Introduction
Bladder cancers (BCs) are mixtures of heterogeneous
cell populations, and multiple factors are involved in determining
their recurrence, progression and survival (1). Whereas the overall survival rate of
patients with non-muscle-invasive bladder cancer (NMIBC) is
excellent compared with that in other malignancies, several of
these patients exhibit a high risk of recurrence and a variable
risk of progression despite administration of local therapies
(2,3).
A total of 10–20% of patients with NMIBCs subsequently develop
invasive or metastatic cancer (4,5).
Muscle-invasive BC (MIBC) is often a life-threatening disease with
short curable period, and the oft-cited 50% overall survival at 5
years for MIBC has remained relatively unchanged in 20 years
(6,7).
Accordingly, identifying patients at risk of developing MIBC is
essential for appropriate disease management in patients with
NMIBC. Clinical risk factors for progression include: Invasion of
the lamina propria, high grade based on the World Health
Organization/International Society of Urologic Pathology consensus
classification (8,9), tumor size, occurrence of carcinoma in
situ (CIS) and multiplicity or recurrence of high-risk tumors
(10–12). Two multivariate risk assessment tools
for predicting the outcome of NMIBC have been developed (13,14).
However, currently none of the predictive tools based on
conventional clinical and pathological parameters are sufficiently
sensitive or specific to detect, monitor and determine the
prognosis of BC (15). To overcome
these limitations, studies have focused on identifying molecular
markers that enable clinicians to classify BCs in more detail,
thereby enabling selection of the optimal treatment regimen
(16,17).
Previously, microarray technology has facilitated
the development of numerous cancer classifiers, identification of
tumor subclasses, discovery of progression markers and prediction
of disease outcome in numerous types of cancer (18). Molecular staging may provide increased
accurate predictions of patient outcome compared with the currently
employed histopathological staging, and may also improve treatment
outcomes by enabling treatment to be tailored to the severity of
the disease (19). Our previous study
performed a microarray analysis of specimens derived from 103
primary NMIBCs and identified an eight-gene progression-associated
gene classifier (Table I) (20). The original study defined progression
as TNM upstaging, and included several cases of progression between
Ta and T1. In the present study, our previously published
progression-associated gene classifier (20) was validated in the prediction of
muscle invasion in NMIBC over an extended follow-up period.
 | Table I.Eight-gene progression-associated
molecular classifiers for non-muscle-invasive bladder cancer. |
Table I.
Eight-gene progression-associated
molecular classifiers for non-muscle-invasive bladder cancer.
| Gene symbol | Sense primer
(5′-3′) | Antisense primer
(5′-3′) | Hazard ratio | Cox regression
coefficients |
|---|
| COCH | AGA AAG CAG ATG TCC
TCT GC | TCC CCC TGA GTT GCT
GAT TA | 1.190 | 0.173953307 |
| CELSR3 | CTC CAT GTT GGT GAC
TGT CAC | TCC TGC CAC ATG TTC
TCA AG | 1.246 | 0.21993842 |
| HMOX1 | AAC TTT CAG AAG GGC
CAG GT | CTT GTT GCG CTC AAT
CTC CT | 1.251 | 0.223943231 |
| KIF1A | AAG AAC AAG GGC AAC
CTT CG | CTC CAT TCA TGT TGG
TGG CC | 1.060 | 0.058268908 |
| MGC17624 | GTC CTG AAC GAC AAG
CAC CT | AGG CTT CTG GGT CGA
TTT CT | 0.670 | −0.400477567 |
| MTAP | TCC TTG AGG GAG GAG
ATT CA | TCC TCT GGC ACA AGA
ATG AC | 0.906 | −0.098715973 |
| PFKB4 | ACT GAA CCC CCT GAA
GAA GA | ATG AGA GTT GGG CAG
TTG GT | 1.400 | 0.336472237 |
| S100A8 | CAT CGA CGT CTA CCA
CAA GT | GAA TGA GGA ACT CCT
GGA AG | 1.176 | 0.162118849 |
Materials and methods
Patients and tissue samples
The present study was performed in agreement with
applicable laws and regulations, good clinical practices and
ethical principles as described in the Declaration of Helsinki. The
Ethics Committee of Chungbuk National University (Cheongju, Korea)
approved this protocol (IRB approval no. 2010-01-001), and written
informed consent was obtained from each patient. Collection and
analysis of all samples was approved by the Institutional Review
Board of Chungbuk National University.
The present study utilized previously published gene
expression profiles from 176 consecutive primary NMIBC tumor
specimens with histologically verified transitional cell carcinoma.
The patients from whom these tumors were derived underwent
transurethral resection (TUR) at Chungbuk National University
Hospital in South Korea between January 1995 and December 2009. To
make the study population more homogeneous, patients with
concomitant CIS or who had undergone radical cystectomy were
excluded. To avoid the risk of under-staging, or when a high-grade
tumor was detected, a second TUR was performed 2–4 weeks after
initial resection if the original BC specimen did not include
proper muscle tissue. Patients with a T1 tumor, multiple tumors,
large tumors (≥3 cm in diameter) or a high-grade tumor received one
cycle of intravesical Bacillus Calmette-Guerin (BCG) treatment.
Following initial TUR, each patient was monitored according to
standard guidelines (21). Response
to treatment was assessed by cystoscopy and urinary cytology.
Patients who were disease-free within 3 months subsequent to
treatment were assessed every 3 months for the first 2 years and
subsequently every 6 months. Tumors were staged and graded
according to the 2002 tumor-node-metastasis (TNM) classification
and the 1973 World Health Organization grading system (22,23). A
follow-up period of ≥6 months was required (unless recurrence
and/or progression occurred within 6 months). Progression was
defined as muscular invasion or metastatic disease. The clinical
and pathological progression risk was calculated using two
multivariate risk assessment tools, the European Organization for
Research and Treatment of Cancer (EORTC) risk tables and the
Spanish Urological Club for Oncological Treatment (CUETO)
prognostic scoring model (13,14).
All tumors were macrodissected within 15 min of
surgical resection, fresh-frozen in liquid nitrogen and stored at
−80°C until use. Each cancer specimen was confirmed as
representative by pathological confirmation of adjacent tissue in
fresh-frozen sections from TUR specimens. Resected tumors were
fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer for
24 h at room temperature. The sections were processed on a
computerized tissue processor (Tissue-Tek VIP; Sakura Finetek USA,
Inc., Torrance, CA, USA) and embedded in paraffin (Paraplast
Medium; Leica Biosystems, Wetzlar, Germany) on a tissue embedding
console system (Tissue-Tek TEC; Sakura Finetek USA, Inc.). All
paraffin embedded tissue blocks were sectioned at 4 µm thickness
with a microtome, and slides were then prepared. Deparaffinization
and rehydration of the sections was performed using 4 incubation
steps of xylene and a series of 100% ethanol, 95% ethanol and twice
with 70% ethanol. Each step was performed for 5 min at room
temperature. The sections were deionized using water for 15 min at
room temperature. Subsequently, the sections were stained with
hematoxylin for 5 min and eosin for 10 sec at room temperature.
Final stages of processing were performed with an automated slide
stainer (Tissue-Tek Prisma; Sakura Finetek USA, Inc.) and automated
coverslip (Tissue-Tek Glas; Sakura Finetek USA, Inc.). Sections
were examined under a light microscope under magnifications, ×100
and ×400 (Olympus BX53; Olympus Corp., Tokyo, Japan).
Selection of prognosis-associated gene
classifiers using microarray gene expression profiling
The experimental and statistical method for
determining the eight-gene progression-associated classifier was
described previously (20). The full
microarray data set is available online (http://www.ncbi.nlm.nih.gov/geo/) under accession
number GSE13507.
RNA extraction and construction of
cDNA
RNA was isolated from tissue using 1 ml TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and homogenized in a 5 ml glass tube. The homogenate was then
transferred to a 1.5 ml tube and mixed with 200 µl chloroform.
Following 5 min incubation at 4°C, the homogenate was centrifuged
for 13 min at 13,000 × g at 4°C. The upper aqueous phase was
transferred to a clean tube and 500 µl isopropanol was added. The
mixture was incubated for 60 min at 4°C, and the tube was
centrifuged for 8 min at 13,000 × g at 4°C. The upper aqueous phase
was then mixed with 500 µl 75% ethanol, and centrifuged for 5 min
at 13,000 × g at 4°C. After discarding the upper aqueous layer, the
pellet was dried at room temperature, dissolved in
diethylpyrocarbonate-treated water, and then stored at −80°C. The
quality and integrity of the RNA were confirmed by agarose gel
electrophoresis and ethidium bromide staining, followed by visual
inspection under ultraviolet light. cDNA was prepared from 1 µg
total RNA using a First-Strand cDNA Synthesis kit (Amersham; GE
Healthcare, Chicago, IL, USA) according to the manufacturer's
protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
qPCR amplification was performed using a Rotor-Gene
6000 instrument (Corbett Life Science; Qiagen, Inc., Valencia, CA,
USA) to quantify gene expression. qPCR assays were carried out in
micro-reaction tubes (Corbett Life Science; Qiagen, Inc.) using
SYBR-Premix Ex Taq (Takara Biotechnology Co., Ltd., Dalian, China).
The primers used in the amplification are listed in Table I. The PCR reaction was performed in a
final volume of 10 µl consisting of 5 µl 2X SYBR-Premix Ex Taq
buffer, 0.5 µl each sense and antisense primers (10 pmol/µl) and 1
µl cDNA. The product was purified with a QIAquick Extraction kit
(Corbett Life Science; Qiagen, Inc.), quantified with a
spectrophotometer (MBA2000; Perkin Elmer, Inc., San Jose, CA, USA),
and then sequenced with an automated laser fluorescence sequencer
(ABI PRISM 3100 Genetic Analyzer; Applied Biosystems; Thermo Fisher
Scientific, Inc.). Ten-fold serial dilutions of a known
concentration of the product (between 100 and 0.1 pg/µl) were used
to establish the standard curve for qPCR. The qPCR conditions were
as follows: 1 cycle for 20 sec at 96°C, followed by 40 cycles of 2
sec at 96°C for denaturation, 15 sec at 60°C for annealing, and 15
sec at 72°C for extension. The melting program was performed at
72–95°C with a heating rate of 1°C per 45 sec. Spectral data were
captured and analyzed using Rotor-Gene Real-Time Analysis Software
6.0 Build 14 (Corbett Life Science; Qiagen, Inc.). All of the
samples were run in triplicate. GAPDH was analyzed as an endogenous
RNA reference gene and gene expression was normalized to the
expression of GAPDH.
Statistical analysis
To develop an easy-to-use risk score, a previously
developed strategy using the Cox regression coefficient was adopted
for the eight genes in the progression-associated classifier
[coagulation factor C homolog (COCH), EGF LAG seven-pass
G-type receptor 3 (CELSR3), heme oxygenase 1 (HMOX1),
kinesin family member 1A (KIF1A), chromosome 16 open reading
frame 74 (MGC17624), methylthioadenosine phosphorylase
(MTAP), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase
4 (PFKFB4) and S100 calcium binding protein A8
(S100A8)] (20). The risk
score for each patient was calculated as the sum of each genes
score, which was derived by multiplying the expression level of the
gene by its corresponding coefficient (Risk score = ∑ Cox
coefficient of gene Gi × expression value of gene Gi). Molecular
progression risk scores were presented as mean ± standard deviation
and differences across dichotomous categories were assessed using
the Mann-Whitney U test or Kruskal-Wallis test. The Kaplan-Meier
method was used to estimate time to development of invasive tumor,
and differences were assessed using the log-rank test. The
prognostic value of the molecular progression risk score for
prediction of development of invasive tumor was analyzed using
multivariate Cox proportional hazard regression models. The
molecular progression risk score was estimated as either a
continuous or categorical variable, in which classified as having a
good-prognosis or poor-prognosis signature, with the 50th
percentile (median) of the progression risk score in regression
analysis. P<0.05 was considered to indicate a statistically
significant difference, and all reported P-values are two-sided.
Analyses were performed using the SPSS 21.0 software (IBM SPSS,
Armonk, NY, USA).
Results
Table II summarizes
the baseline characteristics of the 176 primary patients with NMIBC
included in the analysis. Median follow-up subsequent to surgery
was 72.8 months (interquartile range, 37.0–118.7), ~30 months
longer than in the original study (20). The majority of patients (76.1%) had
stage T1 tumors, and the histological grade distribution was 30.1%
for grade 1, 52.8% for grade 2 and 17.1% for grade 3. Of the 176
patients, 73 (43.2%) experienced recurrence, and 26 (14.8%)
progressed to MIBC.
 | Table II.Baseline characteristics of the
patients. |
Table II.
Baseline characteristics of the
patients.
| Parameters | No. of patients, n
(%) |
|---|
| Number of
patients | 176 |
| Mean ± SD age, years
(range) | 63.45±12.72
(24–89) |
| Median follow-up,
months (IQR) | 72.0
(37.0–118.7) |
| Gender |
|
| Male | 146 (83.0) |
|
Female | 30
(17.0) |
| Tumor size |
|
| ≤3
cm | 94
(53.4) |
| ≥3
cm | 82
(46.6) |
| Multiplicity |
|
|
Single | 96
(54.5) |
| 2–7 | 56
(31.8) |
| ≥8 | 24
(13.6) |
| BCG induction
therapy | 126 (71.6) |
| Stage |
|
| Ta | 42
(23.9) |
| T1 | 134 (76.1) |
| Grade |
|
| 1 | 53
(30.1) |
| 2 | 93
(52.8) |
| 3 | 30
(17.1) |
| Recurrence | 73
(43.2) |
| Progression to
MIBC | 26
(14.8) |
Molecular progression risk scores were calculated
for each patient (Risk score = Σ Cox coefficient of gene Gi ×
expression value of gene Gi, as determined by RT-qPCR). To
determine whether molecular grade represented the tumor
aggressiveness phenotype, the association between the molecular
progression risk score and clinicopathological variables was
evaluated. Patients with higher grade, T1 stage or multiple tumors
had an increased molecular progression risk score compared with
those with lower grade, Ta stage or a single tumor (P<0.05 for
all comparisons). Higher progression risk groups according to the
EORTC or CUETO risk tables tended toward higher molecular
progression risk scores (Table
III).
 | Table III.Progression risk score according to
clinicopathological variables. |
Table III.
Progression risk score according to
clinicopathological variables.
| Variables | Number, n (%) | Molecular progression
risk score (mean ± standard deviation) | P-value |
|---|
| Gender |
|
| 0.159a |
|
Male | 146 (83.0) | 10.82±2.53 |
|
|
Female | 30 (17.0) | 11.53±2.36 |
|
| Tumor size |
|
| 0.199a |
| ≤3
cm | 94 (53.4) | 10.72±2.53 |
|
| ≥3
cm | 82 (46.6) | 11.21±2.47 |
|
| Multiplicity |
|
| 0.001b |
|
Single | 96 (54.5) | 10.34±2.48 |
|
|
2–7 | 56 (31.8) | 11.22±2.23 |
|
| ≥8 | 24 (13.6) | 12.22±2.57 |
|
| Stage |
|
| 0.025a |
| Ta | 42 (23.9) | 10.20±2.76 |
|
| T1 | 134 (76.1) | 11.18±2.38 |
|
| Grade |
|
|
<0.001b |
| 1 | 53 (30.1) | 10.09±2.64 |
|
| 2 | 93 (52.8) | 10.91±2.48 |
|
| 3 | 30 (17.1) | 12.55±1.34 |
|
| EORTC progression
risk score |
|
| 0.001b |
| 0 | 18 (10.2) | 9.02±2.00 |
|
|
2–6 | 53 (30.1) | 10.65±2.79 |
|
|
7–13 | 100 (56.8) | 11.41±2.32 |
|
|
14–23 | 5 (2.8) | 11.73±0.39 |
|
| CUETO progression
risk scorec |
|
|
<0.001b |
|
0–4 | 68 (38.6) | 10.21±2.14 |
|
|
5–6 | 27 (15.3) | 10.42±2.47 |
|
|
7–9 | 18 (10.2) | 11.83±1.70 |
|
|
10–14 | 13 (7.4) | 12.91±1.46 |
|
| Recurrence |
|
| 0.009a |
| No | 100 (56.8) | 10.52±2.48 |
|
|
Yes | 76 (43.2) | 11.51±2.44 |
|
| Progression to
MIBC |
|
|
<0.001a |
| No | 150 (85.2) | 10.57±2.41 |
|
|
Yes | 26 (14.8) | 13.11±1.87 |
|
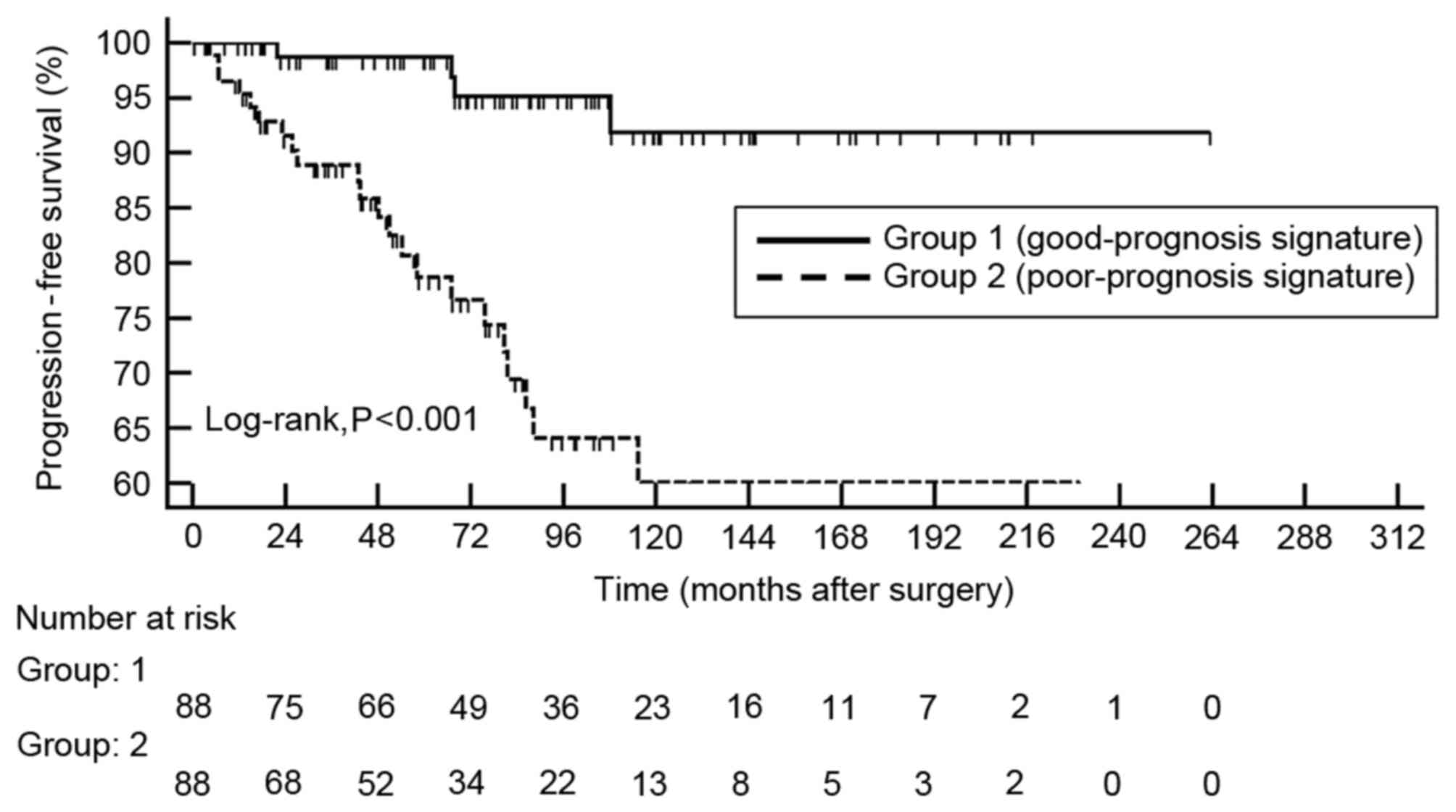
Kaplan-Meier survival curves revealed significant
differences in time to development of MIBC between the
good-prognosis and poor-prognosis signature groups (log-rank,
P<0.001; Fig. 1). Multivariate Cox
regression analysis demonstrated that molecular progression risk
score was an independent predictor of tumor invasion, either as a
continuous variable [hazard ratio (HR), 1.489; 95% confidence
interval (CI), 1.216–1.823; P<0.001] or as a categorical
variable (HR, 5.026; 95% CI, 1.619–15.608; P=0.005; Table IV).
 | Table IV.Multivariate Cox regression models
for the risk of development of invasive tumors in primary
non-muscle-invasive bladder cancer. |
Table IV.
Multivariate Cox regression models
for the risk of development of invasive tumors in primary
non-muscle-invasive bladder cancer.
|
| Multivariate
analysis as a continuous variable | Multivariate
analysis as a categorical variable |
|---|
|
|
|
|
|---|
| Parameter | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age
(continuous) | 1.031
(0.991–1.072) | 0.136 | 1.033
(0.995–1.072) | 0.094 |
| Gender
(female) | 0.634
(0.205–1.962) | 0.429 | 0.822
(0.271–2.487) | 0.728 |
| Size (>3
cm) | 2.178
(0.889–5.336) | 0.088 | 2.371
(0.948–5.930) | 0.065 |
| Multiplicity
(multiple) |
|
|
|
|
|
Single | – |
| – |
|
|
2–7 | 1.265
(0.496–3.224) | 0.622 | 1.229
(0.497–3.037) | 0.655 |
| ≥8 | 1.138
(0.368–3.516) | 0.823 | 1.493
(0.487–4.577) | 0.483 |
| Grade |
|
|
|
|
| 1 | – |
| – |
|
| 2 | 2.182
(0.435–10.948) | 0.343 | 1.918
(0.474–7.756) | 0.361 |
| 3 | 2.731
(0.454–16.428) | 0.272 | 2.663
(0.521–13.601) | 0.239 |
| Stage (T1) | 1.873
(0.369–9.504) | 0.449 | 2.116
(0.504–8.877) | 0.306 |
| Progression risk
score (continuous) | 1.489
(1.216–1.823) | <0.001 | – |
|
| Progression risk
score (high) | – |
| 5.026
(1.619–15.608) | 0.005 |
Discussion
The present study aimed to obtain long-term
validation of a previously reported progression-associated gene
classifier in the prediction of muscle-invasive disease. The
present findings revealed that the molecular progression risk
score, based on an eight-gene progression-associated classifier,
represented biological aggressiveness in the absence of clinical
information. Furthermore, the molecular progression risk score was
an independent predictor of development of muscular invasion. The
molecular progression risk score may aid in patient counseling and
selection of optimal treatments.
In our previous study, a clinically applicable qPCR
gene signature was developed to predict progression of NMIBC
(20,24). Our original study identified a
progression-associated classifier in patients with NMIBC, including
the following eight genes: COCH, CELSR3,
HMOX1, KIF1A, MGC17624, MTAP,
PFKFB4 and S100A8. This eight-gene signature was
successfully validated in the original and independent cohorts. No
cancer progressed in any patients in the good-prognosis signature
group (20).
The original study by Kim et al (20) was a comprehensive attempt to identify
genetic signatures associated with disease prognosis in BC. By
contrast, the present study focused on identifying a molecular
predictor of muscle invasion in NMIBC over an extended follow-up
period. During the follow-up period, 14.8% of patients progressed
to muscle invasion; therefore, there is a demonstrable requirement
for extended follow-up validation. In addition, the original study
was unable to conduct multivariate analysis, since no patients with
NMIBC in the good-prognosis signature group experienced cancer
progression. In the present study, these molecular signatures were
validated by multivariate Cox regression analysis and the
independent value of the molecular progression risk score in
predicting development of tumor invasion was confirmed. In a
previous international validation study by Dyrskjøt et al
(10), a four-gene classifier was
used to predict disease recurrence and progression. Combined
analysis with an 88-gene progression classifier and a 68-gene CIS
signature yielded a strong hazard ratio of 4.6 in a multivariate
Cox regression analysis for prediction of muscle-invasive disease.
The main strength of the molecular progression risk score is that
it was calculated from the expression levels of only eight genes,
making it cost-effective. Another advantage of the progression risk
score is that it may be employed not only as a categorical model,
but also as a continuous risk score model. To the best of our
knowledge, no previous study has reported an independent molecular
predictor of muscle invasion in NMIBC that may be employed as a
continuous risk score.
It was also investigated whether the molecular
progression risk score represented tumor aggressiveness.
Furthermore, a significant concordance between pathological
aggressive phenotype and molecular progression risk score was
observed, and patients with aggressive bladder tumors exhibited
increased molecular progression risk scores. Notably, the molecular
progression risk score was significantly associated with
clinicopathological multivariate progression models, including the
EORTC and CUETO risk tables (13,14). Thus,
a molecular progression risk score based on gene expression may
represent biological aggressiveness, even in the absence of
clinical information.
One possible limitation of the present study is that
data obtained from a previous study population was used, without
adding new cases. However, the present study sought to validate the
previously established progression-associated classifier with
extended follow-up and a different study design. External
validation or collaborative studies are required to confirm the
clinical utility of the molecular progression risk score in
predicting muscle invasion in NMIBC. Another concern is that BCG
maintenance therapy was not considered, since only a small
percentage of patients were able to complete the maintenance
schedule due largely to BCG-associated side effects. In addition,
patients diagnosed with a concomitant CIS, which frequently
resembles muscle-invasive disease due to its aggressive biological
features, were also excluded from the study. Consequently, the
present results may not be reflective of all patients with NMIBC,
particularly those with high-risk tumors. Notwithstanding these
limitations, the present study represents an important step toward
the clinical use of the molecular progression risk score in NMIBC.
Introduction of the present molecular progression risk score into
routine clinical practice will require additional external
validation in a prospective study using a larger number of
samples.
In conclusion, the present results identified a
molecular progression risk score, based on RT-qPCR analysis, which
may represent biological aggressiveness in the absence of clinical
information. In the clinic, this progression-associated gene
classifier may be used to guide selection of treatment regimen for
patients initially diagnosed with NMIBC. Frequent cystoscopic
follow-up, adjuvant intravesical instillations or early timing of
radical treatment are recommended in patients with a molecular
signature associated with poor prognosis.
Acknowledgements
The present study was supported by the Functional
Districts of the Science Belt support program, and the National
Research Foundation of Korea (grant nos. NRF-2014R1A2A1A09006983
and NRF-2015R1A2A2A03004100) of Ministry of Science, ICT and Future
Planning. The authors wish to thank Ms. Eun-Ju Shim from the
National Biobank of Korea at Chungbuk National University Hospital
for sample preparation and technical assistance. This work was
supported by a research grant from Chungbuk National University,
2012. The specimens for the present study were provided by Chungbuk
National University Hospital, a member of the National Biobank of
Korea, which is supported by the Ministry of Health, Welfare and
Family Affairs.
References
|
1
|
Kim WJ, Park S and Kim YJ: Biomarkers in
bladder cancer: Present status and perspectives. Biomark Insights.
2:95–105. 2007.PubMed/NCBI
|
|
2
|
Gregg JR, Dahm P and Chang SS:
Guideline-based management of non-muscle invasive bladder cancer.
Indian J Urol. 31:320–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clark P, Agarwal N and Biagioli M: NCCN
clinical practice guideline in oncology: Bladder cancer. Version I.
2013, simpleNCCN.orgJanuary 10–2013
|
|
4
|
Ahn JH, Jung SI, Yim SU, Kim SW, Hwang EC
and Kwon DD: Impact of glycemic control and metformin use on the
recurrence and progression of non-muscle invasive bladder cancer in
patients with diabetes mellitus. J Korean Med Sci. 31:1464–1471.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yun SJ, Kim SK and Kim WJ: How do we
manage high-grade T1 bladder cancer? Conservative or aggressive
therapy? Investig Clin Urol. 57 Suppl 1:S44–S51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Milowsky MI, Rumble RB, Booth CM, Gilligan
T, Eapen LJ, Hauke RJ, Boumansour P and Lee CT: Guideline on
muscle-invasive and metastatic bladder cancer (European Association
of Urology guideline): American society of clinical oncology
clinical practice guideline endorsement. J Clin Oncol.
34:1945–1952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Witjes JA, Compérat E, Cowan NC, De Santis
M, Gakis G, Lebret T, Ribal MJ, Van der Heijden AG and Sherif A;
European Association of Urology, : EAU guidelines on
muscle-invasive and metastatic bladder cancer: Summary of the 2013
guidelines. Eur Urol. 65:778–792. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Epstein JI, Amin MB, Reuter VR and Mostofi
FK: The World Health Organization/International Society of
Urological Pathology consensus classification of urothelial
(transitional cell) neoplasms of the urinary bladder. Am J Surg
Pathol. 22:1435–1448. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
May M, Brookman-Amissah S, Roigas J,
Hartmann A, Störkel S, Kristiansen G, Gilfrich C, Borchardt R,
Hoschke B, Kaufmann O and Gunia S: Prognostic accuracy of
individual uropathologists in noninvasive urinary bladder
carcinoma: A multicentre study comparing the 1973 and 2004 World
Health Organisation classifications. Eur Urol. 57:850–858. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dyrskjøt L, Zieger K, Real FX, Malats N,
Carrato A, Hurst C, Kotwal S, Knowles M, Malmström PU, de la Torre
M, et al: Gene expression signatures predict outcome in
non-muscle-invasive bladder carcinoma: A multicenter validation
study. Clin Cancer Res. 13:3545–3551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Rhijn BW, Burger M, Lotan Y, Solsona
E, Stief CG, Sylvester RJ, Witjes JA and Zlotta AR: Recurrence and
progression of disease in non-muscle-invasive bladder cancer: From
epidemiology to treatment strategy. Eur Urol. 56:430–442. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van den Bosch S and Witjes JA: Long-term
cancer-specific survival in patients with high-risk,
non-muscle-invasive bladder cancer and tumour progression: A
systematic review. Eur Urol. 60:493–500. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sylvester RJ, van der Meijden AP,
Oosterlinck W, Witjes JA, Bouffioux C, Denis L, Newling DW and
Kurth K: Predicting recurrence and progression in individual
patients with stage Ta T1 bladder cancer using EORTC risk tables: A
combined analysis of 2596 patients from seven EORTC trials. Eur
Urol. 49:466–477. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fernandez-Gomez J, Madero R, Solsona E,
Unda M, Martinez-Piñeiro L, Gonzalez M, Portillo J, Ojea A, Pertusa
C, Rodriguez-Molina J, et al: Predicting nonmuscle invasive bladder
cancer recurrence and progression in patients treated with bacillus
Calmette-Guerin: The CUETO scoring model. J Urol. 182:2195–2203.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim WJ and Bae SC: Molecular biomarkers in
urothelial bladder cancer. Cancer Sci. 99:646–652. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Knowles MA and Hurst CD: Molecular biology
of bladder cancer: New insights into pathogenesis and clinical
diversity. Nat Rev Cancer. 15:25–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kamat AM, Hegarty PK, Gee JR, Clark PE,
Svatek RS, Hegarty N, Shariat SF, Xylinas E, Schmitz-Dräger BJ,
Lotan Y, et al: ICUD-EAU international consultation on bladder
cancer 2012: Screening, diagnosis, and molecular markers. Eur Urol.
63:4–15. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kang HW, Yoon HY, Ha YS, Kim WT, Kim YJ,
Yun SJ, Lee SC and Kim WJ: FAM70B as a novel prognostic marker for
cancer progression and cancer-specific death in muscle-invasive
bladder cancer. Korean J Urol. 53:598–606. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Netto GJ: Molecular biomarkers in
urothelial carcinoma of the bladder: Are we there yet? Nat Rev
Urol. 9:41–51. 2012. View Article : Google Scholar
|
|
20
|
Kim WJ, Kim EJ, Kim SK, Kim YJ, Ha YS,
Jeong P, Kim MJ, Yun SJ, Lee KM, Moon SK, et al: Predictive value
of progression-related gene classifier in primary non-muscle
invasive bladder cancer. Mol Cancer. 9:32010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brausi M, Witjes JA, Lamm D, Persad R,
Palou J, Colombel M, Buckley R, Soloway M, Akaza H and Böhle A: A
review of current guidelines and best practice recommendations for
the management of nonmuscle invasive bladder cancer by the
International Bladder Cancer Group. J Urol. 186:2158–2167. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Greene FL: The American Joint Committee on
Cancer: Updating the strategies in cancer staging. Bull Am Coll
Surg. 87:13–15. 2002.PubMed/NCBI
|
|
23
|
Kirkali Z, Chan T, Manoharan M, Algaba F,
Busch C, Cheng L, Kiemeney L, Kriegmair M, Montironi R, Murphy WM,
et al: Bladder cancer: Epidemiology, staging, and grading and
diagnosis. Urology. 66 6 Suppl 1:S4–S34. 2005. View Article : Google Scholar
|
|
24
|
Jeong P, Ha YS, Cho IC, Yun SJ, Yoo ES,
Kim IY, Choi YH, Moon SK and Kim WJ: Three-gene signature predicts
disease progression of non-muscle invasive bladder cancer. Oncol
Lett. 2:679–684. 2011.PubMed/NCBI
|