Introduction
Malignant glioma is an aggressive type of cancer and
the most frequent form of primary brain tumor, which is mainly
accompanied by a poor prognosis. Despite advanced therapeutic
strategies, the median overall survival time of newly diagnosed
glioblastoma patients remains at 1 to 2 years (1,2). The
primary treatment strategy for patients with malignant glioma
consists of surgical resection followed by adjuvant radiation and
chemotherapy. Therapeutic options for these patients are limited
and the prognosis remains dismal (3,4).
Therefore, suppressing the aggressiveness of glioma cell
proliferation and cell migration could be a novel and effective
therapeutic strategy that deserves further research.
Thioredoxin-interacting protein (TXNIP), also known
as vitamin D3 upregulating protein 1 or thioredoxin-binding protein
2 (5), is a multifunctional protein
that is involved in cell proliferation, differentiation, and
apoptosis (6,7). TXNIP has been identified as a major
redox regulator and a tumor suppressor gene in various solid tumors
and hematological malignancies (8–10), but its
role in malignant glioma cells or in glioma tissues has not been
investigated prior to the present study.
Materials and methods
Patients and tissue samples
The study was approved by the Research Ethics
Committee of Tangdu Hospital, the Fourth Military Medical
University (Xi'an, China). All patients willingly consented to
participate in the study by signing the written informed consent
form. All specimens and data were handled according to the relevant
ethical and legal standards.
A total of 54 glioma specimens (24 male and 30
female), resected between October 2008 and Octover 2010 and stored
in liquid nitrogen, were retrieved from the archives of the
Department of Pathology, Tangdu Hospital (Xi'an, China). The age of
these patients ranged from 6 to 75 (mean age of 42.74 and median
age of 42.5 years). These retrieved specimens were directly used
for reverse transcription- quantitative PCR (RT-qPCR) analysis, and
formalin-fixed and paraffin-embedded for immunohistochemical (IHC)
analysis. Conferring to the World Health Organization (WHO)
classification (11), the
histopathological sections were evaluated by two pathologists, with
differences resolved by careful discussion. Accordingly, a total of
29/54 gliomas were classified as low-grade gliomas [7 pilocytic
astrocytomas (WHO I) and 22 diffuse astrocytomas (WHO II)] and
25/54 as high-grade gliomas [20 anaplasia astrocytomas (WHO III)
and 5 primary glioblastomas (WHO IV)]. None of the patients had
been treated with chemotherapy or radiotherapy prior to surgery.
The clinicopathological features of all patients are summarized in
Table I.
 | Table I.Clinicopathological characteristics of
glioma patients. |
Table I.
Clinicopathological characteristics of
glioma patients.
|
|
| TXNIP expression |
|---|
|
|
|
|
|---|
| Clinicopathological
features | Patient number | High, n (%) | Low, n (%) |
|---|
| WHO grade |
|
|
|
| I | 7 | 6
(86) | 1
(14) |
| II | 22 | 18 (82) | 4
(18) |
| III | 20 | 4
(20) | 16 (80) |
| IV | 5 | 0
(0) | 5
(100) |
| Age, years |
|
|
|
|
<55 | 42 | 23 (55) | 19 (45) |
|
≥55 | 12 | 5
(42) | 7
(58) |
| Sex |
|
|
|
|
Male | 24 | 10 (42) | 14 (58) |
|
Female | 30 | 18 (60) | 12 (40) |
The patient's survival time was assessed by
calculating the days from the date of the initial glioma-related
surgery to mortality. Patients who succumbed to diseases not
directly associated with the gliomas or those who succumbed for
other reasons were excluded from this study. The association
between the overall survival time and the TXNIP expression level
was established using the Kaplan-Meier method.
IHC analysis
Formalin-fixed, paraffin-embedded, sectioned tissues
(4-µm thick) were stained using the Elivision Plus/horseradish
peroxidase (HRP) detection system (Fuzhou Maixin Biotech Co., Ltd.,
Fuzhou, China) and counterstained with hematoxylin. In brief,
following a peroxidase block with 3%
H2O2/methanol (Fuzhou Maixin Biotech Co.,
Ltd.) for 30 min, specimens were blocked with 5% normal goat serum.
The slides were then incubated overnight with mouse polyclonal
anti-human TXNIP primary antibody (cat. no. K0205-3; Medical &
Biological Laboratories Co., Ltd., Nagoya, Japan) at 1:50 dilution,
at 4°C. An IHC antibody diluent (cat. no. ZLI-9029; ZSGB-BIO,
Beijing, China) at 1:50 dilution replaced the primary antibody for
negative control slides. The specimens were then briefly washed
three times with phosphate-buffered saline (PBS) and incubated at
room temperature with Solution A for 20-min, followed by a 30-min
incubation with Solution B (EnVision Plus/HRP mouse/Rabbit IHC kit;
cat. no. KIT-9902; Fuzhou Maixin Biotech Co., Ltd.) at 37°C. The
signal visualization was performed by treatment with DAB chromogen
for 2 to 3 min. After washing with water, specimens were
counterstained with Meyer's hematoxylin (Maixin Biotech, Fuzhou,
China). The WHO I neoplastic brain tissues of 54 glioma specimens,
which were obtained from the archives of the Department of
Pathology, Tangdu Hospital, were used as control tissues.
The evaluation standards of the pathologists were as
follows: i) The intensity of the cell staining; non-staining as
negative (−), light brown staining as weakly positive (+), brown
staining as medium positive (++) and sepia staining as strong
positive (+++/++++); ii) the number of positive cells; the numbers
of positive cells <25% (+), the numbers of positive cells among
25–49% (++) and the numbers of positive cells >50% (+++).
Qualitative and half-quantitative staining results were achieved by
combined the analysis results of the two experiments. A total of
5–10 high power fields were randomly observed and the averages were
taken. Negative (−) and weakly positive (+) was regarded as low
expression and the others were regarded as high expression.
Cell culture
The glioma U251MG cell line was purchased from the
Chinese Academy of Sciences Cell Bank (Shanghai, China) and was
stored in liquid nitrogen until use. The cell line was
authenticated by short tandem repeat profiling. Under culture, the
cells were maintained at 37°C in high-glucose Dulbecco's Modified
Eagle's Medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in a humidified 95%
air and 5% CO2 incubator.
TXNIP downregulation and
transfection
A total of 3 sequences of human lentiviral TXNIP
short hairpin RNA (shRNA) (Table II)
and the non-specific control shRNA were purchased and packaged into
the lentivirus by Shanghai GenePharma Co., Ltd. (Shanghai, China).
U251-MG cells were infected with 0.5 ml shRNA lentiviruses
(1×106/ml) and cultured at 37°C in conventional DMEM
supplemented with 2 µg/ml puro (Sigma-Aldrich; Merck KGaA) in a
humidified 95% air and 5% CO2 incubator for 1 week
following transfection. These four cell lines were labeled as
LV3-TXNIP shRNA1-U251, LV3-TXNIP shRNA2-U251, LV3-TXNIP shRNA3-U251
and LV3-NC-U251. The TXNIP expression level was evaluated by
RT-qPCR and western blotting.
 | Table II.Sequences of shRNAs and reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Sequences of shRNAs and reverse
transcription-quantitative polymerase chain reaction.
| Identity | Sequence
(5′-3′) | (Refs.) |
|---|
| TXNIP shRNA1 |
ATCAGTCAGAGGCAATCATAT |
|
| TXNIP shRNA2 |
GTGGAGGTGTGTGAAGTTA |
|
| TXNIP shRNA3 |
CTCAAGACAGCCCTATCTTTA |
|
| TXNIP-qRT forward
primer |
TCATGGTGATGTTCAAGAAGATC | (13) |
| TXNIP-qRT reverser
primer |
ACTTCACACACCTCCACTATC | (13) |
| GAPDH forward
primer |
GTATTGGGCGCCTGGTCAC | (13) |
| GAPDH reverse
primer |
CTCCTGGAAGATGGTGATGG | (13) |
Western blot analysis and RT-qPCR
analysis
For western blot analysis, cells (LV3-TXNIP
shRNA1-U251, LV3-TXNIP shRNA2-U251, LV3-TXNIP shRNA3-U251 and
LV3-NC-U251) were cultured to cell densities of 2–3×106
in 60 mm dishes. Subsequently, the cells were harvested and lysed
with radio immunoprecipitation assay buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China). Total protein
concentrations were determined using the bicinchoninic acid protein
assay kit (Boster Systems, Inc., Pleasanton, CA, USA). Total
proteins (20 µg) were divided by 12% SDS-PAGE (Genshare Biological,
Shaanxi, China) and transferred onto polyvinylidene fluoride
membranes (EMD Millipore, Billerica, MA, USA). Following a 2-h
blocking period with TBST buffer (TBS plus 0.1% Tween-20)
supplemented with 5% w/v non-fat milk, the membranes were incubated
overnight in TXNIP antibody (cat. no. K0205-3, Medical &
Biological Laboratories Co., Ltd.) at a 1:1,500 dilution at 4°C.
Subsequently, the membranes were incubated with HRP-conjugated goat
anti-mouse secondary antibody (cat. no. BS50350, 1:5,000; Bioworld
Technology, Inc., St. Louis Park, MN, USA) at room temperature for
1 h. Following washing, proteins were visualized using an ECL
detection system (Genshare Biological).
For RT-qPCR, RNA samples were extracted from the
cells and 54 glioma tissues. Typically, 1 µg total RNA was used to
generate cDNA using SuperScript II RT (Takara Bio, Inc., Otsu,
Japan) with an oligo-dT primer. RT-qPCR was performed using the
Power SYBR-Green PCR Master mix, as per the manufacturer's
instruction (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Thermo cycling for all reactions was initiated with a denaturation
step at 95°C for 10 min, followed by 40 cycles at 95°C for 5 sec
and 60°C for 30 sec. Each assay was performed in triplicate. GAPDH
was used as the internal control. SDS 2.2.1 software (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to perform
relative quantification of target genes using the comparative cycle
threshold (2−∆∆Cq) method (12). The primer sequences are described in
Table II (13).
Cell proliferation assay
Cell proliferation was analyzed in vitro
using tetrazolium salt
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT).
A total of five identical 96-well plates were seeded simultaneously
with the four types of cells aforementioned, at a concentration of
2×103 cells/well in 200 µl DMEM. Each cell type was
seeded into seven wells in each plate. After a designated culture
time (12, 24, 48, 72 and 96 h), the medium was replaced with 200 µl
medium supplemented with 0.5 mg/ml MTT (Sigma-Aldrich; KGaA)
reagent and incubated at 37°C for 4 h. Next, the supernatant was
extracted, and the cells were lysed in 200 µl dimethyl sulfoxide
(Sigma-Aldrich; KGaA) for 10 min at 37°C. The optical density
values were measured by recording the absorbance at 490 nm using a
plate reader. The obtained values were presented as the fold
increase with respect to the control group.
Wound-healing assays
U251-MG cells were seeded in 6-well plates and
cultured until they reached confluence. A wound was then created by
manually scraping the cell layer with a 200-µl pipette tip. The
floating cells were removed by washing with PBS three times. The
cells were then incubated in DMEM supplemented with 1% FBS for 24
h. For each group, the number of cells that had migrated into the
simulated wound was counted by imaging eight randomly selected
microscopic fields at 0, 12 and 24 h after wound creation. Images
were acquired with a Nikon DS-5 M Camera System (Nikon Corporation,
Tokyo, Japan) mounted on a phase-contrast Leitz microscope (Leica
Microsystems GmbH, Wetzlar, Germany) and were processed using Adobe
Photoshop 7.0 (Adobe Systems, Inc., San Jose, CA, USA).
In vitro migration and invasion
assays
Migration and invasion experiments were performed
using a QCM 24-Well Cell Invasion assay kit (Cell Biolabs, Inc.,
San Diego, CA, USA). The top chamber of the Transwell system
received 2×103 cells, plated directly on the
polycarbonate Transwell filters (for the Transwell migration assay)
or on the Matrigel-coated polycarbonate Transwell filter (for the
Transwell matrix-penetration invasion assay). For the Transwell
migration assay, the top (containing the cells) and bottom chambers
received serum-free medium. However, for the invasion assay, cells
in the top chamber were suspended in serum-free medium, whereas the
bottom chamber received medium supplemented with 10% FBS, serving
as a chemoattractant. The cells were incubated at 37°C for 8 h (for
the migration assay) or 16 h (for the invasion assay). The
non-migratory or non-invading cells on the upper surface of the
polycarbonate membrane in the top chamber were wiped away using
cotton swabs. The migrated and invaded cells on the lower surface
of the membrane were fixed in 95% ethanol for 5 min, air-dried and
stained with 4 g/l crystal violet prior to counting under a
microscope. A total of three independent experiments were conducted
and the data are presented as the mean ± standard error of the mean
(SEM).
Statistical analysis
All computations were performed using SPSS version
13.0 software, for Windows (SPSS, Inc., Chicago, IL, USA). Data are
expressed as the mean ± SEM throughout. Statistical differences
between the levels of TXNIP expression in different pathological
grades were evaluated with the non-parametric Kruskal-Wallis test,
and those differences in the binominal clinical categories were
evaluated with the non-parametric Mann-Whitney test. The survival
time was calculated according to the Kaplan-Meier method.
Differences between values obtained from in vitro
experiments of the treatment groups were compared to the untreated
control group by paired Student's t-test. P<0.05 was considered
to indicate statistical significance.
Results
Low expression of TXNIP in human
glioma tissues
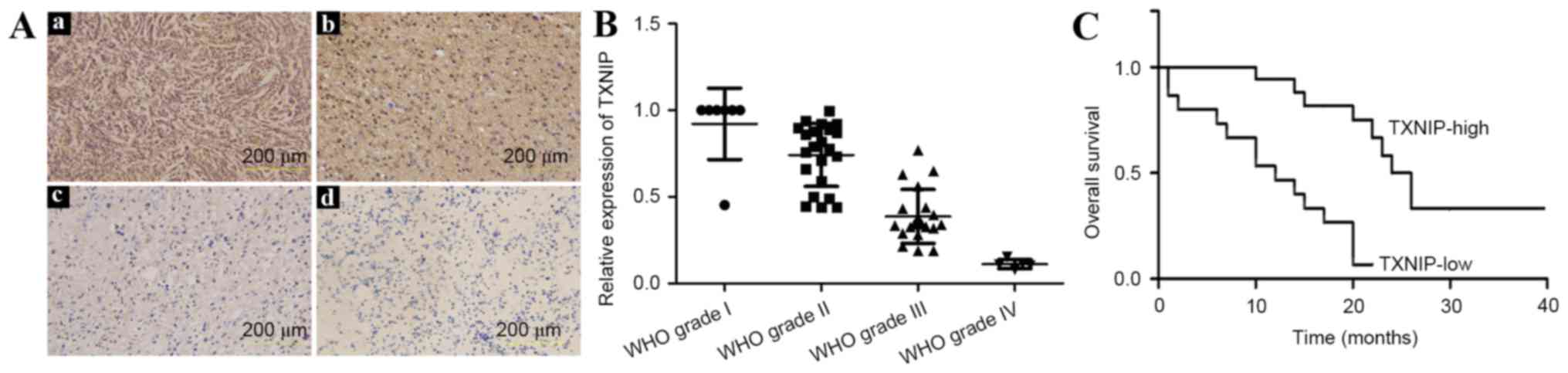
In the 54 patients with gliomas, IHC analysis
revealed high TXNIP expression, localized mainly in the nucleus in
51.9% (28/54) of samples (Fig. 1A),
the majority of which were from low-grade glioma patients. The
remaining 48.1% (26/54) of samples demonstrated lower TXNIP
expression, which was found to be associated with a higher
histopathological glioma grade (P<0.01).
To validate the specificity of the
immunohistochemical results, RT-qPCR analysis was performed. The
primers are listed in Table II.
TXNIP was strongly expressed in low-grade glioma samples, but was
not detected or was only weakly detected in high-grade gliomas,
particularly in glioblastoma (grade IV) (Fig. 1B). These results were consistent with
those of the IHC analysis.
To analyze the overall survival time, the patients
were followed up for a total of 40 months from the initiation of
the study, during which 52/54 patients succumbed to glioma. The
Kaplan-Meier analysis showed a strong association of TXNIP
expression with the overall survival of glioma patients (Fig. 1C); high expression was associated with
increased overall survival time.
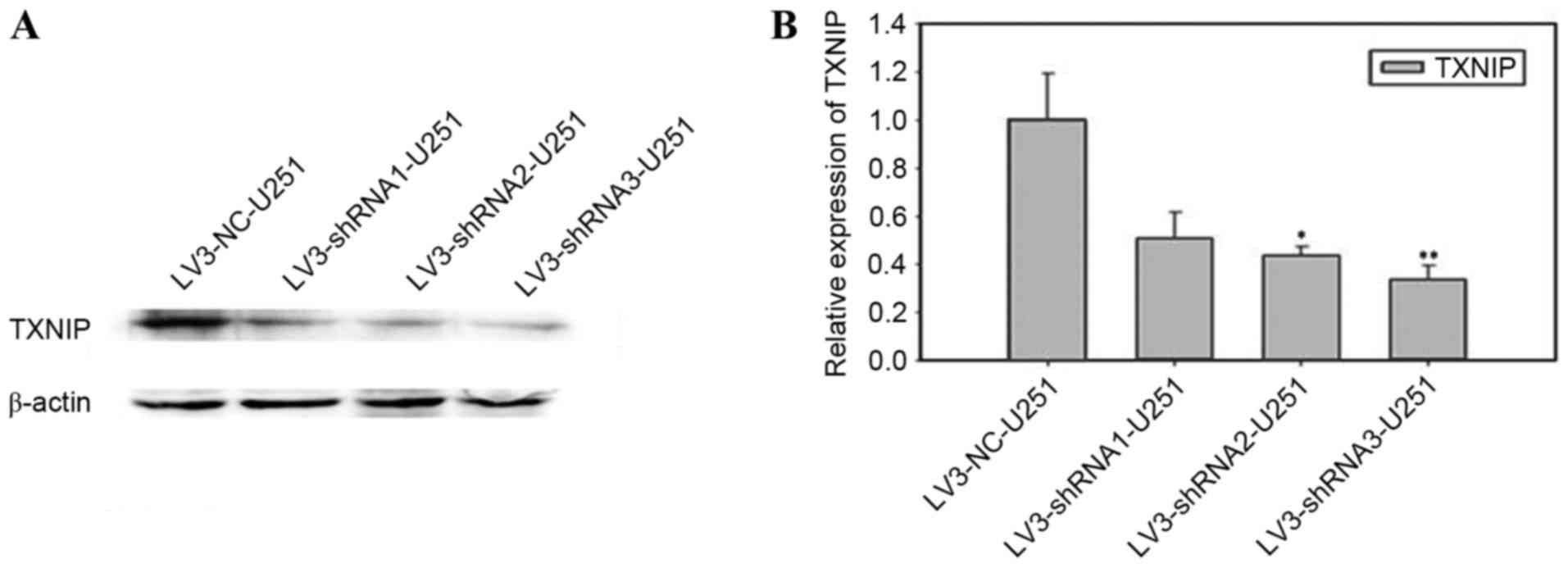
Construction and identification of
TXNIP shRNA-stabilized cell line
U251-MG cells were transfected with one of the TXNIP
shRNA lentiviruses (shRNA1-3) or the scrambled shRNA lentivirus.
The results of the western blot analysis (Fig. 2A) and RT-qPCR (Fig. 2B) revealed that transfection of shRNA2
and shRNA3 was more efficient than shRNA1, and that shRNA3
transfection was more efficient (P<0.01) than shRNA2
transfection (P<0.05).
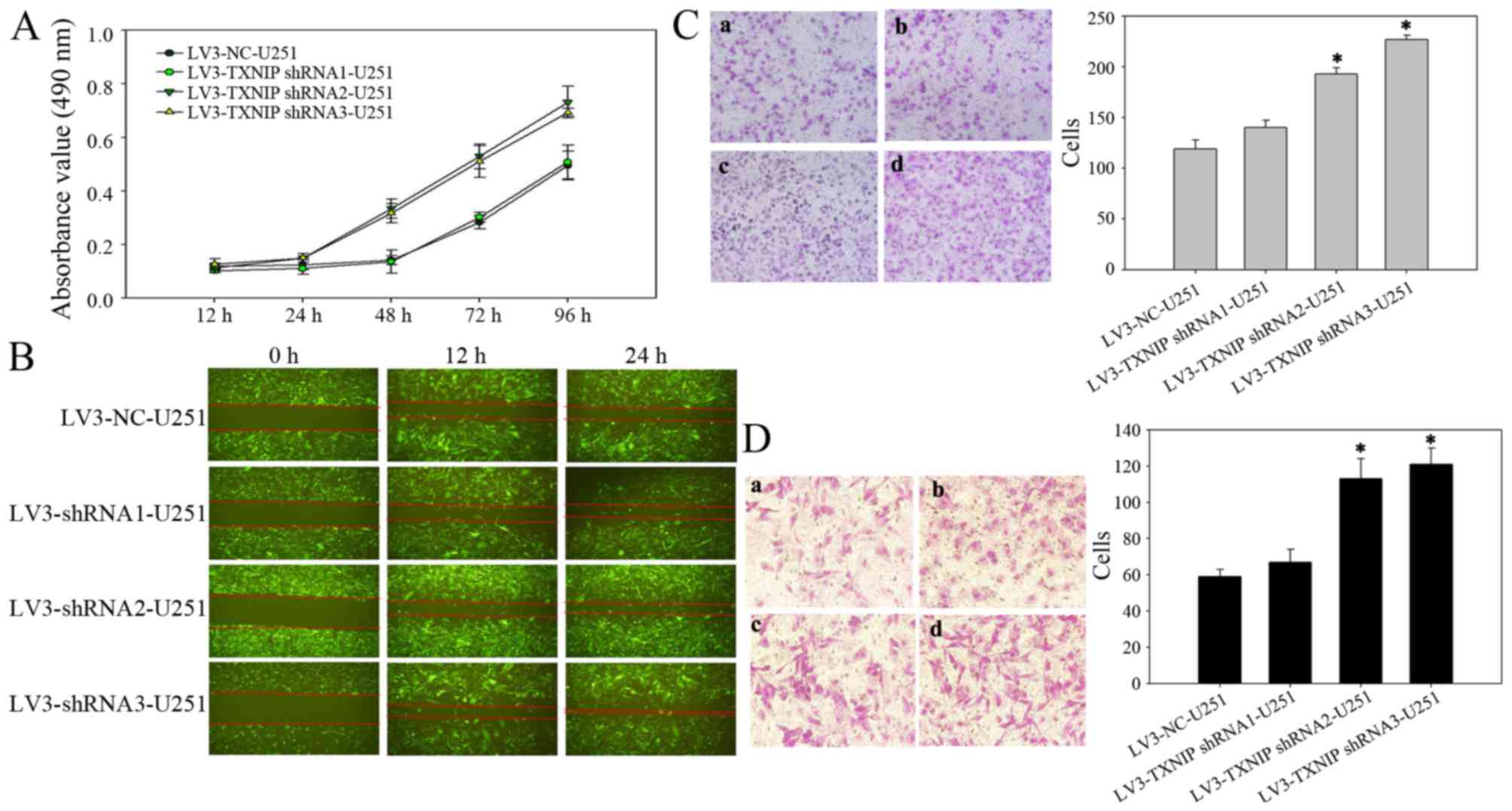
Downregulation of TXNIP promotes
glioma cell invasion, migration and proliferation
Several studies have shown that TXNIP is strongly
downregulated in several types of clinical tumor samples and tumor
cell lines (10,14–16). It
was hypothesized that the downregulation of TXNIP would induce a
more aggressive behavior in cultured glioma cells. To test this
hypothesis, the glioma U251-MG cell line was infected with a
lentivirus encoding an shRNA sequence targeted at TXNIP. First, an
MTT assay was used to examine the effect of TXNIP on cell
proliferation. The cell proliferation was increased in the TXNIP
shRNA group compared with the control groups at 24 h after
transfection, indicating that downregulation of TXNIP could
significantly promote the proliferation of glioma cells (Fig. 3A). A wound-healing assay was then used
to examine the effect of TXNIP on cell migration. Compared with the
LV-NC cells, the TXNIP shRNA cells exhibited rapid migration
(Fig. 3B). Furthermore, Transwell
assays revealed that the downregulation of TXNIP significantly
enhanced the invasion (Fig. 3C) and
migration (Fig. 3D) of glioma
cells.
Discussion
Cancer development is a highly-orchestrated process
that requires complex transcriptional and post-transcriptional
regulation of gene expression (17).
TXNIP has recently been established as a tumor suppressor gene, the
downregulation of which is associated with tumorigenesis and
metastasis (8–10). Low expression of TXNIP has been
observed in breast, liver, stomach and other cancer types (10,14–16). TXNIP
deficiency may promote tumor cell proliferation and inhibit
apoptosis (9,10,18). In
relation to its antitumor mechanisms, TXNIP is essential in a
variety of normal cellular functions, including natural killer cell
development, cellular growth inhibition and apoptosis,
thioredoxin-mediated antioxidant function and the modulation of
other molecules, including mediator complex subunit 23, KiSS-1
metastasis-suppressor and exportin 1 (9,10,19–21).
Although TXNIP expression is frequently suppressed
in human cancer, genetic alterations such as translocation,
deletion or somatic point mutation are uncommon (22). This lack of genetic alteration
suggests that aberrant TXNIP expression in cancer is mainly
regulated via post-transcriptional and translational mechanisms.
TXNIP promoter hypermethylation was first observed in renal cell
carcinoma using methylation-specific PCR, induced by ferric
nitrilotriacetate (23). Ahsan et
al (24) proposed that DNA
methylation could be implicated in the silencing of TXNIP
expression in leukemia. The process of DNA methylation is an
established method of silencing various other tumor suppressor
genes (25–27). In addition to this, histone
deacetylation, regulation by microRNAs and protein translation may
also serve important roles in repressing TXNIP expression (14,28–30).
In the present study, IHC analysis of the expression
level of TXNIP was conducted in surgically resected glioma tissues,
which revealed the marked downregulation of TXNIP in high-grade
human glioma compared with low-grade human glioma. Furthermore,
under in vitro conditions, shRNAs were used to downregulate
expression of TXNIP in the U251-MG glioma cell line, which enhanced
the invasion, migration and proliferation of the glioma cells.
Downregulation of TXNIP converted the cells to an aggressive cancer
phenotype, which may be representative of cells in high-grade human
glioma. The results of the present study established a strong
inverse association between TXNIP expression level and the glioma
aggressiveness and histopathological grading of the disease, and a
direct association with patient survival time.
In conclusion, from the results of the present study
it is apparent that the tumor suppressor activity of TXNIP is
strongly associated with glioma development and progression. These
data highlight the importance of TXNIP as a potential therapeutic
target and prognostic marker in advanced human gliomas.
Acknowledgements
This study was supported by grants from the National
Natural Scientific Foundation of China (nos. 81272419 and
81572983), the Social Development of Technology Research Projects
in Shaanxi Province (no. 2015SF027), the Natural Scientific
Foundation of Shaanxi Province (no. 2014JM4148) and the Beijing Key
Laboratory of Brain Major Diseases Open Project (2015).
References
|
1
|
Chang L, Su J, Jia XZ and Ren H: Treating
malignant glioma in Chinese patients: Update on temozolomide. Onco
Targets Ther. 7:235–244. 2014.PubMed/NCBI
|
|
2
|
Davis FG and McCarthy BJ: Current
epidemiological trends and surveillance issues in brain tumors.
Expert Rev Anticancer Ther. 1:395–401. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang P, Wang Y, Peng X, You G, Zhang W,
Yan W, Bao Z, Wang Y, Qiu X and Jiang T: Management and survival
rates in patients with glioma in China (2004–2010): A retrospective
study from a single-institution. J Neurooncol. 113:259–266. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen C, Xu T, Lu Y, Chen J and Wu S: The
efficacy of temozolomide for recurrent glioblastoma multiforme. Eur
J Neurol. 20:223–230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen KS and DeLuca HF: Isolation and
characterization of a novel cDNA from HL-60 cells treated with 1,
25-dihydroxyvitamin D-3. Biochim Biophys Acta. 1219:26–32. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim SY, Suh HW, Chung JW, Yoon SR and Choi
I: Diverse functions of VDUP1 in cell proliferation,
differentiation, and diseases. Cell Mol Immunol. 4:345–351.
2007.PubMed/NCBI
|
|
7
|
Patwari P, Chutkow WA, Cummings K,
Verstraeten VL, Lammerding J, Schreiter ER and Lee RT:
Thioredoxin-inde-pendent regulation of metabolism by the
alpha-arrestin proteins. J Biol Chem. 284:24996–25003. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou J, Yu Q and Chng WJ: Txnip (vdup-1,
tbp-2): A major redox regulator commonly suppressed in cancer by
epigenetic mechanisms. Int J Biochem Cell Biol. 43:1668–1673. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou JB and Chng WJ: Roles of thioredoxin
binding protein (TXNIP) in oxidative stress, apoptosis and cancer.
Mitochondrion. 13:163–169. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoshihara E, Masaki S, Matsuo Y, Chen Z,
Tian H and Yodoi J: Thioredoxin/Txnip: Redoxisome, as a redox
switch for the pathogenesis of diseases. Front Immunol. 4:5142014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou J, Bi C, Cheong LL, Mahara S, Liu SC,
Tay KG, Koh TL, Yu Q and Chng WJ: The histone methyltransferase
inhibitor, DZNep, up-regulates TXNIP, increases ROS production, and
targets leukemia cells in AML. Blood. 118:2830–2839. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Butler LM, Zhou X, Xu WS, Scher HI,
Rifkind RA, Marks PA and Richon VM: The histone deacetylase
inhibitor SAHA arrests cancer cell growth, up-regulates
thioredoxin-binding protein-2, and down-regulates thioredoxin. Proc
Natl Acad Sci. 99:pp. 11700–11705. 2002; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamaguchi F, Takata M, Kamitori K, Nonaka
M, Dong Y, Sui L and Tokuda M: Rare sugar d-allose induces specific
up-regulation of txnip and subsequent G1 cell cycle arrest in
hepatocellular carcinoma cells by stabilization of p27kip1. Int J
Oncol. 32:377–385. 2008.PubMed/NCBI
|
|
16
|
Song H, Cho D, Jeon JH, Han SH, Hur DY,
Kim YS and Choi I: Vitamin d (3) up-regulating protein 1 (vdup1)
antisense DNA regulates tumorigenicity and melanogenesis of murine
melanoma cells via regulating the expression of fas ligand and
reactive oxygen species. Immunol Lett. 86:235–247. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ying Z, Li Y, Wu J, Zhu X, Yang Y, Tian H,
Li W, Hu B, Cheng SY and Li M: Loss of miR-204 expression enhances
glioma migration and stem cell-like phenotype. Cancer Res.
73:990–999. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang P, Pang X and Tu Y:
Thioredoxin-interacting protein as a common regulation target for
multiple drugs in clinical therapy/application. Cancer Transl Med.
1:26–30. 2015. View Article : Google Scholar
|
|
19
|
Han SH, Jeon JH, Ju HR, Jung U, Kim KY,
Yoo HS, Lee YH, Song KS, Hwang HM, Na YS, et al: VDUP1 upregulated
by TGF-beta1 and 1,25-dihydorxyvitamin D3 inhibits tumor cell
growth by blocking cell-cycle progression. Oncogene. 22:4035–4046.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Le Jan S, Le Meur N, Cazes A, Philippe J,
Le Cunff M, Léger J, Corvol P and Germain S: Characterization of
the expression of the hypoxia-induced genes neuritin, txnip and
IGFBP3 in cancer. FEBS Lett. 580:3395–3400. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee KN, Kang HS, Jeon JH, Kim EM, Yoon SR,
Song H, Lyu CY, Piao ZH, Kim SU, Han YH, et al: Vdup1 is required
for the development of natural killer cells. Immunity. 22:195–208.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Erkeland SJ, Palande KK, Valkhof M, Gits
J, Danen-van Oorschot A and Touw IP: The gene encoding
thioredoxin-interacting protein (TXNIP) is a frequent integration
site in virus-induced mouse leukemia and is overexpressed in a
subset of AML patients. Leuk Res. 33:1367–1371. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dutta KK, Nishinaka Y, Masutani H,
Akatsuka S, Aung TT, Shirase T, Lee WH, Yamada Y, Hiai H, Yodoi J
and Toyokuni S: Two distinct mechanisms for loss of
thioredoxin-binding protein-2 in oxidative stress-induced renal
carcinogenesis. Lab Invest. 85:798–807. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ahsan MK, Masutani H, Yamaguchi Y, Kim YC,
Nosaka K, Matsuoka M, Nishinaka Y, Maeda M and Yodoi J: Loss of
interleukin-2-dependency in HTLV-I-infected T cells on gene
silencing of thioredoxin-binding protein-2. Oncogene. 25:2181–2191.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng YD, Geng H, Cheng SH, Liang P, Bai
Y, Li J, Srivastava G, Ng MH, Fukagawa T, Wu X, et al: The KRAB
Zinc finger protein ZNF382 is a general, proapoptotic tumor
suppressor repressing multiple oncogenes and frequently silenced in
multiple carcinomas. Cancer Res. 70:6516–6526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng YD, Liang P, Geng H, Wang Z, Li L,
Cheng SH, Ying J, Su X, Ng KM, Ng MH, et al: A novel 19q13
nucleolar zinc finger protein suppresses tumor cell growth through
inhibiting ribosome biogenesis and inducing apoptosis but is
frequently silenced in multiple carcinomas. Mol Cancer Res.
10:925–936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang S, Cheng Y, Du W, Lu L, Zhou L, Wang
H, Kang W, Li X, Tao Q, Sung J and Yu J: Zinc-finger protein 545 is
a novel tumour suppressor that acts by inhibiting ribosomal RNA
transcription in gastric cancer. Gut. 62:833–841. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kato T, Shimono Y, Hasegawa M, Jijiwa M,
Enomoto A, Asai N, Murakumo Y and Takahashi M: Characterization of
the HDAC1 complex that regulates the sensitivity of cancer cells to
oxidative stress. Cancer Res. 69:3597–3604. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ungerstedt JS, Sowa Y, Xu WS, Shao Y,
Dokmanovic M, Perez G, Ngo L, Holmgren A, Jiang X and Marks PA:
Role of thioredoxin in the response of normal and transformed cells
to histone deacetylase inhibitors. Proc Natl Acad Sci USA. 102:pp.
673–678. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan GR, Xu SH, Tan ZL, Liu L and He QY:
Global identification of miR-373-regulated genes in breast cancer
by quantitative proteomics. Proteomics. 11:912–920. 2011.
View Article : Google Scholar : PubMed/NCBI
|