Gargett T, Melville EL, Peng K, D'Andrea RJ, Glonek
GG, et al: Genome-wide identification of human FOXP3 target genes
in natural regulatory T cells. J Immunol. 185:1071–1081. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamamoto M, Kondo E, Takeuchi M, Harashima
A, Otani T, Tsuji-Takayama K, Yamasaki F, Kumon H, Kibata M and
Nakamura S: miR-155, a modulator of FOXO3a protein expression, is
underexpressed and cannot be upregulated by stimulation of HOZOT, a
line of multifunctional Treg. PLoS One. 6:e168412011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
National Research Council, . Guide for the
Care and Use of Laboratory Animals. National Academies Press;
Washington, DC: pp. 41–48. 1974
|
|
18
|
Mestdagh P, Van Vlierberghe P, De Weer A,
Muth D, Westermann F, Speleman F and Vandesompele J: A novel and
universal method for microRNA RT-qPCR data normalization. Genome
Biol. 10:R642009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kraan MC, Smith MD, Weedon H, Ahern MJ,
Breedveld FC and Tak PP: Measurement of cytokine and adhesion
molecule expression in synovial tissue by digital image analysis.
Ann Rheum Dis. 60:296–298. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shiozawa E, Yamochi-Onizuka T, Takimoto M
and Ota H: The GCB subtype of diffuse large B-cell lymphoma is less
frequent in Asian countries. Leuk Res. 31:1579–1583. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Oki Y, Yamamoto K, Kato H, Kuwatsuka Y,
Taji H, Kagami Y and Morishima Y: Low absolute lymphocyte count is
a poor prognostic marker in patients with diffuse large B-cell
lymphoma and suggests patients' survival benefit from rituximab.
Eur J Haematol. 81:448–453. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jabłońska J, Jesionek-Kupnicka D, Potemski
P, Kowalik A, Sygut J and Kordek R: Comparison of two different
immunohistochemical algorithms identifying prognostic subgroups of
DLBCL. Pol J Pathol. 61:124–132. 2010.PubMed/NCBI
|
|
23
|
Qian BZ and Pollard JW: Macrophage
diversity enhances tumor progression and metastasis. Cell.
141:39–51. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kono K, Kawaida H, Takahashi A, Sugai H,
Mimura K, Miyagawa N, Omata H and Fujii H: CD4(+)CD25high
regulatory T cells increase with tumor stage in patients with
gastric and esophageal cancers. Cancer Immunol Immunother.
55:1064–1071. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Badoual C, Hans S, Rodriguez J, Peyrard S,
Klein C, Nel H Agueznay, Mosseri V, Laccourreye O, Bruneval P,
Fridman WH, et al: Prognostic value of tumor-infiltrating CD4+
T-cell subpopulations in head and neck cancers. Clin Cancer Res.
12:465–472. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salama P, Phillips M, Grieu F, Morris M,
Zeps N, Joseph D, Platell C and Iacopetta B: Tumor-infiltrating
FOXP3+ T regulatory cells show strong prognostic significance in
colorectal cancer. J Clin Oncol. 27:186–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Khattri R, Cox T, Yasayko SA and Ramsdell
F: An essential role for Scurfin in CD4+CD25+ T regulatory cells.
Nat Immunol. 4:337–342. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rai D, Karanti S, Jung I, Dahia PL and
Aguiar RC: Coordinated expression of microRNA-155 and predicted
target genes in diffuse large B-cell lymphoma. Cancer Genet
Cytogenet. 181:8–15. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eis PS, Tam W, Sun L, Chadburn A, Li Z,
Gomez MF, Lund E and Dahlberg JE: Accumulation of miR-155 and BIC
RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 102:pp.
3627–3632. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kohlhaas S, Garden OA, Scudamore C, Turner
M, Okkenhaug K and Vigorito E: Cutting edge: The Foxp3 target
miR-155 contributes to the development of regulatory T cells. J
Immunol. 182:2578–2582. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bayer AL, Yu A and Malek TR: Function of
the IL-2R for thymic and peripheral CD4+CD25+Foxp3+ T regulatory
cells. J Immunol. 178:4062–4071. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu LF, Thai TH, Calado DP, Chaudhry A,
Kubo M, Tanaka K, Loeb GB, Lee H, Yoshimura A, Rajewsky K and
Rudensky AY: Foxp3-dependent microR-NA155 confers competitive
fitness to regulatory T cells by targeting SOCS1. Immunity.
30:80–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X
and Liao YH: MicroRNA-155 modulates Treg and Th17 cells
differentiation and Th17 cell function by targeting SOCS1. PLoS
One. 7:e460822012. View Article : Google Scholar : PubMed/NCBI
|