Introduction
Ovarian cancer is the fifth most common type of
disease in females, and in the United States in 2014 there were an
estimated 21,290 new cases and 14,180 mortalities due to ovarian
cancer (1). Although the 5-year
survival rate of females with ovarian cancer has improved, it is
only ~20% (2). Platinum-based
combinations of chemoresistance is one of the obstacles limiting
the success of cancer drug treatments and minimizing the
effectiveness of chemotherapy in a large number of patients
(3). Cisplatin, one of the most
common forms of platinum, is often used as one of the first-lines
of treatment following surgical resection of visible nidus in
ovarian cancer (3). In order to
improve patient outcomes, it is critical to overcome cisplatin
resistance of ovarian cancer cells (4).
Epigenetic changes at the molecular and cellular
levels contributing to cisplatin-resistance have previously been
reported, including alterations of platinum-DNA adducts, impairment
in the apoptotic response of cells to adduct products, DNA
methylation status transformation, histone modification and
microRNAs (miRs) (5,6). miRs are reported to be involved in the
regulation of various biological processes, including embryonic
development, cellular proliferation, differentiation, migration and
apoptosis (7,8). Studies have suggested that aberrant miR
expression levels have been associated with tumor biology,
including resistance to various chemotherapeutic agents (9,10). For
example, let-7b suppression induces resistance to cisplatin by the
upregulation of cyclin D1 in glioblastoma (11). miRs overexpression has also been
demonstrated to result in resistance to drugs in colorectal and
prostate cancer (12,13). miR-522 expression level was reduced in
doxorubicin (DOX) resistant colon HT29 cell line and affected the
sensitivity of the cells to DOX treatment by targeting ABCB5
(14). miR-200b has been shown to
enhance chemosensitivity in prostate cancer via the regulation of
Bmi-1 (15). miR-20a, a member of the
miR-17-92 cluster, acts as a transforming growth factor β receptor
2 suppressor for reverting cisplatin-resistance and inhibiting
metastasis in non-small cell lung cancer (16). Of note, miR-20a also inhibited the
pro-apoptotic activity and induced chemoresistance in leukemia
cells (17). Our previous study
demonstrated that miR-20a promoted proliferation and invasion by
targeting the amyloid precursor protein in the ovarian cancer
OVCAR3 cell line (18). The present
study hypothesized that miR-20a may be involved in in ovarian
cancer resistance to cisplatin and aimed to investigate the
underlying mechanism of chemoresistance in OVCAR3 cells. A
cisplatin-resistant subline, OVCAR3/DDP, was established from the
OVCAR3 ovarian cell line. miR-20a facilitated OVCAR3 cells
resistance to cisplatin and contributed to OVCAR3/DDP cell
migration. The enhanced migration ability of OVCAR3/DDP cells may
be due to epithelial-mesenchymal transition (EMT) induced by
miR-20a.
Materials and methods
Cell culture and transfection
Cells were routinely cultured in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 IU/ml penicillin and 100
µg/ml streptomycin, and incubated at 37°C in a humidified chamber
supplemented with 5% CO2 until confluence reached
70–80%. Transfection was performed using Lipofectamine™ 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol.
Plasmid construction
To construct the overexpression and control
plasmids, the sequences of miR-20a precursor (sh-miR-20a) and
control (NC-miR-20a) were subcloned into pcDNA3.1 polyclone sites
with HindIII and BamHI sites, pri-miR-10a and pcDNA3, respectively.
To knockdown the miR-20a expression, the sequence of miR-20a
inhibitor (ASO-miR-20a) and control were synthesized (Gene Pharma,
Shanghai, China). The sequences used are indicated in Table I.
 | Table I.Primer sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer | Sequence |
|---|
| miR-20a | F |
GCTGCCGTAAAGTGCTTATAGTG |
|
| R |
CAGAGCAGGGTCCGAGGTA |
| miR-20a
inhibitor |
|
CTACCTGCACTATAAGCACTTTA |
| miR-20a control |
|
GACTACACAAATCAGCGATTT |
| sh-miR- | F |
UAAAGUGCUUAUAGUGCAGGUAGTT |
| 20a | R |
CUACCUGCACUAUAAGCACUUUATT |
| sh-NC | F |
UUCUCCGAACGUGUCACGUTT |
|
| R | ACG
UGACACGUUCGGAGAATT |
| U6 | F |
ATTGGAACGATACAGAGAAGATT |
|
| R |
GGAACGCTTCACGAATTTG |
Establishment of OVCAR3/DDP cell
lines
OVCAR3/DDP cells were induced using a progressive
concentration of cisplatin. Briefly, OVCAR3 that were sourced from
Tianjin Medical University (Tianjin, China) cells in the
logarithmic growth phase were treated with 2.5 µmol/l cisplatin.
Following 48–72 h, cisplatin was removed and the cells were
cultured at 37°C without cisplatin for ~3 weeks until they
recovered. Then the cells were treated with 5 µmol/l and 10 µmol/l
cisplatin, respectively. The cisplatin resistant OVCAR3/DDP cell
line was successfully established when cells survived in 10 µmol/l
cisplatin for ~2 months with a normal activity.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using CCK-8 (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan). The absorbance at
450 nm was detected using a Thermo Scientific Mulyiskan FC
Microplate Spectrophotometer. For the inhibition detection, the
OVCAR3 and OVCAR3/DDP cells were treated with 5 different
concentrations (0, 0.25, 2.5, 25, 250, 2,500 µmol/l) for 48 h, and
then added with CCK-8. Inhibition (%) = {1 - [OVCAR3/DDP optical
density (OD) - Blank OD]/(OVCAR3 OD - Blank OD)} × 100%. Resistance
index = OVCAR3/DDP half-maximal inhibitory concentration
(IC50)/OVCAR3 IC50.
Cell cycle analysis
Cells were seeded in 25 cm2 tissue
culture flasks at a density of 25×104/well in the
corresponding growth medium, and harvested when they reached 80%
confluence. Subsequently, cells were washed with PBS twice and
fixed in ice-cold 70% ethanol in PBS for 24 h at 4°C. Subsequent to
washing, the fixed cells were treated with 0.01% RNase for 10 min
at 37°C and then stained with 0.05% propidium iodide (PI) for 20
min at 4°C in the dark. The cell cycle was determined using a
FACScanto flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA)
and analyzed using ModFit software v.4.1 (Verity Software House,
Inc., Topsham, ME, USA).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA of OVCAR3/DDP cells was isolated using
EASYspin tissue/cell total RNA extraction kit (Aidlab
Biotechnologies, Ltd., Beijing, China) and the cDNA was synthesized
using the First-Strand cDNA Synthesis kit including DNase (Takara
Bio., Inc., Otsu, Japan) from 3 µg total RNA with a thermocycler
(Arktik 96, Thermo Fisher Scientific, Inc.). miR expression level
was analyzed by stem-loop RT-qPCR using Hairpin-itä miRs RT-PCR
Quantitation kit (Shanghai GenePharma Co., Ltd., Shanghai, China)
and SYBR Premix Ex Taq (Takara Bio, Inc.) on a PikoReal 96
Real-Time PCR System (Thermo Fisher Scientific, Inc.). The qPCR
assay was performed as follows: 95°C for 3 min for 40 cycles of
95°C for 12 sec and 62°C for 40 sec. The fold change was
normalizing to U6 by using 2−ΔΔCq (19). The primer sequences are provided in
Table I. All experiments were
performed in triplicate.
Western blot analysis
Protein samples were obtained by lysing ovarian
cancer OVCAR3/DDP cells in a standard sample buffer (50 mM
Tris-HCl; pH 6.8; 2% SDS; 10% glycerol) with level of shaking for
20 min at 4°C, with gentle triturating 4 times. Lysates were
collected and cleared by centrifugation at 12,830 × g for 5 min at
4°C. Then, 30 µg protein of each group was loaded into 10%
SDS-PAGE. The membranes were blocked with 5% bovine serum albumin
(BSA; Amresco, LLC, Solon, OH, USA) at 37°C for 1 h and incubated
with the specific antibodies diluted with 5% BSA at 4°C overnight.
The membranes were washed with TBST 3 times for 5 min, and then
incubated with goat anti-rabbit peroxidase-conjugated secondary
antibody for 2 h at 37°C. The following primary antibodies were
used at the indicated dilutions: Anti-E-cadherin pAb (dilution,
1:500; cat. no. sc-7870), anti-N-cadherian pAb (dilution, 1:500;
cat. no. sc-393933; both from Santa Cruz Biotechnology Inc.,
Dallas, TX, USA), anti-Vimentin (dilution, 1:500; cat. no.
bs-8533R) and β-tubulin pAb (dilution, 1:1,000; cat. no. bs-14263R,
both from Bioss Biological Technology Co., Ltd., Beijing,
China).
Boyden chamber transwell migration
assay
The migratory capability of cells was determined
using a Transwell chamber culture system (8 µm pore; Corning
Incorporated, Corning, NY, USA). Cells were seeded in a Boyden
chamber Transwell without matrigel-coated inserts
(2×104/well) with serum-free growth medium Opti-MEM
(Gibco; Thermo Fisher Scientific, Inc.). Complete growth medium
supplemented with 10% FBS was added to the lower chamber. Following
incubation at 37°C for 24 h, the cells attached to the lower
surface of the insert filter were counted following 0.1% crystal
violet (Solarbio Science & Technology Co., Ltd, Beijing, China)
staining for 10 mins at room temperature.
Statistical analysis
All experiments were repeated independently ≥3
times, and the results are presented as the mean ± standard
deviation. A Student t-test was used to evaluate the statistical
differences between the two groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Establishment of cisplatin-resistant
human ovarian cancer cell line OVCAR3/DDP
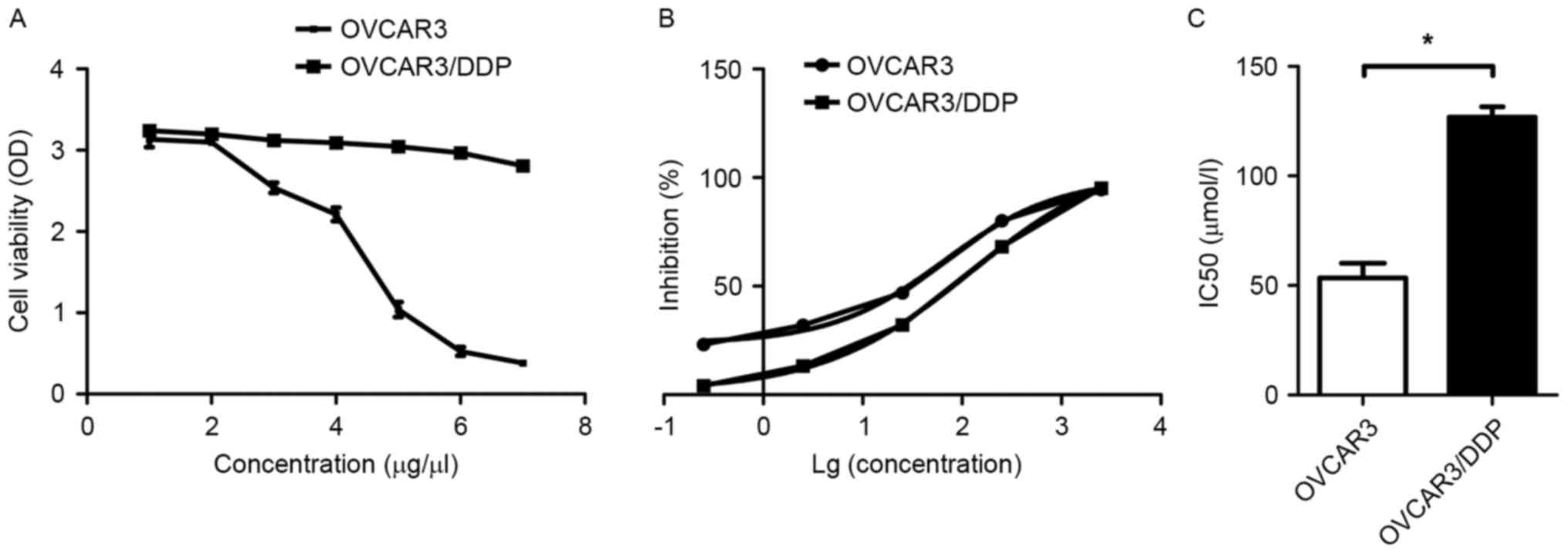
Cisplatin-resistant capability of OVCAR3/DDP cells
and the parental cells was assessed using a CCK-8 assay at various
concentrations of cisplatin. As presented in Fig. 1A, OVCAR3 cell viability was gradually
decreased and almost completely inhibited following treatment with
7 µmol/l cisplatin. OVCAR3/DDP cell viability was slightly
decreased following the same concentration treatment. Subsequently,
the IC50 values for the two types of cells were detected
using CCK-8 assay with 5 concentrations of cisplatin (0.25, 2.5,
25, 250 and 2,500 µmol/l). The growth inhibition rate of OVCAR3/DDP
cells was lower than that of OVCAR3 cells at each concentration,
except 2,500 µmol/l, at which the two cell lines were almost
entirely inhibited (Fig. 1B). The
IC50 of OVCAR3/DDP and OVCAR3 cells were 122.30±2.83 and
66.08±5.25, respectively, and the index (RI) of OVCAR3/DDP cells
was 1.87 (Fig. 1C). These results
indicated that OVCAR3/DDP cells had increased resistance to
cisplatin compared with OVCAR3 cells.
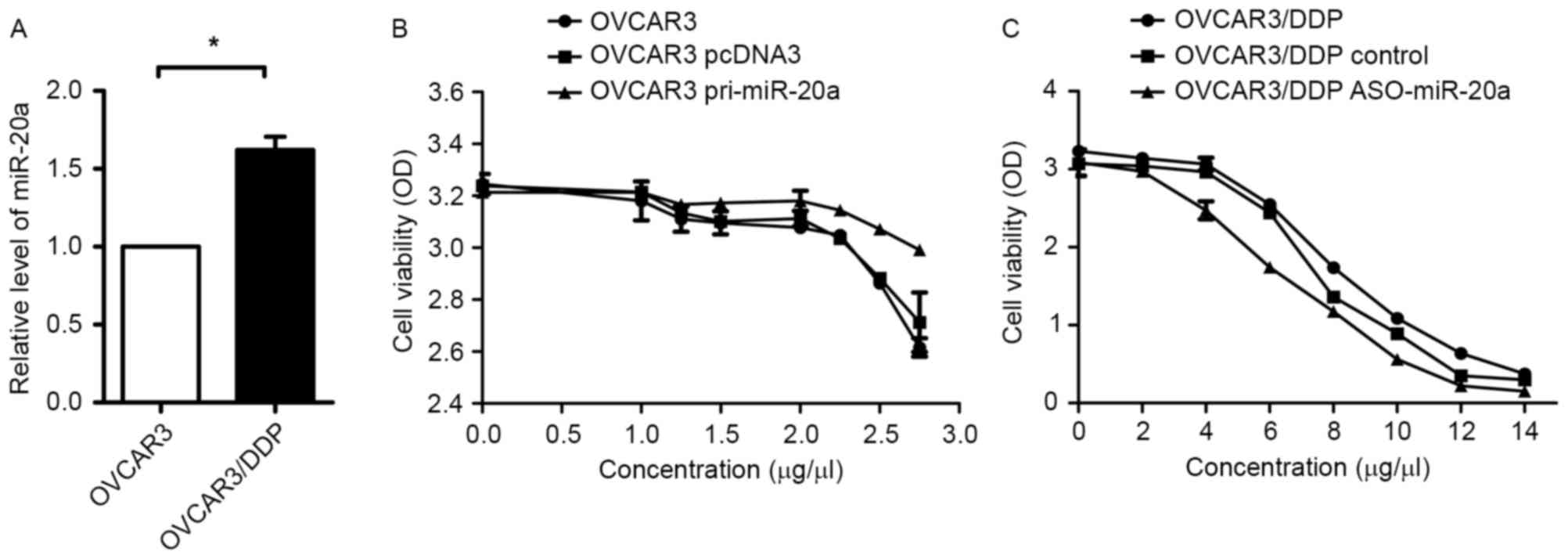
Increased expression level of miR-20a
contributes to cisplatin-resistance of OVCAR3/DDP cells
It was reported that miR-20a was involved in
drug-metastasis in various types of cancer cells (16,20). In
order to investigate whether miR-20a performs a role in
cisplatin-resistant OVCAR3 cells, mRNA expression levels of miR-20a
in OVCAR3 and OVCAR3/DDP cells were evaluated using RT-qPCR. As
presented in Fig. 2A, miR-20a was
overexpressed in OVCAR3/DDP cells compared with in the parent
cells. In addition, overexpression of miR-20a (pri-miR-20a) in
OVCAR3 could enhance resistance to cisplatin compared with in the
control group (pcDNA3; Fig. 2B).
Knockdown of miR-20a by ASO-miR-20a could reduce resistant
capability to cisplatin compared with the control in OVCAR3/DDP
cells (Fig. 2C). These results
demonstrated that miR-20a enhanced cisplatin resistance in
OVCAR3/DDP cells.
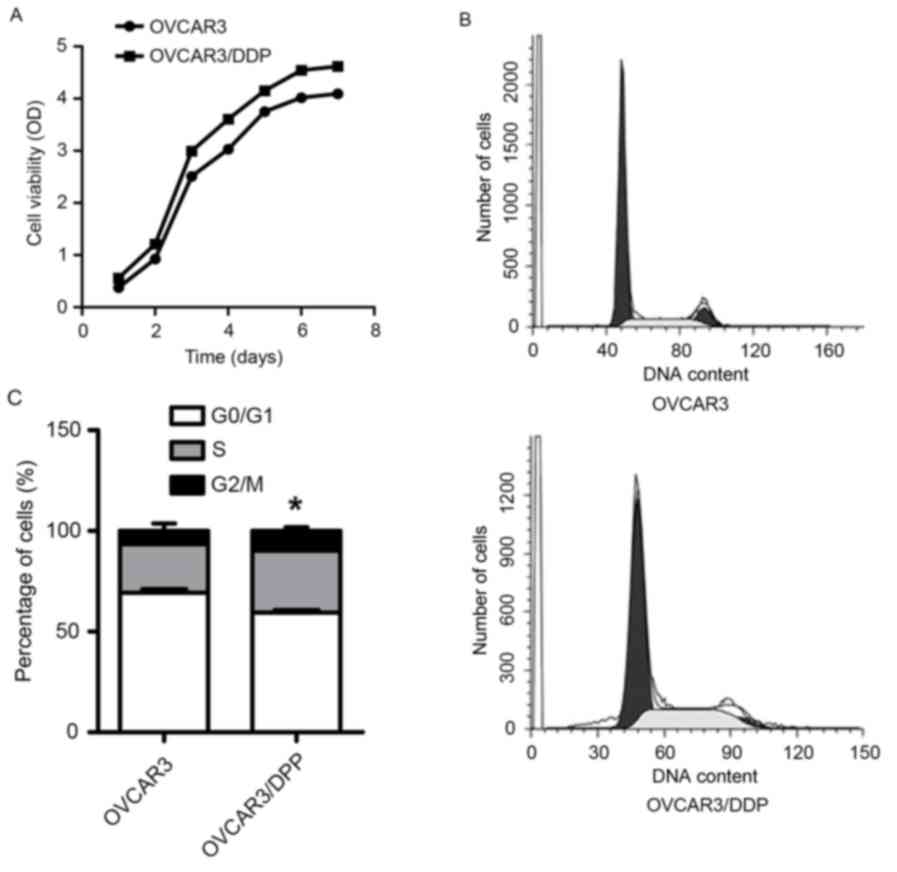
The proliferation of OVCAR3/DDP cells
is increased
In order to determine the proliferation of
OVCAR3/DDP cells, a CCK-8 assay was used to detect the cell
viability of OVCAR3 and OVCAR3/DDP cells for 7 consecutive days.
The population doubling time was analyzed according to
Td (doubling time) = Δtxlg2/(lgNt -
lgN0); where N0 is the cell number at the
beginning and Nt is the cell number at the end, and Δt
is the time from N0 to Nt. Td
(OVCAR3) and Td (OVCAR3/DDP) were ~88 and 69 h,
respectively (Fig. 3A), which
suggested that OVCAR3/DDP cells acquired a reinforced proliferation
compared with OVCAR3 cells. In addition, flow cytometry assay
results demonstrated that there were fewer OVCAR3/DDP cells in the
G0/G1 cell cycle phase and an increased
number of cells in the S and G2M phases (Fig. 3B and C). The proliferation index of
OVCAR3/DDP and OVCAR3 cells was 40.59 and 30.73%, respectively,
which suggested that miR-20a may promote OVCAR3/DDP cellular
proliferation.
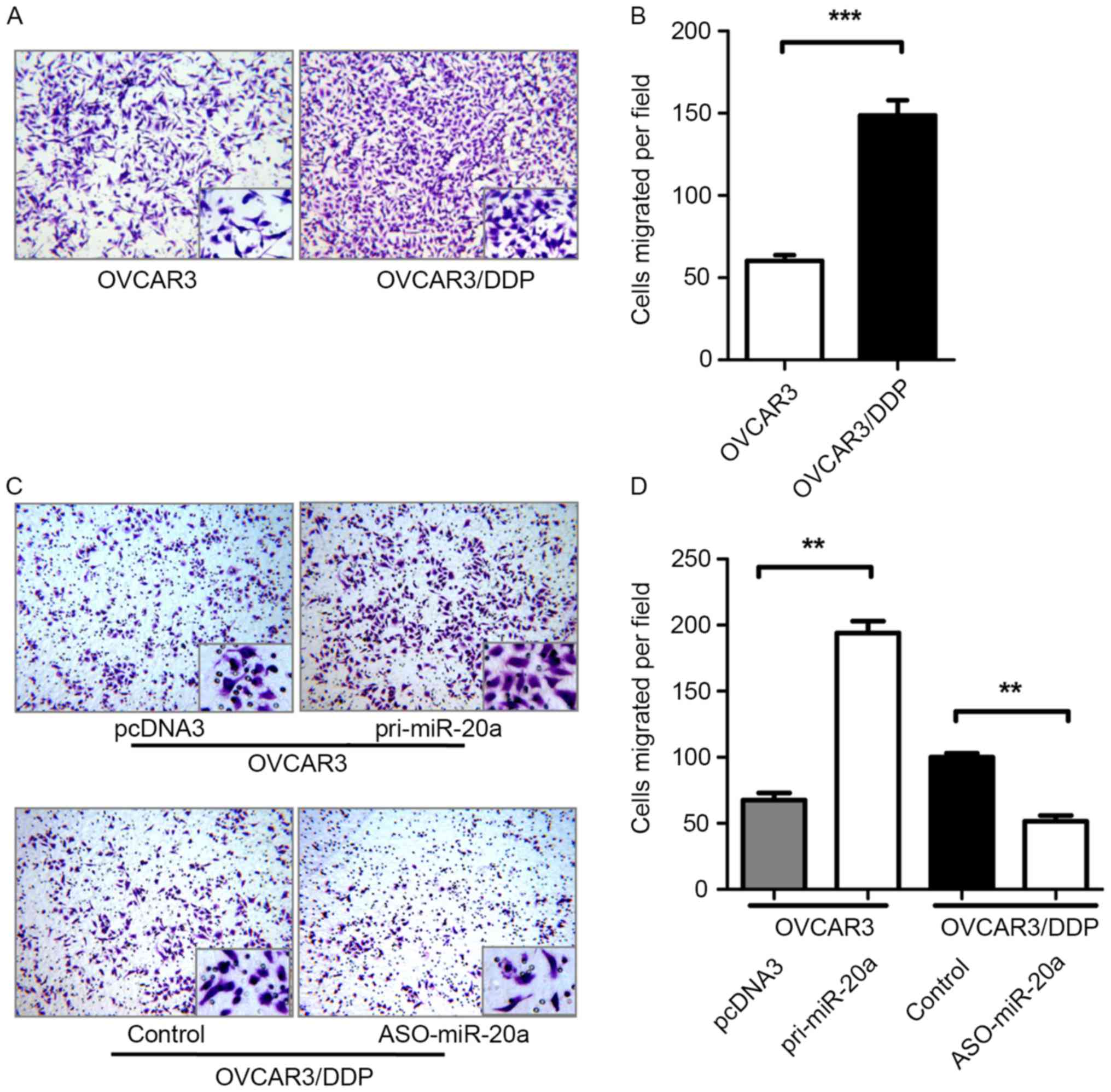
miR-20a regulates OVCAR3 and
OVCAR3/DDP cell migration
The present study verified the migration abilities
of OVCAR3 and OVCAR3/DDP cells. The migration ability of OVCAR3/DDP
cell was enhanced 155% compared with OVCAR3 cells (Fig. 4A and B). To determine whether miR-20a
had an effect on the migration of cells, a Transwell assay was
performed to determine the migration of OVCAR3 cells transfected
with pri-miR-20a or pcDNA3 and OVCAR3/DDP cells transfected with
ASO-miR-20a or the control. An increased or decreased number of
migrated cells were observed corresponding to upregulation or
downregulation of miR-20a (Fig. 4C and
D).
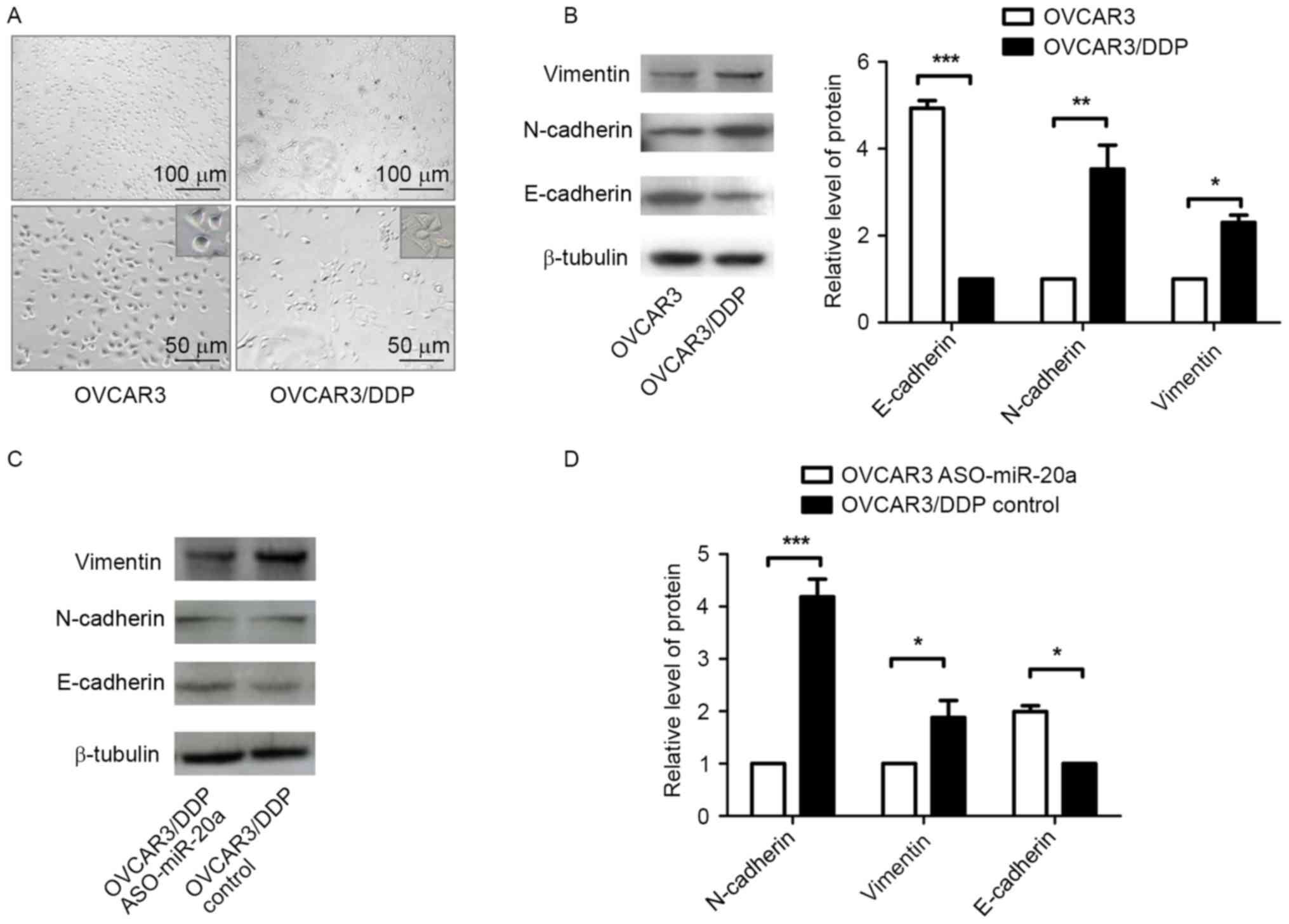
OVCAR3/DDP cells undergo EMT
During the cisplatin-resistant cells development,
OVCAR3 cells exhibited protrusion and mesenchymal phenotypes
(Fig. 5A). Thus, the present study
evaluated the EMT markers of OVCAR3 and OVCAR3/DDP cells by western
blot analysis. OVCAR3/DDP cells expressed increased levels of
Vimentin and N-cadherin and decreased levels of E-cadherin compared
with OVCAR3 cells (Fig. 5B), which
suggested that EMT occurred during the process of
cisplatin-resistance. In addition, OVCAR3/DDP cells transfected
with ASO-miR-20a exhibited decreased expression levels of
N-cadherin and Vimentin protein and increased expression levels of
E-cadherin protein, compared with the control group (Fig. 5C and D). These results suggested that
miR-20a may induce EMT in OVCAR3/DDP cells.
Discussion
Ovarian cancer is the most lethal type of
gynecological cancer (21). Although
the international standard of care is surgery supplemented with
paclitaxel and platinum based chemotherapy, the majority of
patients will relapse in one to two years due to chemoresistance
(22). Cisplatin is one of the most
commonly used platinum based chemotherapy treatments for ovarian
cancer; however, the resistant mechanism underlying cisplatin
cytotoxicity to cancer cells remains unclear. Additional
understanding of the mechanism underlying ovarian cancer resistance
to cisplatin may aid the development of extending the survival rate
and increasing the percentage of females who are cured from ovarian
cancer. In the present study, a cisplatin-resistant subline,
OVCAR3/DDP, was established from OVCAR3 ovarian cancer cells. The
proliferation and migration abilities of OVCAR3/DDP were enhanced
compared with those of OVCAR3 cells.
Dysregulation of miRs has been widely documented in
almost all types of human malignancies (23). Certain miRs may improve chemotherapy
sensitivity, whereas others may induce chemoresistance of cancer
cells (24–26). In ovarian cancer, miR-130a and
miR-374a were demonstrated to function as negative regulators of
cisplatin resistance in A2780 cells (27), whereas miR-31 positively regulated
cisplatin resistance in numerous ovarian cancer cells (28). In the present study, miR-20a was
identified as a novel positive regulator for OVCAR3 cells cisplatin
resistance and migration. Previous studies have suggested that miRs
may induce EMT development, drug resistance and metastasis
(29–32). During EMT, epithelial cells acquire a
mesenchymal phenotype that is characterized by the loss of
intercellular junctions and increased cell migration (33). In the present study, OVCAR3/DDP cells
morphology was altered due to cisplatin, and the cells expressed
increased levels of E-cadherin and decreased levels of Vimentin and
N-cadherin, compared with the parental cells, which suggested that
EMT occurred during the cells cisplatin-resistance. In addition,
the effects of the EMT markers were reversed by inhibition of the
expression of miR-20a. These results indicated that EMT in
OVCAR3/DDP cells may be activated by miR-20a, which accelerated the
ovarian cancer malignant development.
The present study provided a novel insight into
understanding the mechanism underlying chemoresistance of ovarian
cancer cells. It also suggested that miR-20a may be promising as a
novel therapeutic target for overcoming drug resistance and
metastasis of ovarian cancer.
Acknowledgements
The authors would like to thank of Department of
Pathogen Biology of Tianjin Medical University for donating the
OVCAR3 cell line. The present study was supported by the National
Natural Science Foundation of China (grant no. 81301779).
References
|
1
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Colombo N, Peiretti M, Parma G, Lapresa M,
Mancari R, Carinelli S, Sessa C and Castiglione M; ESMO Guidelines
Working Group, : Newly diagnosed and relapsed epithelial ovarian
carcinoma: ESMO clinical practice guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 21 Suppl 5:v23–v30. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foster T, Brown TM, Chang J, Menssen HD,
Blieden MB and Herzog TJ: A review of the current evidence for
maintenance therapy in ovarian cancer. Gynecol Oncol. 115:290–301.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morgan RJ Jr, Alvarez RD, Armstrong DK,
Burger RA, Chen LM, Copeland L, Crispens MA, Gershenson DM, Gray
HJ, Hakam A, et al: Ovarian cancer, version 2.2013. J Natl Compr
Canc Netw. 11:1199–1209. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen DW, Pouliot LM, Hall MD and Gottesman
MM: Cisplatin resistance: A cellular self-defense mechanism
resulting from multiple epigenetic and genetic changes. Pharmacol
Rev. 64:706–721. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Borley J and Brown R: Epigenetic
mechanisms and therapeutic targets of chemotherapy resistance in
epithelial ovarian cancer. Ann Med. 47:359–369. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Anglicheau D, Muthukumar T and
Suthanthiran M: MicroRNAs: Small RNAs with big effects.
Transplantation. 90:105–112. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu C, Xie S, Song C, Huang L and Jiang Z:
Lin28 mediates cancer chemotherapy resistance via regulation of
miRNA signaling. Hepatogastroenterology. 61:1138–1141.
2014.PubMed/NCBI
|
|
10
|
Niu J, Xue A, Chi Y, Xue J, Wang W, Zhao
Z, Fan M, Yang CH, Shao ZM, Pfeffer LM, et al: Induction of
miRNA-181a by genotoxic treatments promotes chemotherapeutic
resistance and metastasis in breast cancer. Oncogene. 35:1302–1313.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo Y, Yan K, Fang J, Qu Q, Zhou M and
Chen F: Let-7b expression determines response to chemotherapy
through the regulation of cyclin D1 in glioblastoma. J Exp Clin
Cancer Res. 32:412013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie T, Huang M, Wang Y, Wang L, Chen C and
Chu X: MicroRNAs as regulators, biomarkers and therapeutic targets
in the drug resistance of colorectal cancer. Cell Physiol Biochem.
40:62–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kopczynska E: Role of microRNAs in the
resistance of prostate cancer to docetaxel and paclitaxel. Contemp
Oncol (Pozn). 19:423–427. 2015.PubMed/NCBI
|
|
14
|
Yang G, Jiang O, Ling D, Jiang X, Yuan P,
Zeng G, Zhu J, Tian J, Weng Y and Wu D: MicroRNA-522 reverses drug
resistance of doxorubicin-induced HT29 colon cancer cell by
targeting ABCB5. Mol Med Rep. 12:3930–3936. 2015.PubMed/NCBI
|
|
15
|
Yu J, Lu Y, Cui D, Li E, Zhu Y, Zhao Y,
Zhao F and Xia S: miR-200b suppresses cell proliferation, migration
and enhances chemosensitivity in prostate cancer by regulating
Bmi-1. Oncol Rep. 31:910–918. 2014.PubMed/NCBI
|
|
16
|
Jiang Z, Yin J, Fu W, Mo Y, Pan Y, Dai L,
Huang H, Li S and Zhao J: MiRNA 17 family regulates
cisplatin-resistant and metastasis by targeting TGFbetaR2 in NSCLC.
PloS One. 9:e946392014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weng H, Huang H, Dong B, Zhao P, Zhou H
and Qu L: Inhibition of miR-17 and miR-20a by oridonin triggers
apoptosis and reverses chemoresistance by derepressing BIM-S.
Cancer Res. 74:4409–4419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan X, Liu Y, Jiang J, Ma Z, Wu H, Liu T,
Liu M, Li X and Tang H: miR-20a promotes proliferation and invasion
by targeting APP in human ovarian cancer cells. Acta Biochim
Biophys Sin (Shanghai). 42:318–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Zheng L, Ding Y, Li Q, Wang R,
Liu T, Sun Q, Yang H, Peng S, Wang W and Chen L: MiR-20a induces
cell radioresistance by activating the PTEN/PI3K/Akt signaling
pathway in hepatocellular carcinoma. Int J Radiat Oncol Biol Phys.
92:1132–1140. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jayson GC, Kohn EC, Kitchener HC and
Ledermann JA: Ovarian cancer. Lancet. 384:1376–1388. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Markman M: Current standards of care for
chemotherapy of optimally cytoreduced advanced epithelial ovarian
cancer. Gynecol Oncol. 131:241–245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:pp. 2999–3004. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li N, Yang L, Wang H, Yi T, Jia X, Chen C
and Xu P: MiR-130a and MiR-374a function as novel regulators of
cisplatin resistance in human ovarian cancer A2780 cells. PloS One.
10:e01288862015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Samuel P, Pink RC, Caley DP, Currie JM,
Brooks SA and Carter DR: Over-expression of miR-31 or loss of
KCNMA1 leads to increased cisplatin resistance in ovarian cancer
cells. Tumour Biol. 37:2565–2573. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song J and Li Y: miR-25-3p reverses
epithelial-mesenchymal transition via targeting Sema4C in
cisplatin-resistance cervical cancer cells. Cancer Sci. 108:23–31.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Raza U, Saatci Ö, Uhlmann S, Ansari SA,
Eyüpoğlu E, Yurdusev E, Mutlu M, Ersan PG, Altundağ MK, Zhang JD,
et al: The miR-644a/CTBP1/p53 axis suppresses drug resistance by
simultaneous inhibition of cell survival and epithelial-mesenchymal
transition in breast cancer. Oncotarget. 7:49859–49877.
2016.PubMed/NCBI
|
|
31
|
Singh R, Yadav V, Kumar S and Saini N:
MicroRNA-195 inhibits proliferation, invasion and metastasis in
breast cancer cells by targeting FASN HMGCR, ACACA and CYP27B1. Sci
Rep. 5:174542015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pavelic S Kraljevic, Sedic M, Bosnjak H,
Spaventi S and Pavelic K: Metastasis: New perspectives on an old
problem. Mol Cancer. 10:222011. View Article : Google Scholar : PubMed/NCBI
|