Introduction
The increasing use of palliative chemotherapy with
novel targeted agents and improving supportive care during
chemotherapy have prolonged the survival and maintained the quality
of life of patients with metastatic colorectal cancer (mCRC)
(1,2).
Based on recent clinical trials (3),
the median expected survival time of patients with mCRC is
estimated to be >2 years. Thus, there is a requirement for
techniques to improve the survival of patients with mCRC. Although
some patients possess sufficient performance status (PS) to receive
further treatment following standard chemotherapy, including
oxaliplatin/irinotecan with targeted agents, therapeutic options
following standard chemotherapy are lacking. Recently, regorafenib
and TAS-102 have been used for third-line chemotherapy (4,5); however,
in practice it is difficult to use these drugs due to cost
effectiveness. Therefore, physicians often use chemotherapeutic
agents that the patient has previously received as a salvage
treatment.
Several studies have investigated the reuse of
chemotherapeutic drugs, including capecitabine, mitomycin C and
gemcitabine, and have revealed limited effectiveness (6–10). Certain
types of chemotherapeutic agents, including S-1 and oxaliplatin
combined with 5-fluorouracil (5-FU) or capecitabine, have
demonstrated modest efficacy with response rates of 15–20%
(11–14). However, there are no parameters to
predict the benefit from rechallenge treatment subsequent to
standard chemotherapy.
The optimal methods for determining tumor response
have been widely studied previously. The Response Evaluation
Criteria in Solid Tumors (RECIST) (15) is the most popular method to assess
tumor responses. However, the objective response rate fails to
identify strong surrogate markers for overall survival (OS) and
progression-free survival (PFS), and is also limited in its
prediction of OS (16,17). Therefore, early tumor shrinkage (ETS)
and depth of response (DoR) have been developed to overcome these
problems (16). ETS and DoR have been
associated with prolonged PFS, post-progression survival (PPS) and
OS (18,19). ETS and DoR were initially developed to
predict the efficacy of targeted agents (19), but their roles in conventional
chemotherapy have not yet been fully evaluated (20). As a biomarker to predict tumor
response, several single-nucleotide polymorphisms (SNPs) have been
evaluated as prognostic and predictive markers for the
effectiveness of chemotherapy (21–23),
although genome-wide association studies are being actively studied
in various tumors. The methylenetetrahydrofolate reductase (MTHFR)
polymorphism is known to be associated with the metabolism of
folate and response of 5-FU treatment in CRC (24–31).
Furthermore, the X-ray repair cross-complementing 1 (XRCC1) gene is
known to be associated with the response of patients to
platinum-based anticancer therapy in CRC (32–36). 5-FU
and platinum are basic chemotherapies for MCRC. Thus, such SNPs
could be predictive biomarkers of clinical outcome in mCRC.
The present study evaluated tumor response following
first-line chemotherapy based on the RECIST, ETS, DoR and SNPs
associated with response to chemotherapy, to evaluate their roles
as prognostic indicators of survival outcomes. With these results,
the SNPs as a biomarker, and ETS and DoR as clinical parameters,
were analyzed to define the prognostic significance of 5-FU
rechallenge chemotherapy as a third-line of treatment for patients
with mCRC.
Materials and methods
Patients
The data of patients who were diagnosed with mCRC
and who underwent chemotherapy at Chonnam National University
Hwasun Hospital (Gwangju, Korea) between April 2004 and December
2012 was retrospectively reviewed using medical records and imaging
materials. The present study was conducted to evaluate genetic
polymorphisms and treatment outcomes in gastrointestinal cancer and
was approved by the Institutional Review Board of Chonnam National
University Hwasun Hospital. All patients provided written informed
consent.
Patients were considered eligible if they had
histologically diagnosed adenocarcinoma in the colon or rectum,
were initially diagnosed with stage IV (37) or recurrent mCRC, and were suitable for
RECIST, ETS and DoR evaluations using target lesion measurements
following first-line chemotherapy. In addition, these patients
received palliative chemotherapies. Exclusion criteria included no
target lesions (based on RECIST), another primary malignancy or
severe combined illness that affected chemotherapy treatment. In
total, 242 patients were consecutively enrolled from March 2004,
and survival data was evaluated until mortality or June 2015. Blood
samples from 171/242 patients were available and SNPs known to be
associated with chemotherapy response were analyzed in these 171
patients.
Tumor assessment and definition
Tumor assessment by computed tomography (CT) scans
were conducted every 8–10 weeks to evaluate disease progression.
Undetermined regions were further assessed using magnetic resonance
imaging or positron emission tomography CT. Tumor response was
assessed according to RECIST (version 1.1) and ETS was defined as
an ≥20% decrease in the sum of RECIST target lesions' longest
diameters at week 8 compared with the baseline. DoR was defined as
the relative change in the sum of the longest diameters of RECIST
target lesions at the nadir, in the absence of new lesions or the
progression of non-target lesions, compared with the baseline. The
outcomes on PFS, PPS and OS were compared. PFS was defined as the
time from the beginning of the first-line treatment until
documented tumor progression or mortality. OS was defined as the
time from the beginning of the first-line treatment until
mortality, or the time of the last assessment of disease status.
PPS was defined as the time from tumor progression subsequent to
first-line treatment until mortality, or the time of the last
assessment of disease status. Overall response (OR) was defined as
the proportion of patients who achieve a complete or partial
response per RECIST criteria. Disease control rate (DCR) was
defined as the proportion of patients who achieved a complete
response, partial response or stable disease per RECIST
criteria.
Genotyping
Genomic DNA was extracted from peripheral blood
using a QIAamp DNA Blood Mini kit (Qiagen, Valencia, CA), according
to the manufacturer's protocol. Genotyping to analyze polymorphisms
was performed using a TaqMan allelic discrimination assay,
polymerase chain reaction (PCR)-restriction fragment length
polymorphism, high-resolution melting (HRM) analysis and
electrophoresis methods. The primer sequences, methods, and
references used in the present study are presented in Table I. Genotyping of MTHFR C677T using
quantitative PCR was performed with allelic discrimination using
dual-labeled probes containing locked nucleic acids (LNA). PCR
primers and LNA probes were designed and synthesized by Integrated
DNA Technologies (Coralville, IA, USA). Quantitative PCR was
performed using a Rotor-Gene 3000 multiplex system (Qiagen) in a
10-µl reaction volume containing 200 nM PCR primer, 10 nM of each
probe, 0.5 U f-taq polymerase (Solgent, Daejeon, Korea) and 40 ng
of genomic DNA. The thermocycling conditions were as follows:
Initial 5-min hold at 95°C; followed by 40 cycles at 95°C for 5 sec
and 64°C for 30 sec. HRM genotyping for glutathione S-transferase
pi 1 (GSTP1) rs1695, XRCC1 rs25487 and excision repair 1
endonuclease non-catalytic subunit (ERCC1) rs11615 was performed in
10-µl reaction volumes with 200 nM PCR primer, 1 µMSyto 9
fluorescent dye, 0.5 U f-Taq polymerase, and 40 ng of genomic DNA,
using a Rotor-Gene 6000 high-resolution system (Qiagen). Genotyping
for 28-bp thymidylate synthetase variable number of tandem repeat
(2R→3R) (TS VNTR) genotypes in the 5′untranslated region of the TS
gene was conducted using a protocol described by Horie et al
(21). The TSSNP, rs34743033,
involving the 12th nucleotide of the second repeat of the 3R
alleles in TS VNTR was genotyped by digesting the TS VNTR PCR
products with HaeIII (Takara Bio, Inc., Otsu, Japan) followed by
electrophoresis on a 6% polyacrylamide gel and visualization with
ethidium bromide. TSSNP rs34743033 genotypes of the patients were
classified in two groups: High expression type (2R3G, 3C3G and
3G3G) and low expression type (2R2R, 2R/3C and 3C3C) (38).
 | Table I.Sequences of primers and method used
by genotype. |
Table I.
Sequences of primers and method used
by genotype.
| Gene and rs
number | Genotype | Primers and
probe | Method | (Refs.) |
|---|
| MTHFR677
rs1801133 | C/T,
Ala222Val | F,
5′-CTTTGAGGCTGACCTGAAGC-3′ | TaqMan RQ | (46) |
|
|
| R,
5′-TCACAAAGCGGAAGAATGTG-3′ |
|
|
|
|
| aLNA probes for C allele: 5′-FAM-ATG
GcT ccc-BHQ1-3′ |
|
|
|
|
| LNA probes for T
allele: 5′-cy5-cgA CTc cCg C-BHQ2-3′ |
|
|
| GSTP1
rs1695 | A/G,
Val105Ile | F,
5′-TGGTGGACATGGTGAATGAC-3′ | HRM | (47) |
|
|
| R,
5′-TGCAGATGCTCACATAGTTGG-3′ |
|
|
| XRCC1
rs25487 | G/A,
Arg399Gln | F,
5′-TAAGGAGTGGGTGCTGGACT-3′ | HRM | Present |
|
|
| R,
5′-ATTGCCCAGCACAGGATAAG-3′ |
|
|
| ERCC1
rs11615 | C/T,
Asn118Asn | F,
5′-TCCCTATTGATGGCTTCTGC-3′ | HRM | Present |
|
|
| F,
5′-GAGCTCACCTGAGGAACAGG-3′ |
|
|
| TS 2R3R | VNTR | F,
5′-GTGGCTCCTGCGTTTCCCCC-3′ | PCR, EP | (21) |
|
|
| R,
5′-CCAAGCTTGGCTCCGAGC |
|
|
|
|
|
CGGCCACAGGCATGGCGCGG-3′ |
|
|
|
TSrs34743033 | 3R/G/C |
| RFLP:HaeIII,
EP |
|
Statistical analysis
Continuous variables were expressed as the mean ±
standard deviation. χ2 or Fisher's exact tests were used
to compare categorical variables, and Student's t-test was used to
compare continuous variables.
ETS, DoR and RECIST responses were examined for
their prognostic impact on PFS, PPS and OS. DoR was evaluated as
five levels based on quintile distribution with increments of 20%.
The cut-off value of DoR >60% was achieved through this
categorizing. The Kaplan-Meier estimator method was used to examine
cumulative survival and time to mortality or progression, and the
log-rank test was used to compare differences between groups. The
factors identified as significant by univariate analysis were
subjected to stepwise multivariate analysis (forward selection) to
determine which factors retained statistical significance, and
which were merely dependent on other factors. Multivariate analyses
were performed using the Cox proportional hazards model and
logistic regression analysis to identify independent prognostic
variables. P<0.05 (two-tailed) was considered to indicate a
statistically significant difference. All analyses were performed
using SPSS software (version 21.0; IBM SPSS, Armonk, NY, USA).
Results
Baseline characteristics and tumor
responses following first-line chemotherapy
The baseline patient characteristics and types of
first-line chemotherapies are illustrated in Table II. The median patient age was 67
years, and 139 (57.4%) patients were >65 years of age. Of the
patients in the study, 42 (17.4%) patients had previously received
adjuvant chemotherapy prior to recurrence, and 14 (5.8%) patients
received a target agent combined with systemic chemotherapy.
First-line chemotherapy regimens included FOLFOX (oxaliplatin with
5-FU; 51.7%), FOLFIRI (irinotecan with 5-FU; 34.7%) and
capecitabine (13.6%). In total, 187 (76.9%) patients received
second-line chemotherapy and 119 (49.2%) patients received
third-line chemotherapy (data not shown).
 | Table II.Baseline patient characteristics
according to the number of chemotherapy treatments (n=242). |
Table II.
Baseline patient characteristics
according to the number of chemotherapy treatments (n=242).
| Characteristic | Number of patients
(%) |
|---|
| Age (years) |
|
<65 | 103 (42.6) |
|
>65 | 139 (57.4) |
| Sex |
|
Male | 151 (62.4) |
|
Female | 91 (37.6) |
| ECOG PS |
| 0 | 88 (36.4) |
| 1 | 119 (49.2) |
| 2 | 30 (12.4) |
| 3 | 5 (2.0) |
| Primary tumor
site |
|
Colon | 195 (80.6) |
|
Rectum | 47 (19.4) |
| Tumor grade |
| 1 | 67 (27.7) |
| 2 | 126 (52.1) |
| 3 | 28 (11.6) |
| NA | 21 (8.6) |
| Stage at
diagnosis |
|
Metastatic | 166 (68.6) |
|
Recurred | 68 (28.1) |
| NA | 8 (3.3) |
| Metastatic
site |
|
Liver | 157 (64.9) |
|
Lung | 75 (31.0) |
|
Bone | 10 (4.1) |
| LN | 48 (19.8) |
| Previous adjuvant
chemotherapy | 42 (17.4) |
| Use of target
agent | 14 (5.8) |
| First-line
regimen |
|
5-FU+oxaliplatin | 125 (51.7) |
|
5-FU+irinotecan | 84 (34.7) |
|
Capecitabine | 33 (13.6) |
Based on RECIST, the OR following first-line
chemotherapy was 41.3% (n=100) (data not shown). The median
percentage of tumor shrinkage at week 8 was 26.47±24.94%, and the
percentage of ETS was 42.6% (n=103) in patients without
progression. The median DoR was 38.53±30.08% in patients without
progression. The median OS, PPS and PFS of all enrolled patients
were 18.00±18.09 [95% confidence interval (CI), 20.13–24.66],
11.13±14.78 (95% CI, 12.93–16.63) and 5.95±7.12 (95% CI, 6.74–8.58)
months, respectively (data not shown).
Association between tumor response and
survival parameters
An OR was associated with a significantly longer PFS
(9.7 vs. 3.1 months; P<0.001) and OS (23.7 vs. 16.9 months;
P=0.008) compared with stable or progressive disease. However,
there was no association between OR and PPS (12.8 vs. 11.7 months;
P=0.364) (data not shown).
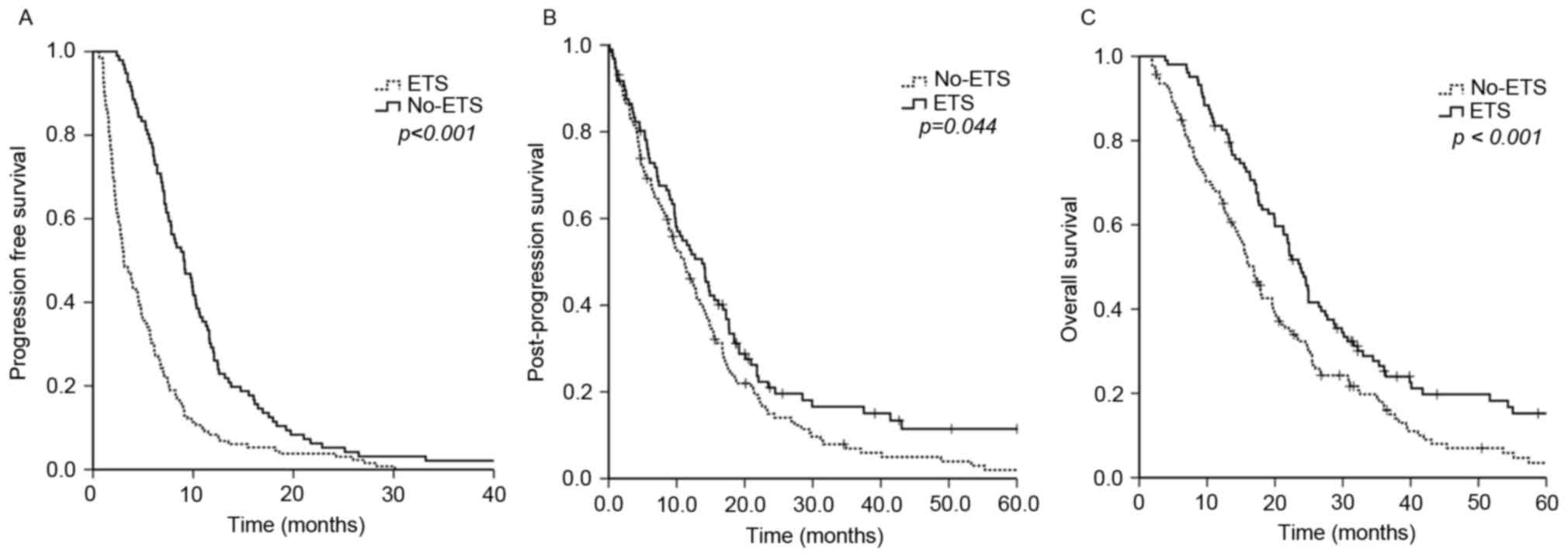
Patients who achieved ETS had a significantly longer
PFS compared with non-ETS-achieving patients (9.1 vs. 3.1 months;
P<0.001), PPS (13.7 vs. 11.2 months; P=0.044), and OS (23.7 vs.
16.8 months; P<0.001) (Fig. 1). To
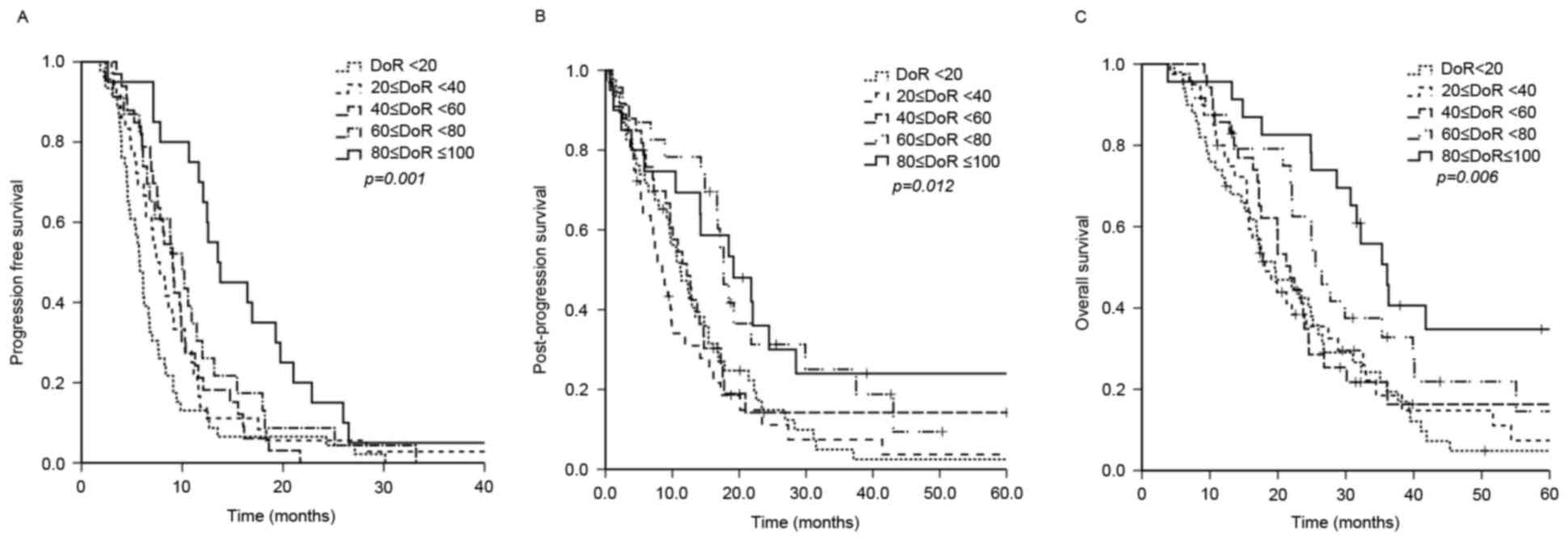
determine the association between DoR and survival, patients were
randomly divided into five groups (I–V). The median OS of each DoR
group was 19.6, 18.1, 21.8, 25.5 and 36.1 months in groups I–V,
respectively (P=0.006). In addition, PFS (5.77, 7.43, 9.07, 10.00
and 13.53 months in groups I–V, respectively; P=0.001) and PPS
(11.20, 8.50, 12.17, 17.67 and 19.17 months in group I–V,
respectively; P=0.012) were also significantly associated with DoR
(Fig. 2). Patients with a DoR ≥60%
had significantly improved PFS (11.6 vs. 4.8 months; P<0.001),
PPS (18.4 vs. 10.1 months; P<0.001) and OS (31.6 vs. 17.2
months; P<0.001) compared with patients with a DoR <60%.
Association between genotyping of SNPs
and survival parameters
Of the patients in the current study, 171 were
genotyped for SNPs known to be associated with chemotherapy
responses. Genotype and haplotype distributions of the patients are
illustrated in Table III. According
to univariate analyses, the GSTP1 (Ile105Val) AA genotype,
lymph node metastasis, liver metastasis, OR, ETS and a DoR ≥60%
were significantly associated with PFS. Using multivariate
analyses, GSTP1 (Ile105Val) AA, a DoR ≥60% and OR were
independent prognostic factors for an improved PFS, and liver
metastasis for a worse PFS (Table
IV). The XRCC1 (G399A) AA/AG genotype, ECOG PS 0–1,
stage IV cancer at diagnosis, bone metastasis, ETS and a DoR ≥60%
were associated with improved OS using univariate analyses. The
XRCC1 (G399A) AA/AG genotype, ECOG PS 0–1, a DoR ≥60% and
the absence of bone metastasis were independent factors associated
with an improved OS.
 | Table III.Genotype and haplotype distribution
of single-nucleotide polymorphisms (n=171). |
Table III.
Genotype and haplotype distribution
of single-nucleotide polymorphisms (n=171).
| Genotype | Number of patients
(%) |
|---|
| MTHFR
(C677T) |
| CC | 66
(38.8) |
| CT,
TT | 164 (61.2) |
| XRCC1
(G399A) |
| GG | 98 (57.6) |
| AA,
AG | 72 (42.4) |
| GSTP1
(Ile105Val) |
| AA | 81 (47.6) |
| AG,
GG | 89 (52.4) |
| ERCC1
(T19007C) |
| CC | 89 (52.4) |
| TT,
TC | 81 (47.6) |
| TS2R3R |
|
3R/3R | 117 (68.8) |
| 2R/2R,
2R/3R | 53 (31.2) |
|
TSrs34743033a |
| High
expression | 122 (71.8) |
| Low
expression | 48 (28.2) |
 | Table IV.Multivariate analysis of the
association between genotyping of single-nucleotide polymorphisms
and survival parameters (n=171). |
Table IV.
Multivariate analysis of the
association between genotyping of single-nucleotide polymorphisms
and survival parameters (n=171).
| Variable | HR | 95% CI | P-value |
|---|
| PFS |
| Liver
metastasis | 0.549 | 0.384–0.784 | 0.001 |
|
GSTP1 (Ile105Val)
AA | 1.390 | 1.007–1.918 | 0.045 |
| DoR
≥60% | 1.639 | 1.074–2.500 | 0.022 |
| OR | 2.424 | 1.705–3.445 | <0.0001 |
| OS |
| Bone
metastasis | 0.310 | 0.147–0.655 | 0.002 |
|
XRCC1 (G399A) AA,
AG | 1.681 | 1.190–2.375 | 0.003 |
| ECOG PS
0–1 | 1.862 | 1.212–2.857 | 0.005 |
| DoR
≥60% | 2.195 | 1.443–3.341 | <0.0001 |
Analysis of tumor response and SNPs as
prognostic markers for benefiting from third-line chemotherapy
Amongst the 171 patients whose SNPs were analyzed,
88 received third-line chemotherapy following the second
progression. Third-line chemotherapy included therapy using 5-FU
[n=63, 71.6%; capecitabine (n=47); S-1 (n=8); 5-FU with leucovorin
(n=8)], 5-FU with oxaliplatin (n=18, 20.5%), 5-FU with irinotecan
(n=6, 6.8%) and 5-FU with other drugs (n=1, 1.1%) (data not shown).
Patients receiving third-line chemotherapy were younger compared
with patients not receiving third-line chemotherapy (median age
62.5 vs. 68.0 years, respectively; P=0.023). There was no
significant difference in PS between the two groups (PS 0–1 87.5%
vs. 76.8%; P=0.074). The response rate was 17.0% (n=15) and the
disease control rate was 45.5% (n=40). However, the number of
patients in each treatment group was small and the efficacies
according to the agents used for treatment were not evaluated. The
response duration from the start of chemotherapy to progression was
18.7±19.0 weeks (data not shown).
To identify patients that benefited from additional
chemotherapy subsequent to the second progression, patients were
grouped according to the average OS of all patients (≥ or <23
months). The longer OS group included 50 patients, and the shorter
OS group included 38 patients. No significantly different clinical
characteristics between the longer and shorter OS groups were
identified (data not shown).
Univariate analyses revealed that a XRCC1
(G399A) AA/AG genotype, ETS, a MTHFR (C677T) CC genotype and
a DoR ≥60% were frequently present in the longer OS group. However,
multivariate analyses demonstrated that a MTHFR (C677T) CC
genotype [hazard ratio (HR), 2.755; 95% CI, 1.057–7.143; P=0.038]
and a DoR ≥60% (HR, 6.469; 95% CI=1.701–24.594; P=0.006) were
significant prognostic factors for improved OS following third-line
chemotherapy (Table V).
 | Table V.Univariate and multivariate analyses
of the benefit of third-line chemotherapy (n=88). |
Table V.
Univariate and multivariate analyses
of the benefit of third-line chemotherapy (n=88).
| Variable | HR | 95% CI | P-value |
|---|
| Univariate
analysis |
|
XRCC1 (G399A) AA,
AG | 2.571 | 1.079–6.130 | 0.033 |
|
ETS | 2.659 | 1.088–6.501 | 0.032 |
|
MTHFR (C677T) CC | 2.801 | 1.126–6.944 | 0.027 |
| DoR
≥60% | 6.562 |
1.766–24.392 | 0.005 |
| Multivariate
analysis |
|
MTHFR (C677T) CC | 2.755 | 1.057–7.143 | 0.038 |
| DoR
≥60% | 6.469 | 1.701–24.594 | 0.006 |
Discussion
The present analysis is the first attempt, to the
best of our knowledge, to evaluate parameters indicating benefit
from 5-FU retreatment as a third-line chemotherapy for mCRC. The
data of 243 patients with mCRC was retrospectively analyzed in the
present study. Due to the fact that targeted treatments were not
adopted and reimbursed at this time in South Korea, the majority of
patients included in the present study received conventional
chemotherapies. Therefore, the median OS in the enrolled patients
was only 18 months. As discussed above, there are a number of
treatment options for refractory mCRC, and rechallenge chemotherapy
may be used as salvage chemotherapy. In the current study, 36%
(n=88) patients received third-line chemotherapy, including 5-FU
following the second progression. Retreatment with 5-FU resulted in
an OR of 17.0% and a DCR of 45.5% following 18 weeks of response.
These results are slightly improved compared with those of previous
studies (9–11,13,22).
Although there is a lack of evidence to determine
what molecular mechanisms underlie the effects of rechallenge
treatments, a recent clinical result suggests a rationale for this
treatment approach (19,23). In the updated results of the Triplet
Plus Bevacizumab study, 5-FU, oxaliplatin and irinotecan
[FOLFOXIRI] with bevacizumab as a first-line treatment revealed
promising efficacy compared with FOLFIRI with bevacizumab (23). Notably, in the present study of
FOLFOXIRI with bevacizumab as a first-line treatment was effective
and improved PPS to >17 months. Taking into account the fact
that patients had already been exposed to four agents (FOLFOXIRI
with bevacizumab) and that 80% of the patients received second-line
chemotherapy, including previous chemotherapy with or without
targeted agents, subsequent rechallenge treatment may be beneficial
to prolong survival.
Tyrosine kinase inhibitors for patients with lung
cancer exhibiting mild and asymptomatic progression were continued
subsequent to initial evidence of progressive disease in previous
studies (20,39). Radiological progression does not
always involve all tumor sites possessing the same cause of
resistance; certain tumor sites could be sensitive to previously
used chemotherapy at the time of RECIST progression. These results
suggest that rechallenge or retreatment with previously used agents
could prolong the survival of patients with mCRC. However, further
studies are required to verify the effectiveness of retreatment
with 5-FU as a third-line chemotherapy for the treatment of
patients with mCRC.
When considering the lack of current indicators for
retreatment benefit, the results from the current study could aid
in identifying patients who may benefit from 5-FU rechallenge
chemotherapy on the basis of tumor response and tumor sensitivity.
Multivariate analyses in the present study revealed that the
MTHFR (C677T) CC genotype and a DoR ≥60% were associated
with improved OS following receiving third-line treatment using
5-FU, regardless of age or PS score.
The reduced activity of MTHFR in allele T
carriers increases the availability of
5,10-methylenetetrahydrofolate (5,10-MTHF), a necessary cofactor
for 5-FU inhibition of TYMS. Using an in vitro model, 5-FU
demonstrated increased activity in the presence of the MTHFR
677T allele in CRC (40).
However, in vivo studies here revealed conflicting results.
Several studies demonstrated that the 677T allele had a
positive or no effect on survival (24–27), while
other studies revealed a negative effect on the treatment response
(30,31). The present study revealed that the
MTHFR (C677T) CC genotype was associated with improved OS
during additional 5-FU chemotherapy. Possible explanations for
these conflicting results may involve the cellular availability of
5,10-MTHF, which depends on the MTHFR genotype and other
factors, including dietary folate (31,39,41).
The association between SNPs and tumor responses
have been evaluated in a number of previous studies. XRCC1
repairs single-strand DNA breaks by encoding a protein that defends
breaks (32), and the XRCC1 protein
is important for repairing the DNA damage induced by platinum-based
anticancer drugs (42). Several
studies have reported the efficacy of platinum-based anticancer
therapy as a predictive marker in CRC (32–36).
Similar to previous investigations, the current study also
demonstrated good prognosis in terms of OS in patients with the
XRCC1 (G399A) AA/AG genotype; however, this genotype was not
associated with an improved PFS.
Although the individual development and survival
mechanisms of cancer cells have been studied, human cancer exhibits
intratumoral heterogeneity in phenotypic features. These include
cellular morphology, gene expression, metabolism, motility, and
angiogenic, proliferative, immunogenic and metastatic potential
(43). Opposed to SNPs as markers for
innate tumor sensitivity, ETS and DoR could be surrogate markers
covering various responses according to heterogeneous tumor
characteristics, as they measure changes to the size of the tumor
mass following chemotherapy.
ETS and DoR have been demonstrated to be associated
with OS and PFS in several previous studies (18,19,44,45).
In the present study, the majority of patients were treated with
conventional chemotherapy to evaluate ETS and DoR as prognostic
markers when using targeted agents, unlike previous studies.
Although the current study was retrospective, it suggested that ETS
and DoR can be used as prognostic factors for survival outcomes,
including PFS, PPS and OS, in patients with mCRC who receive
conventional cytotoxic chemotherapy. The cutoff value of DoR ≥60%
was achieved by categorizing the patients into five DoR groups
based on increments of 20%, with similar results to a previous
study showing an optimal DoR of 62.4% (18).
In conclusion, the present study demonstrated that
DoR and XRCC1 (AG/AA) are associated with more improved
prognoses in patients with refractory mCRC receiving palliative
chemotherapy. In particular, patients with a DoR >60% following
first-line chemotherapy and an MTHFR (C677T) CC genotype
exhibited a survival benefit from 5-FU retreatment as a form of
third-line chemotherapy. Therefore, DoR and MTHFR genotype
are potential clinical markers for selecting patients with
refractory mCRC that would benefit from 5-FU rechallenge
therapy.
Acknowledgements
The English in this document has been checked by at
least two professional editors, both native speakers of English.
For a certificate, please see http://www.textcheck.com/certificate/BuYyzv.
References
|
1
|
Van Cutsem E, Cervantes A, Nordlinger B
and Arnold D; ESMO Guidelines Working Group, : Metastatic
colorectal cancer: ESMO clinical practice guidelines for diagnosis,
treatment and follow-up. Ann Oncol. 25 Suppl 3:iii1–iii9. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Benson AB III, Venook AP, Bekaii-Saab T,
Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ,
Fuchs CS, et al: Colon cancer, version 3.2014. J Natl Compr Canc
Netw. 12:1028–1059. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Cutsem E, Köhne CH, Láng I, Folprecht
G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D,
Tejpar S, et al: Cetuximab plus irinotecan, fluorouracil, and
leucovorin as first-line treatment for metastatic colorectal
cancer: Updated analysis of overall survival according to tumor
KRAS and BRAF mutation status. J Clin Oncol. 29:2011–2019. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grothey A, Van Cutsem E, Sobrero A, Siena
S, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C, et
al: Regorafenib monotherapy for previously treated metastatic
colorectal cancer (CORRECT): An international, multicentre,
randomised, placebo-controlled, phase 3 trial. Lancet. 381:303–312.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mayer RJ, Van Cutsem E, Falcone A, Yoshino
T, Garcia-Carbonero R, Mizunuma N, Yamazaki K, Shimada Y, Tabernero
J, Komatsu Y, et al: Randomized trial of TAS-102 for refractory
metastatic colorectal cancer. N Engl J Med. 372:1909–1919. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nielsen DL, Palshof JA, Larsen FO, Jensen
BV and Pfeiffer P: A systematic review of salvage therapy to
patients with metastatic colorectal cancer previously treated with
fluorouracil, oxaliplatin and irinotecan +/− targeted therapy.
Cancer Treat Rev. 40:701–715. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scartozzi M, Falcone A, Pucci F, Braconi
C, Pierantoni C, Cavanna L, Franciosi V, Berardi R, Beretta G, Masi
G, et al: Capecitabine and mitomycin C may be an effective
treatment option for third-line chemotherapy in advanced colorectal
cancer. Tumori. 92:384–388. 2006.PubMed/NCBI
|
|
8
|
Vormittag L, Kornek GV, Gruhsmann B,
Lenauer A, Föger A, Depisch D, Lang F and Scheithauer W:
UFT/leucovorin and mitomycin C as salvage treatment in patients
with advanced colorectal cancer-a retrospective analysis.
Anticancer Drugs. 18:709–712. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gubanski M, Naucler G, Almerud A,
Lideståhl A and Lind PA: Capecitabine as third line therapy in
patients with advanced colorectal cancer. Acta Oncol. 44:236–239.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee S, Kwon HC, Kim SHOHYS, Lee HJ, Yoon
HH, Choi JH and Kim JH: Capecitabine monotherapy, and the clinical
significance of neutrophil-lymphocyte ratio versus
platelet-lymphocyte ratio in patients with metastatic colorectal
cancer. ASCO Annual Meeting Proceedings. pp. 6602012;
|
|
11
|
Jeung HC, Rha SY, Cho BC, Yoo NC, Roh JK,
Roh WJ, Chung HC and Ahn JB: A phase II trial of S-1 monotherapy in
metastatic colorectal cancer after failure of irinotecan- and
oxaliplatin-containing regimens. Br J Cancer. 95:1637–1641. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun Jin Sym, Junshik Hong, Hee Kyung Ahn,
Jinny Park, Eun Kyung Cho, Jae Hoon Lee, Won-Suk Lee, Jeong-Heum
Baek, Ho Park Yeon, Bok Shin Dong, et al: A phase II trial of
salvage treatment with gemcitabine and S-1 combination in heavily
pretreated patients with metastatic colorectal cancer. J Clin
Oncol. 31 Suppl 4:S4882013. View Article : Google Scholar
|
|
13
|
Santini D, Vincenzi B, La Cesa A, Caricato
M, Schiavon G, Spalletta B, Di Seri M, Coppola R, Rocci L and
Tonini G: Continuous infusion of oxaliplatin plus chronomodulated
capecitabine in 5-fluorouracil- and irinotecan-resistant advanced
colorectal cancer patients. Oncology. 69:27–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Townsend AR, Bishnoi S, Broadbridge V,
Beeke C, Karapetis CS, Jain K, Luke C, Padbury R and Price TJ:
Rechallenge with oxaliplatin and fluoropyrimidine for metastatic
colorectal carcinoma after prior therapy. Am J Clin Oncol.
36:49–52. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grothey A, Hedrick EE, Mass RD, Sarkar S,
Suzuki S, Ramanathan RK, Hurwitz HI, Goldberg RM and Sargent DJ:
Response-independent survival benefit in metastatic colorectal
cancer: A comparative analysis of N9741 and AVF2107. J Clin Oncol.
26:183–189. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Buyse M, Thirion P, Carlson RW,
Burzykowski T, Molenberghs G and Piedbois P: Relation between
tumour response to first-line chemotherapy and survival in advanced
colorectal cancer: A meta-analysis. Meta-analysis group in cancer.
Lancet. 356:373–378. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piessevaux H, Buyse M, Schlichting M, Van
Cutsem E, Bokemeyer C, Heeger S and Tejpar S: Use of early tumor
shrinkage to predict long-term outcome in metastatic colorectal
cancer treated with cetuximab. J Clin Oncol. 31:3764–3775. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cremolini C, Loupakis F, Antoniotti C,
Lonardi S, Masi G, Salvatore L, Cortesi E, Tomasello G, Spadi R,
Zaniboni A, et al: Early tumor shrinkage and depth of response
predict long-term outcome in metastatic colorectal cancer patients
treated with first-line chemotherapy plus bevacizumab: Results from
phase III TRIBE trial by the Gruppo Oncologico del Nord Ovest. Ann
Oncol. 26:1188–1194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ettinger DS, Wood DE, Akerley W, Bazhenova
LA, Borghaei H, Camidge DR, Cheney RT, Chirieac LR, D'Amico TA,
Demmy TL, et al: Non-small cell lung cancer, version 6.2015. J Natl
Compr Canc Netw. 13:515–524. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Horie N, Aiba H, Oguro K, Hojo H and
Takeishi K: Functional analysis and DNA polymorphism of the
tandemly repeated sequences in the 5′-terminal regulatory region of
the human gene for thymidylate synthase. Cell Struct Funct.
20:191–197. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee DJ, Lee J, Lee HY, Lim T, Lee SJ, Yi
SY, Park SH, Park JO, Lim HY, Kang WK and Park YS: Salvage S-1
monotherapy in metastatic colorectal cancer patients who failed
irinotecan-based or oxaliplatin-based chemotherapy. Med Oncol. 28
Suppl 1:S291–S294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cremolini C, Loupakis F, Antoniotti C,
Lupi C, Sensi E, Lonardi S, Mezi S, Tomasello G, Ronzoni M,
Zaniboni A, et al: FOLFOXIRI plus bevacizumab versus FOLFIRI plus
bevacizumab as first-line treatment of patients with metastatic
colorectal cancer: Updated overall survival and molecular subgroup
analyses of the open-label, phase 3 TRIBE study. Lancet Oncol.
16:1306–1315. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Etienne-Grimaldi MC, Milano G,
Maindrault-Goebel F, Chibaudel B, Formento JL, Francoual M, Lledo
G, André T, Mabro M, Mineur L, et al: Methylenetetrahydrofolate
reductase (MTHFR) gene polymorphisms and FOLFOX response in
colorectal cancer patients. Br J Clin Pharmacol. 69:58–66. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jakobsen A, Nielsen JN, Gyldenkerne N and
Lindeberg J: Thymidylate synthase and methylenetetrahydrofolate
reductase gene polymorphism in normal tissue as predictors of
fluorouracil sensitivity. J Clin Oncol. 23:1365–1369. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cohen V, Panet-Raymond V, Sabbaghian N,
Morin I, Batist G and Rozen R: Methylenetetrahydrofolate reductase
polymorphism in advanced colorectal cancer: A novel genomic
predictor of clinical response to fluoropyrimidine-based
chemotherapy. Clin Cancer Res. 9:1611–1615. 2003.PubMed/NCBI
|
|
27
|
Castillo-Fernández O, Santibáñez M, Bauza
A, Calderillo G, Castro C, Herrera R, Serrano A, Arrieta O and
Herrera LA: Methylenetetrahydrofolate reductase polymorphism (677
C>T) predicts long time to progression in metastatic colon
cancer treated with 5-fluorouracil and folinic acid. Arch Med Res.
41:430–435. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Suh KW, Kim JH, Kim DY, Kim YB, Lee C and
Choi S: Which gene is a dominant predictor of response during
FOLFOX chemotherapy for the treatment of metastatic colorectal
cancer, the MTHFR or XRCC1 gene? Ann Surg Oncol. 13:1379–1385.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marcuello E, Altés A, Menoyo A, Rio ED and
Baiget M: Methylenetetrahydrofolate reductase gene polymorphisms:
Genomic predictors of clinical response to fluoropyrimidine-based
chemotherapy? Cancer Chemother Pharmacol. 57:835–840. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Derwinger K, Wettergren Y, Odin E,
Carlsson G and Gustavsson B: A study of the MTHFR gene polymorphism
C677T in colorectal cancer. Clin Colorectal Cancer. 8:43–48. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sharma R, Hoskins JM, Rivory LP, Zucknick
M, London R, Liddle C and Clarke SJ: Thymidylate synthase and
methylenetetrahydrofolate reductase gene polymorphisms and toxicity
to capecitabine in advanced colorectal cancer patients. Clin Cancer
Res. 14:817–825. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu H, Xu C, Chen G and Wang J: X-ray
repair cross-complementing 1 polymorphism and prognosis of
platinum-based chemotherapy in gastric and colorectal cancer: A
meta-analysis. J Gastroenterol Hepatol. 29:926–933. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liang J, Jiang T, Yao RY, Liu ZM, Lv HY
and Qi WW: The combination of ERCC1 and XRCC1 gene polymorphisms
better predicts clinical outcome to oxaliplatin-based chemotherapy
in metastatic colorectal cancer. Cancer Chemother Pharmacol.
66:493–500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ye F, Liu Z, Tan A, Liao M, Mo Z and Yang
X: XRCC1 and GSTP1 polymorphisms and prognosis of oxaliplatin-based
chemotherapy in colorectal cancer: A meta-analysis. Cancer
Chemother Pharmacol. 71:733–740. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lv H, Li Q, Qiu W, Xiang J, Wei H, Liang
H, Sui A and Liang J: Genetic polymorphism of XRCC1 correlated with
response to oxaliplatin-based chemotherapy in advanced colorectal
cancer. Pathol Oncol Res. 18:1009–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang MY, Huang ML, Chen MJ, Lu CY, Chen
CF, Tsai PC, Chuang SC, Hou MF, Lin SR and Wang JY: Multiple
genetic polymorphisms in the prediction of clinical outcome of
metastatic colorectal cancer patients treated with first-line
FOLFOX-4 chemotherapy. Pharmacogenet Genomics. 21:18–25. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawakami K and Watanabe G: Identification
and functional analysis of single nucleotide polymorphism in the
tandem repeat sequence of thymidylate synthase gene. Cancer Res.
63:6004–6007. 2003.PubMed/NCBI
|
|
39
|
Ruzzo A, Graziano F, Kawakami K, Watanabe
G, Santini D, Catalano V, Bisonni R, Canestrari E, Ficarelli R,
Menichetti ET, et al: Pharmacogenetic profiling and clinical
outcome of patients with advanced gastric cancer treated with
palliative chemotherapy. J Clin Oncol. 24:1883–1891. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sohn KJ, Croxford R, Yates Z, Lucock M and
Kim YI: Effect of the methylenetetrahydrofolate reductase C677T
polymorphism on chemosensitivity of colon and breast cancer cells
to 5-fluorouracil and methotrexate. J Natl Cancer Inst. 96:134–144.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kawakami K, Omura K, Kanehira E and
Watanabe G: Methylenetetrahydrofolate reductase polymorphism is
associated with folate pool in gastrointestinal cancer tissue.
Anticancer Res. 21:285–289. 2001.PubMed/NCBI
|
|
42
|
Mohrenweiser HW, Xi T, Vázquez-Matías J
and Jones IM: Identification of 127 amino acid substitution
variants in screening 37 DNA repair genes in humans. Cancer
Epidemiol Biomarkers Prev. 11:1054–1064. 2002.PubMed/NCBI
|
|
43
|
Marusyk A and Polyak K: Tumor
heterogeneity: Causes and consequences. Biochim Biophys Acta.
1805:105–117. 2010.PubMed/NCBI
|
|
44
|
Giessen C, Laubender RP, von Weikersthal L
Fischer, Schalhorn A, Modest DP, Stintzing S, Haas M, Mansmann UR
and Heinemann V: Early tumor shrinkage in metastatic colorectal
cancer: Retrospective analysis from an irinotecan-based randomized
first-line trial. Cancer Sci. 104:718–724. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Heinemann V, Modest D, von Weikersthal LF,
Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T,
Lerchenmüller C, Kahl C, et al: Independent radiological evaluation
of objective response, early tumor shrinkage and depth of response
in FIRE-3 (AIO KRK-0306). Annals Oncol. 25:ii1172014. View Article : Google Scholar
|
|
46
|
Cui LH, Shin MH, Kweon SS, Kim HN, Song
HR, Piao JM, Choi JS, Shim HJ, Hwang JE, Kim HR, et al:
Methylenetetrahydrofolate reductase C677T polymorphism in patients
with gastric and colorectal cancer in a Korean population. BMC
Cancer. 10:2362010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim HN, Kim NY, Yu L, Kim YK, Lee IK, Yang
DH, Lee JJ, Shin MH, Park KS, Choi JS and Kim HJ: Polymorphisms of
drug-metabolizing genes and risk of non-Hodgkin lymphoma. Am J
Hematol. 84:821–825. 2009. View Article : Google Scholar : PubMed/NCBI
|