Introduction
Cutaneous malignant melanoma (CMM) is a type of skin
cancer that originated from uncontrolled proliferation of
melanocytes. CMM accounts for 80% of mortalities arising from skin
cancer (1). Although the incidence of
melanoma is low, the number of new melanoma cases has increased
steadily in the recent years (2). The
prognosis of melanoma remains poor due to its high potential for
invasion and metastasis, and its resistance to conventional
radiotherapy and chemotherapy (3).
Therefore, identifying novel molecular markers for the assessment
of prognoses of cutaneous malignant melanoma, or as novel targeted
therapeutic agents, will be beneficial for patients with
melanoma.
The liver kinase B1 gene (LKB1), also known as
STK11, was originally identified as the causative gene of
Peutz-Jeghers syndrome, an autosomal dominant inherited disorder
characterized by mucosal gastrointestinal polyps and an increased
risk of malignant tumors (4). As a
tumor suppressor gene, LKB1 suppresses tumor cell growth, induces
cell death and inhibits tumor cell metastasis (5). Several studies have demonstrated that
the loss of LKB1 promotes metastasis in a number of tumor cell
types, including melanoma cells (6–8). At
present, LKB1 inactivation mechanisms remain controversial. LKB1
mutations were detected in certain types of sporadic cancers,
including lung adenocarcinoma and cervical carcinoma (9). However, only 10% of patients with
cutaneous melanoma have reported to exhibit mutations in the LKB1
gene, which means that genetic mutation is not the major mechanism
for LKB1 gene inactivation, and there may be other mechanisms that
mediate the lack of LKB1 expression (10–11).
Epigenetic alteration of tumor suppressor genes by aberrant
methylation in promoter CpG islands has been acknowledged as an
important mechanism for inactivation of gene expression (12–16).
Previously, several studies have identified that hypermethylation
of the LKB1 promoter region may be detected in certain cancer cell
lines and primary tumor samples. Correspondingly, LKB1 transcripts
were not detected, and treatment with the demethylating agent
5-aza-2′-deoxycytidine was able to restore LKB1 gene expression in
these cells (17). Whether epigenetic
alteration of the LKB1 gene leads to an inactivation of LKB1 and a
lack of LKB1 expression in melanoma remains unclear.
The aim of the present study was to investigate the
status of LKB1 promoter methylation in human cutaneous melanoma and
to evaluate the associations between aberrant LKB1 promoter
methylation with clinicopathological features and prognosis in
patients with melanoma.
Materials and methods
Tissue specimen preparations
A total of 57 cases of melanoma and 50 cases of
benign lesion controls (including dysplastic nevi and common nevi,
the ratio of males:females was 32/25, mean age of all patients was
59.3±0.4.) were selected from archived formalin-fixed paraffin
embedded (FFPE) tissue blocks from The First Affiliated Hospital of
Nanjing Medical University (Nanjing, China) between January 2003
and December 2010. Prior to sectioning, all archived tissue slides
were reviewed by two experienced pathologists using a multi-head
light microscope (Nikon 80i; Nikon Corporation; Tokyo, Japan). The
tumors were classified according to the American Joint Committee on
Cancer (AJCC) staging system (18–19).
Melanoma Breslow's thicknesses (20)
were divided into 2 categories (< or = 2.00, and >2.00 mm).
Detailed clinicopathological parameters, summarized in Table I, were obtained from the medical
records or pathology examination reports of patients. The follow-up
time was from the date of diagnosis to the date of mortality or the
end of follow-up (5 years). Identifiable patient data (name and
other private data) were anonymized during the study process. The
ethics committee of The First Affiliated Hospital of Nanjing
Medical University approved the study design, including the use of
all tissue blocks from patients.
 | Table I.Liver kinase B1 promoter methylation
in melanoma and benign skin lesions. |
Table I.
Liver kinase B1 promoter methylation
in melanoma and benign skin lesions.
|
| Methylation |
|
|---|
|
|
|
|
|---|
| Group | Positive | Negative | P-value |
|---|
| Melanoma tissue | 12 | 45 | <0.05 |
| Benign skin
lesion | 2 | 48 |
|
Immunohistochemistry (IHC)
detection
The level of LKB1 expression was detected by IHC in
57 FFPE melanoma and 50 benign control tissues. Standard
immunohistochemical protocol was used. Paraffin (thickness, 4 µm)
sections from each specimen were heated at 70°C for 60 min and then
deparaffinized with xylene. The sections were hydrated with a
gradient of ethanol and immersed in 10 mM pH 6.0 boiling citrate
buffer in an autoclave for 15 min for antigen retrieval. Subsequent
to blocking with goat serum for 10 min at room temperature, the
slides were incubated with a monoclonal rabbit anti-LKB1 antibody
(cat. no. ab15095; dilution, 1:250; Abcam, Cambridge, UK) overnight
at 4°C in a humidified chamber. The slides were rinsed, and then
incubated with ready-to-use horseradish peroxidase-conjugated
universal secondary antibody (cat. no. K5007; Dako; Agilent
Technologies, Inc., Santa Clara, CA USA) for 15 min at room
temperature. The chromogenic reaction was developed by incubating
the slides with 3,3′-diaminobenzidine and counterstained with
hematoxylin for 3 min. The slides were also incubated with
non-immune serum (cat. no. ab7481; Abcam) overnight at 4°C as
negative controls. LKB1 staining was distributed in the nucleus and
cytoplasm, and scored by two pathologists based on the percent of
positively stained cells and staining intensity. The percentage of
positive staining was scored as: 0, 0%; 1, 0–20%; 2, 20–50%; or 3,
>50%, and the intensity as: 0, no staining; 1, weak staining and
visible at high magnification (×400; Nikon 50i, Nikon Corporation);
2, moderate staining and visible at low magnification (×100) and 3,
dark staining, visible at low magnification. The total
immunostaining score was calculated by multiplying the staining
intensity score (range, 0–9) by the score for the percentage of
positive staining, as described previously (21). The specimens can be classified into
two groups based on LKB1 expression: Low expression (score, 0–3)
and high expression (score, 4–9).
Extraction of nucleic acids and
quantitative reverse transcription polymerase chain reaction
(RT-qPCR)
QIAamp DNA FFPE Tissue kit (cat. no. 56404) was used
to isolate genomic DNA from FFPE tissues. The hematoxylin and eosin
stained slides were first reviewed by pathologists, and the areas
of interest were circled to guide macro-dissection of the tumor
tissue. In total, ≤8 deparaffinized, unstained sections (thickness,
5–10 µm) were scraped in a polypropylene micro-centrifuge tube, and
subsequently processed according to the manufacturer's instructions
(22). The quality and integrity of
the DNA were measured by electrophoresis on 0.8% agarose gels. The
DNA samples were quantified spectrophotometrically and stored at
−20°C for subsequent testing.
Total RNA from melanoma tissues were extracted using
the QIAamp RNeasy FFPE kit (cat no. 73504; Qiagen, Inc., Valencia,
CA, USA), The isolation was performed according to the
manufacturer's protocol. RNA purity was assessed using the
A260/A280 ratio with a SmartSpec Plus spectrophotometer (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Total RNA (1 µg) was
reverse transcribed into cDNA using the QIAamp QuantiTect Reverse
Transcription kit (cat no. 205313; Qiagen, Inc.). RT-qPCR reaction
was performed with a QuantiNova SYBR Green RT-PCR kit (cat no.
208152; Qiagen, Inc.) using the Agilent 3000 Real-Time PCR system
(Agilent Technologies, Inc.). Human LKB1 and GAPDH primers were as
follows: LKB1 forward, 5′-AGGGATGCTTGAGTACGAACC-3′ and reverse,
5′-GTCCTCCAAGTACGGCACC-3′; GAPDH forward,
5′-CAAGATCATCAGCAATGCCT-3′ and reverse,
5′-TCATGAGTCCTTCCACGATAC-3′. GAPDH was used as an internal control.
The relative mRNA expression of LKB1 was normalized to GAPDH and
calculated by using the comparative cycle threshold (Ct) method
(23). All experiments were
replicated three times.
Methylation-specific PCR
Methylation-specific PCR (MSP) was used to detect
promoter methylation. The QIAamp EpiTect Fast DNA Bisulfite kit
(cat. no. 59824; Qiagen, Inc.) was used to convert DNA samples (≥2
µg). The sequences of the unmethylated template primers were:
forward, 5′-TTTGTGTTTTGATGTTTTAGGTTTTTGT-3′ and reverse primer,
5′-AACTCCACACTCTTCCAAAAACAAAACA-3′. The sequences of the methylated
template primers were: Forward, 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ and
reverse, 5′-GCACTCTTCCGAAAACGAAACG-3′. The PCR amplification
conditions were as follows: An initial denaturation step at 95°C
for 5 min, followed by 35 cycles at 94°C for 50 sec, 59°C for 50
sec, 72°C for 50 sec and a final extension at 72°C for 10 min.
Statistical analysis
Statistical analyses were performed using SPSS
version 19.0 (IBM Corp., Armonk, NY, USA) software. Comparisons of
LKB1 methylation status with clinicopathological characteristics
were made using contingency table χ2 test. The Spearman
rank correlation was used to analyze the LKB1 expression at mRNA
and protein levels with LKB1 methylation status. Survival curves
were estimated using the Kaplan-Meier method, and the log-rank test
was used to compare the difference. Multivariate analysis was
performed using the Cox proportional hazard models with a
confidence interval of 95% to examine whether LKB1 promoter
methylation was an independent prognostic factor for 5-year overall
survival rates (OS). The Kruskal-Wallis test was used to compare
LKB1 expression between melanoma and benign lesions, methylated
melanoma and the unmethylated. All P-values were two-sided, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
LKB1 promoter methylation status in
human cutaneous melanoma tissues
LKB1 promoter methylation was performed successfully
in all cases. It was identified that 12/57 cases exhibited
methylation amplification in melanoma tissues, while 2/50 benign
controls displayed LKB1 promoter methylation. The frequency of LKB1
promoter methylation was significantly increased in melanoma
tissues compared with benign skin controls (χ2=6.747,
P<0.05; Table I).
LKB1 promoter methylation is
correlated with LKB1 mRNA and protein expression
RT-qPCR was performed successfully in 50 (50/57)
melanoma tissues. LKB1 mRNA expression level (LKB1/GAPDH) in the
methylated melanoma tissues (range, 0.112–2.524; mean, 0.675) was
reduced compared with the unmethylated melanoma tissues (range,
0.522–3.311; mean, 1.541; P<0.05). In melanoma tissues, LKB1
mRNA expression was correlated with LKB1 promoter methylation
status (r=0.354; P<0.05; data not shown).
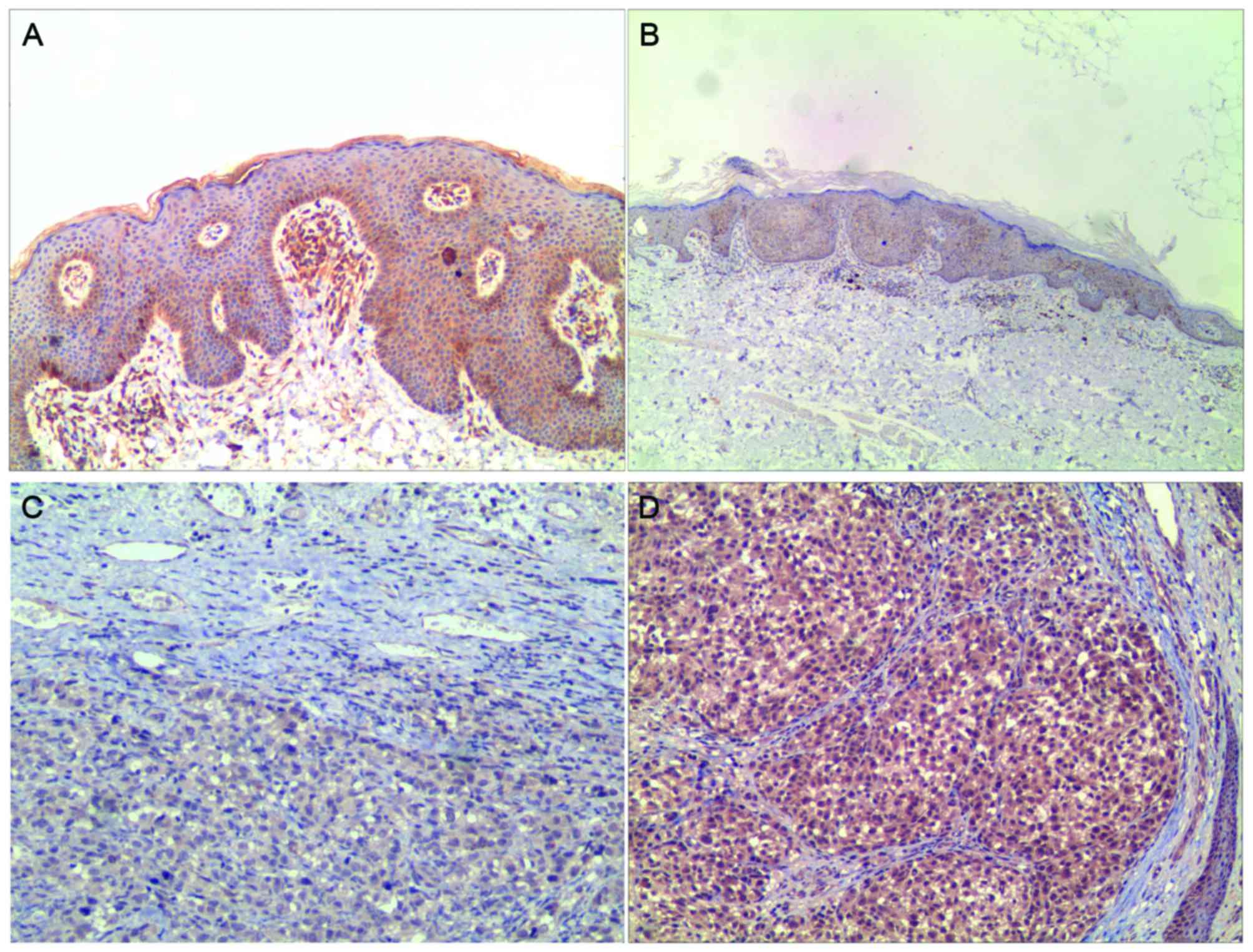
LKB1 protein expression was localized in the
cytoplasm (Fig. 1). Of the 57
melanoma tissues and 50 benign controls, LKB1 expression was
demonstrated in 36 and 44 cases, respectively. In the 12 cases of
LKB1-methylated melanoma, 6 cases demonstrated LKB1 expression.
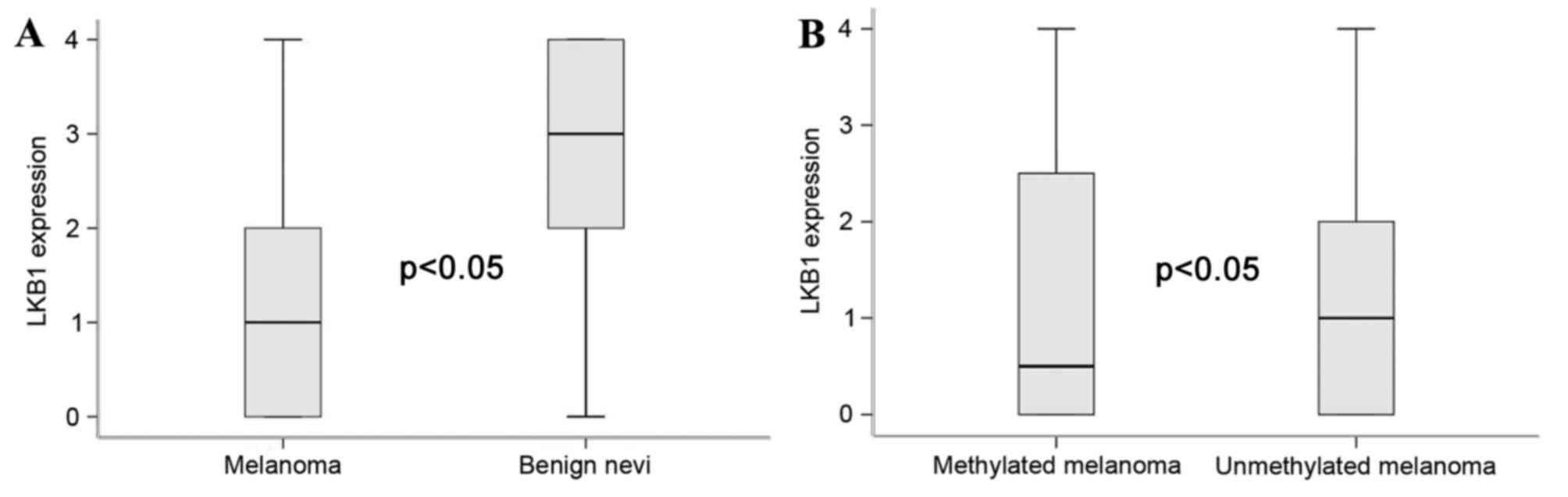
Notably, significant differences in LKB1 expression were observed
between benign lesions with melanomas, and between LKB1 methylated
melanomas and unmethylated melanoma (P<0.05, Kruskal-Wallis
test; Fig. 2A and B, respectively).
In the melanoma cases, LKB1 protein expression determined by IHC
was correlated with the status of LKB1 promoter methylation
(r=0.473; P<0.01; data not shown).
Associations between LKB1 promoter
methylation and clinicopathological factors
Patient information (age, sex, and location of
melanoma) and clinicopathological parameters are detailed in
Table II. Statistical analysis
revealed that LKB1 promoter methylation was closely associated with
tumor Breslow's thickness (P=0.021), the presence of ulceration
(P=0.002) and AJCC stage (P=0.006). In the present study, no
significant correlations between LKB1 promoter methylation and
tumor location, age, sex or tumor subtype were observed (Table II).
 | Table II.Association between LKB1 promoter
methylation and different clinicopathological parameters in 57
melanoma cases. |
Table II.
Association between LKB1 promoter
methylation and different clinicopathological parameters in 57
melanoma cases.
|
|
| LKB1 |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameter | N | Methylated | Unmethylated | χ2 | P-value |
|---|
| Age at diagnosis |
|
|
| 1.201 | 0.273 |
| ≤60 | 30 | 8 | 22 |
|
|
|
>60 | 27 | 4 | 23 |
|
|
| Sex |
|
|
| 0.030 | 0.863 |
| Male | 32 | 7 | 25 |
|
|
|
Female | 25 | 5 | 20 |
|
|
| Location |
|
|
| 0.005 | 0.945 |
|
Sun-protecteda | 28 | 6 | 22 |
|
|
|
Sun-exposedb | 29 | 6 | 23 |
|
|
| Breslow's thickness,
mm |
|
|
| 5.291 | 0.021 |
| ≤2 | 31 | 3 | 28 |
|
|
|
>2 | 26 | 9 | 17 |
|
|
| Ulceration |
|
|
| 9.619 | 0.002 |
|
Absent | 32 | 2 | 30 |
|
|
|
Present | 25 | 10 | 15 |
|
|
| Tumor subtype |
|
|
| 1.679 | 0.642 |
|
ALM | 15 | 2 | 13 |
|
|
|
SSM | 12 | 2 | 10 |
|
|
|
LMM | 14 | 3 | 11 |
|
|
|
Mucosal | 16 | 5 | 11 |
|
|
| AJCC stage |
|
|
| 8.661 | 0.006 |
|
I/II | 39 | 4 | 35 |
|
|
|
III/IV | 18 | 8 | 10 |
|
|
Prognostic analysis of melanoma
patients with LKB1 promoter methylation
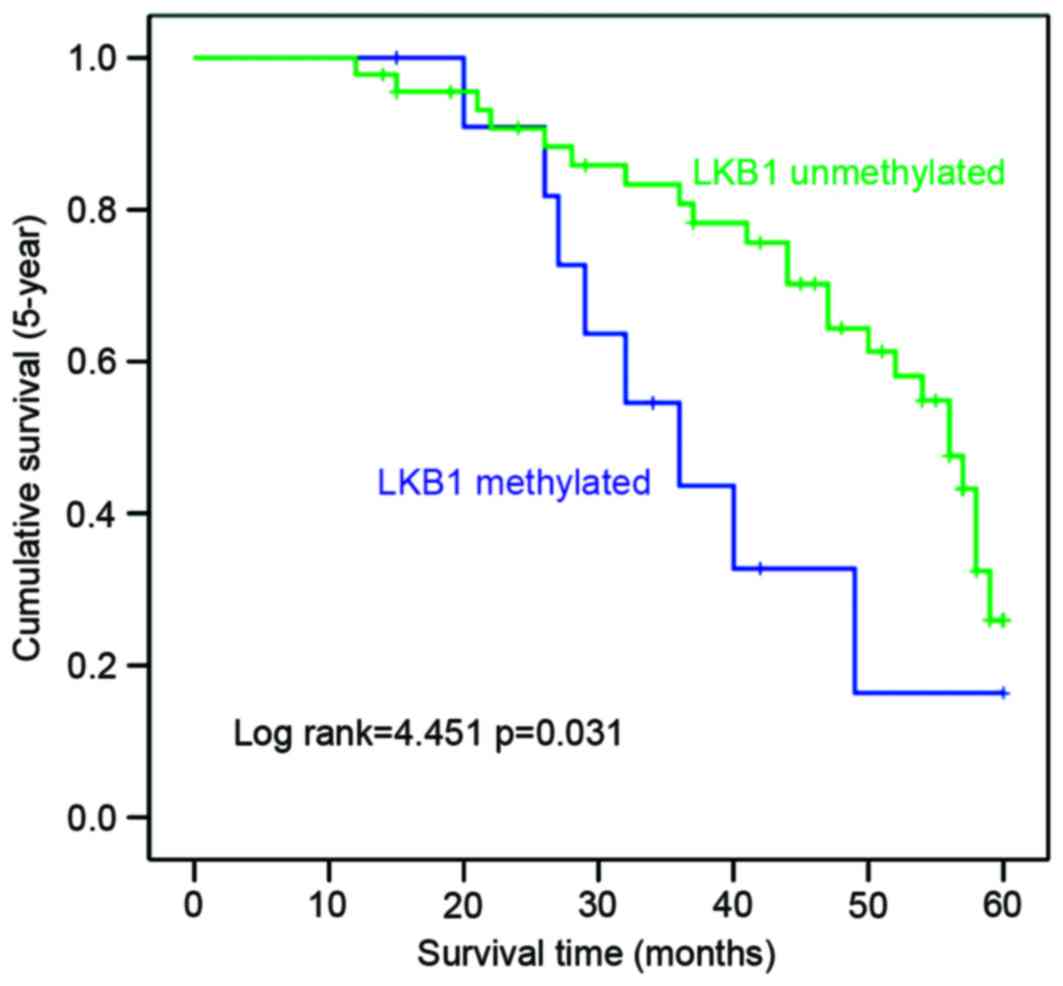
To investigate the prognostic significance of LKB1
promoter methylation in melanoma, Kaplan-Meier survival analysis
and log-rank tests were used to assess the association between LKB1
promoter methylation and information on follow-up of patients. It
was identified that patients with LKB1 methylation exhibited a
significantly shorter overall 5-year OS duration compared with
those with unmethylated LKB1 (P<0.05; Fig. 3). Concurrently, methylation and
presence of ulceration were all associated with prognosis of CMM
(P<0.05; Table III). Age, sex,
location, Breslow's thickness and tumor subtype were not associated
with prognosis of CMM (P>0.05; Table
III). Further multivariate Cox regression analysis indicated
that LKB1 promoter methylation was an independent prognostic
predictor of overall 5-year survival in melanoma patients (Table III).
 | Table III.Multivariate Cox regression analysis
of 5-year overall survival in melanoma patients with LKB1 promoter
methylation. |
Table III.
Multivariate Cox regression analysis
of 5-year overall survival in melanoma patients with LKB1 promoter
methylation.
| Variable | Ba | Standard error | P-value | Hazard ratio | 95% confidence
interval |
|---|
| Age |
0.097 | 0.441 | 0.826 | 1.102 | 0.464–2.615 |
| Sex |
0.495 | 0.383 | 0.196 | 1.641 | 0.774–3.478 |
| Tumor location | −0.326 | 0.399 | 0.415 | 0.722 | 0.330–1.580 |
| Breslow's
thickness | −0.466 | 0.501 | 0.352 | 0.627 | 0.235–1.674 |
| Ulceration | 1.02 | 0.481 | 0.034 | 2.773 | 1.081–7.111 |
| Tumor subtype | −0.065 | 0.196 | 0.74 | 0.937 | 0.639–1.375 |
| AJCC stage | −0.718 | 0.571 | 0.209 | 0.488 | 0.159–1.494 |
| Methylation |
1.305 | 0.606 | 0.031 | 3.688 |
1.124–12.100 |
Discussion
LKB1, also known as serine/threonine kinase, was
identified as a crucial tumor suppressor gene in numerous types of
tumors and cancer cells (4–5). Through activation of multiple signaling
pathways, LKB1 functions to regulate cellular growth, energy
metabolism and polarity in a number of tissues (24,25). A
previous study demonstrated that the loss of LKB1 serves an
important role in tumor initiation and progression (26). At present, the loss of LKB1 in
different human tumors is involved in a combination of three
molecular mechanisms, including mutation, loss of heterozygosity
(LOH) and epigenetic modification (8). Although LKB1 gene mutation was
hypothesized as the primary mechanism of LKBI inactivation in
Peutz-Jeghers syndrome (PJS) or PJS-associated tumors, results from
Avizienyte et al (27)
suggested that mutational inactivation of LKB1 is a rare event in
the majority of sporadic tumor types. As the primary epigenetic
modification in humans, promoter methylation has been demonstrated
in number of tumor and cell types. Trojan et al (28) confirmed that the 5′-CpG islands of the
LKB1 promoter region were methylated when analyzing a large series
of paraffin embedded colorectal cancer specimens. Esteller et
al (29) also demonstrated LKB1
promoter methylation in primary colorectal cancer specimens and
suggested that LKB1 promoter methylation may mediate the
inactivation of LKB1.
To investigate the status of LKB1 promoter
methylation in human cutaneous melanoma, the present study first
examined LKB1 promoter methylation in melanoma tissues and benign
skin lesion tissues by MSP. Of the 57 cases, 12 cases of melanoma
tissues exhibited LKB1 promoter methylation, and the frequency of
LKB1 promoter methylation in melanoma tissues was significantly
increased compared with benign skin lesion tissues. The association
between LKB1 promoter methylation and different clinicopathological
parameters was investigated. It was demonstrated that LKB1
methylation status was significantly associated with Breslow's
thickness (P<0.05), the presence of ulceration (P<0.01) and
AJCC stage (P<0.01). The prognosis survival difference between
the patients with LKB1 promoter methylation and the patients
without methylation was also analyzed. The average duration of
5-year OS in patients with LKB1 methylation was significantly
shorter compared with patients without LKB1 methylation. In
addition, it was identified that LKB1 expression was markedly
reduced in cutaneous melanoma tissues compared with benign skin
lesions. In melanoma tissues, downregulated LKB1 expression was
associated with its methylation status.
In conclusion, the results of the present study
suggest that LKB1 gene promoter methylation in cutaneous melanoma
was associated with clinicopathological parameters, and LKB1 may be
an independent prognosis predicator. It was also demonstrated that
downregulated LKB1 expression may be associated with LKB1
methylation. However, there are several limitations with the
present study, including the use of a small sample size and a
retrospective study design. The data need to be extensively
evaluated in larger sample sizes and in multi-centric studies. The
role of LKB1 mutations and LOH in cutaneous melanoma requires
confirmation, and it is possible that LKB1 loss is caused by a
combination of factors.
References
|
1
|
Cockerell CJ: The pathology of melanoma.
Dermatol Clin. 30:445–468. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wisco OJ and Sober AJ: Prognostic factors
for melanoma. Dermatol Clin. 30:469–485. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tejera-Vaquerizo A, Solís-García E,
Ríos-Martín JJ and Moreno-Ramírez D: Primary cutaneous melanoma:
Prognostic factors not included in the classification of the
american joint committee on cancer. Actas Dermosifiliogr.
102:255–263. 2011.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katajisto P, Vallenius T, Vaahtomeri K,
Ekman N, Udd L, Tiainen M and Mäkelä TP: The LKB1 tumor suppressor
kinase in human disease. Biochim Biophys Acta. 1775:63–75.
2007.PubMed/NCBI
|
|
5
|
Hemminki A, Markie D, Tomlinson I,
Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M,
Höglund P, et al: A serine/threonine kinase gene defective in
Peutz-Jeghers syndrome. Nature. 391:184–187. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Liu J, Li P, Mao X, Li W, Yang J and
Liu P: Loss of LKB1 disrupts breast epithelial cell polarity and
promotes breast cancer metastasis and invasion. J Exp Clin Cancer
Res. 33:702014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng B, Jeong JH, Asara JM, Yuan YY,
Granter SR, Chin L and Cantley LC: Oncogenic B-RAF negatively
regulates the tumor suppressor LKB1 to promote melanoma cell
proliferation. Mol Cell. 33:237–247. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gan RY and Li HB: Recent progress on liver
kinase B1 (LKB1): Expression, regulation, downstream signaling and
cancer suppressive function. Int J Mol Sci. 15:16698–16718. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vaahtomeri K and Mäkelä TP: Molecular
mechanisms of tumor suppression by LKB1. FEBS Lett. 585:944–951.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Partanen JI, Tervonen TA, Myllynen M, Lind
E, Imai M, Katajisto P, Dijkgraaf GJ, Kovanen PE, Mäkelä TP, Werb Z
and Klefström J: Tumor suppressor function of Liver kinase B1
(Lkb1) is linked to regulation of epithelial integrity. Proc Natl
Acad Sci USA. 109:pp. E388–E397. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou W, Zhang J and Marcus AI: LKB1 tumor
suppressor: Therapeutic opportunities knock when LKB1 is
inactivated. Genes Dis. 1:64–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Wilde RF, Ottenhof NA, Jansen M,
Morsink FH, de Leng WW, Offerhaus GJ and Brosens LA: Analysis of
LKB1 mutations and other molecular alterations in pancreatic acinar
cell carcinoma. Mod Pathol. 24:1229–1236. 2012. View Article : Google Scholar
|
|
13
|
Lee SM, Choi JE, Na YK, Lee EJ, Lee WK,
Choi YY, Yoon GS, Jeon HS, Kim DS and Park JY: Genetic and
epigenetic alterations of the LKB1 gene and their associations with
mutations in TP53 and EGFR pathway genes in Korean non-small cell
lung cancers. Lung Cancer. 81:194–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guldberg P, Straten P thor, Ahrenkiel V,
Seremet T, Kirkin AF and Zeuthen J: Somatic mutation of the
Peutz-Jeghers syndrome gene, LKB1/STK11, in malignant melanoma.
Oncogene. 18:1777–1780. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
de Abreu FB, Gallagher TL, Liu EZ and
Tsongalis GJ: Determining methylation status of methylguanine DNA
methyl transferase (MGMT) from formalin-fixed, paraffin embedded
tumor tissue. MethodsX. 1:42–48. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ekizoglu S, Dalay N, Karaman E, Akdeniz D,
Ozaydin A and Buyru N: LKB1 downregulation may be independent of
promoter methylation or FOXO3 expression in head and neck cancer.
Transl Res. 162:122–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He TY, Tsai LH, Huang CC, Chou MC and Lee
H: LKB1 loss at transcriptional level promotes tumor malignancy and
poor patient outcomes in colorectal cancer. Ann Surg Oncol. 21
Suppl 4:S703–S710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Boland GM and Gershenwald JE: Principles
of melanoma staging. Cancer Treat Res. 167:131–148. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Retsas S, Henry K, Mohammed MQ and MacRae
K: Prognostic factors of cutaneous melanoma and a new staging
system proposed by the American Joint Committee on Cancer (AJCC):
Validation in a cohort of 1284 patients. Eur J Cancer. 38:511–516.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moreno-Ramírez D, Ojeda-Vila T,
Ríos-Martín JJ, Nieto-García A and Ferrándiz L: Association between
tumor size and Breslow's thickness in malignant melanoma: A
cross-sectional, multicenter study. Melanoma Res. 25:450–452. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gown AM, Goldstein LC, Barry TS, Kussick
SJ, Kandalaft PL, Kim PM and Tse CC: High concordance between
immunohistochemistry and fluorescence in situ hybridization testing
for HER2 status in breast cancer requires a normalized IHC scoring
system. Mod Pathol. 21:1271–1277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kokkat TJ, Patel MS, McGarvey D, LiVolsi
VA and Baloch ZW: Archived formalin-fixed paraffin-embedded (FFPE)
blocks: A valuable underexploited resource for extraction of DNA,
RNA, and protein. Biopreserv Biobank. 11:101–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chan KT, Asokan SB, King SJ, Bo T, Dubose
ES, Liu W, Berginski ME, Simon JM, Davis IJ, Gomez SM, et al: LKB1
loss in melanoma disrupts directional migration toward
extracellular matrix cues. J Cell Biol. 207:299–315. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shin C and Tallon B: Assessment of tumor
mitotic rate in primary cutaneous malignant melanomas 1 mm or less
in thickness. J Am Acad Dermatol. 72:405–409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu W, Monahan KB, Pfefferle AD, Shimamura
T, Sorrentino J, Chan KT, Roadcap DW, Ollila DW, Thomas NE,
Castrillon DH, et al: LKB1/STK11 Inactivation leads to expansion of
a prometastatic tumor subpopulation in melanoma. Cancer Cell.
21:751–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Avizienyte E, Loukola A, Roth S, Hemminki
A, Tarkkanen M, Salovaara R, Arola J, Bützow R,
Husgafvel-Pursiainen K, Kokkola A, et al: LKB1 somatic mutations in
sporadic tumors. Am J Pathol. 154:677–681. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Trojan J, Brieger A, Raedle J, Esteller M
and Zeuzem S: 5′-CpG island methylation of the LKB1/STK11 promoter
and allelic loss at chromosome 19p13.3 in sporadic colorectal
cancer. Gut. 47:272–276. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Esteller M, Avizienyte E, Corn PG, Lothe
RA, Baylin SB, Aaltonen LA and Herman JG: Epigenetic inactivation
of LKB1 in primary tumors associated with the Peutz-Jeghers
syndrome. Oncogene. 19:164–168. 2000. View Article : Google Scholar : PubMed/NCBI
|