Introduction
Functional human telomerase reverse transcriptase
(hTERT) mRNA is detected in lymphocytes (1) and in the normal brain, liver, prostate,
heart and primary fibroblasts, irrespective of the degree of
telomerase activity (2). The
full-length Yonsei hTERT variant is associated with high telomerase
activity (3). However, the presence
of a full-length hTERT variant is not sufficient to allow
telomerase activity when an abundance of multiple hTERT spliced
variants is concomitantly present (4).
A total of 6 alternative splicing sites (4 insertion
and 2 deletion) have been identified within hTERT mRNA (5). Splice variants, depending on tissue
type, continued to be synthesized even when the full-length variant
was not (6). The presence of
alternatively spliced variants in human preimplantation embryos may
suggest a lack of telomerase activity, resulting in the appearance
of chromosomes with shortened telomeres (7,8). The exact
functions of each alternatively spliced variant of hTERT remain
unknown, although the positions of the spliced sites suggest that
the majority of the variants do not code for functional reverse
transcriptase. Depending on the cell line, this telomerase
inhibition resulted either in cell death or a senescence-like state
(9). The β splice variant did not
reconstitute active telomerase activity in a previous in
vitro transcription and translation study (10). The alteration of hTERT full-length
variant expression levels has previously been reported to
demonstrate various gene expression profiles and genomic copy
number changes in cancer cell lines (11).
In telomerase-positive cell lines, there is a highly
uniform pattern of splicing. In total, ~5% of hTERT mRNA is in
full-length form, 80–90% in β-spliced form, 5–15% in α/β-spliced
form and <1% in α-spliced form (12). However, in contrast, considerable
variation in the total number and quantitative distribution of
spliced variants has been observed in various cancer tissues and in
normal tissues. In renal cell carcinoma, the β variant was the
major type and the α variant was not detected (13). In breast cancer, the α variant was
always coexpressed with the full-length and/or β variant,
otherwise, the full-length and β variant were observed either in
combination with other transcripts or expressed as the only hTERT
transcript (14). Melanoma expressed
full-length transcripts with various combinations of α, β and α/β
variants, although a prevalence of the β variant was observed
(4).
The presence of a full-length variant was correlated
with telomerase activity in the endometrium and myometrium
(15). Two potential mechanisms
underlying the reduction of telomerase activity with
differentiation have previously been suggested: One is the
post-translational alterations in hTERT with stable total
transcription and splicing patterns, and the other is the minimal
change in total transcription with a dramatic decrease of the
full-length variant, predominantly remaining in β-spliced form
(13).
The aim of the present study was to analyze the
hTERT splicing pattern in association with telomerase activity in
patients with colorectal cancer.
Materials and methods
Tissue sampling and RNA isolation
A total of 40 patients who underwent surgery between
May 2002 and May 2003 at Yonsei Cancer Center, Severance Hospital,
Yonsei University Health System (Seoul, Korea) were enrolled in the
present study. Clinical characteristics of the patients are listed
in Table I. Paired tumor tissues and
normal tissues from 30 patients and tumor tissues alone from 10
patients were studied. Tumor tissues (≥70% carcinoma cellularity)
were frozen immediately following surgery. In paired samples,
normal tissues 10 cm from the lateral margin of the primary tumor
were obtained. Total RNA was isolated from cells and tissues using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's protocol.
Approval for the present study was obtained from the institutional
review board of the Severance Hospital (Seoul, Korea; approval no.
IRB-4-2004-0083). Written, informed consent was obtained from all
patients for the use of the samples in the present study.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Clinical factors | Patient number
(%) |
|---|
| Age (median, range;
years) | 66 (44–81) |
| Sex |
|
| Male | 25 (62.5) |
|
Female | 15 (37.5) |
| Eastern Cooperative
Oncology Groupa |
|
| 0–1 | 40 (100) |
| Primary site |
|
| Colon
cancer | 23 (57.5) |
| Rectal
cancer | 17 (42.5) |
| Stage
(Duke)b |
|
| A | 1
(2.5) |
| B | 21 (52.5) |
| C | 13 (32.5) |
| D | 5
(12.5) |
| Stage (TNM) |
|
| I | 3
(7.5) |
| II | 19 (47.5) |
| III | 13 (32.5) |
| IV | 5
(12.5) |
| Operation type |
|
| Low
anterior resection | 26 (65.0) |
| Right
hemicolectomy | 9
(22.5) |
| Left
hemicolectomy | 5
(12.5) |
| Relapse |
|
| No
recurrence | 28 (70.0) |
|
Recurrence | 12 (30.0) |
| Survival rate |
|
|
Survived | 31 (77.5) |
|
Succumbed | 9
(22.5) |
| Follow-up duration
(median, range; months) | 69.5
(7.4–76.3) |
Telomeric repeat amplification
protocol (TRAP) assay
Each tissue sample was washed in ice-cold wash
buffer [10 mM Hepes-KOH (pH 7.5), 1.5 mM MgCl2, 10 mM
KCl, 1 mM dithiothreitol] and then homogenized with 100 µl ice-cold
lysis buffer [10 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM
egtazic acid (EGTA), 0.1 mM phenylmethylsulfonyl fluoride, 5 mM
β-mercaptoethanol, 0.5% CHAPS, 10% glycerol]. The TRAP assay was
performed as follows: A total of 20 µl of each extract was assayed
in a 50 µl reaction mixture supplemented with 20 mM Tris-HCl (pH
8.3), 1.5 mM MgCl2, 63 mM KCl, 0.005% Tween-20, 1 mM
EGTA, 50 µM deoxynucleotide triphosphates, 0.5 µl
(α-32P) deoxycytidine triphosphate (dCTP; 3,000 Ci/mmol;
GE Healthcare Life Sciences, Chalfont, UK), 0.1 µg
template-switching oligonucleotide (5′-AATCCGTCGAGCAGAGTT-3′), 1 µg
T4 gene 32 protein, bovine serum albumin (0.1 mg/ml; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) and 2 U Taq DNA polymerase (Applied
Biosystems; Thermo Fisher Scientific, Inc.) in an assay tube
supplemented with 0.1 µg CX primer
(5′-CCCTTACCCTTACCCTTACCCTAA-3′). The reaction mixture was
amplified for 30 cycles at 94°C for 30 sec, 50°C for 30 sec, 72°C
for 90 sec, and then for 10 min at 72°C. Analysis of 10 µl of each
polymerase chain reaction (PCR) product was performed on 12%
non-denaturing acrylamide gels. The present study also determined
the optical density (OD) of the bands in the film by
computer-assisted densitometry (Vilber Lourmat Vision-Capt software
version 1; Vilber Lourmat, Marne-la-Vallée, France). The OD of the
band with the greatest density in 6 µg protein from the human
normal kidney immortalized cell line which displays telomerase
activity (HEK-293 cell line) was arbitrarily defined as 100 U
(16). In order to compare the ODs of
telomerase activity from each tissue with the same baseline, TRAP
assays were performed using the same amount of tissue extracts (6
µg) in all 40 patients. The bands from each tissue were compared
with the control (HEK-293 cell line) and expressed in arbitrary
units (11,17,18).
Alternative splicing of hTERT
Total RNA was collected from samples using
TRIzol-Reagent (Thermo Fisher Scientific, Inc.). First-strand cDNA
synthesis was performed according to the manufacturer's protocol.
Total RNA (1 µg), oligo(dT) primer, 10 mM dNTPs and M-MuLV reverse
transcriptase (Fermentas; Thermo Fisher Scientific, Inc.) was used
to synthesize cDNA for each sample. In order to evaluate hTERT mRNA
splicing, the present study performed reverse transcription-PCR
using primer sets for the reverse transcriptase domain of the hTERT
transcript (19). The cDNA samples
were amplified in a 5 µl reaction mixture supplemented with 0.25
uCi (α-32P) dCTP (GE Healthcare Life Sciences) and 0.2
µM of each primer. The first hTERT cDNA amplification used
TERT-1784S, 5′-CGGAAGAGTGTCTGGAGCAA-3′ and TERT-1928A,
5′-GGATGAAGCGGAGTCTGGA-3′ oligonucleotides with an initial heating
at 94°C for 90 sec, followed by 33 cycles of 95°C for 20 sec, 68°C
for 40 sec and 72°C for 30 sec. Primers 774 (forward;
5′-GGGAATTCAAAACTGGAACGGTGAAGG-3′) and 775 (reverse;
5′-GGAAGCTTATCAAAGTCCTCGGCCACA-3′) were added at 72°C during cycle
13 as a β-actin internal control. The second hTERT cDNA
amplification used TERT-2164S sense, 5′-GCCTGAGCTGTACTTTGTCAA-3′
and TERT-2620A anti-sense, 5′-CGCAAACAGCTTGTTCTCCATGTC-3′
oligonucleotides (6) with an initial
heating at 94°C for 90 sec, followed by 35 cycles of 95°C for 25
sec, 68°C for 50 sec and 72°C for 50 sec. Primers 774 and 775 were
added at 72°C during cycle 15 as a β-actin internal control. Bands
from each tissue were compared with the control and expressed in
arbitrary units. The HL-60 cell line (American Tissue Type Culture
Collection, Manassas, VA, USA) was used as a positive control for
the expression level of alternatively spliced variants.
Statistical analysis
Clinical data were retrieved from the medical
records of Severance Hospital (Seoul, Korea). The χ2
test or Mann-Whitney U tests were used for categorical variables
and the Student's t-test was used to compare continuous variables.
Progression-free survival (PFS) was defined as the time interval
between the date of surgery and disease progression or mortality
from any cause. Overall survival (OS) was evaluated from the date
of surgery to mortality or last contact (censored observation).
Survival curves were estimated using the Kaplan-Meier method and
differences in survival curves among groups were compared using the
log-rank test. Univariate associations between prognostic factors
and OS and between prognostic factors and PFS were assessed using
the log-rank test. In order to adjust for confounding variables,
Cox proportional hazards models were used to estimate the
simultaneous effects of prognostic factors on survival. All tests
were two-sided and P<0.05 was considered to indicate a
statistically significant difference. All analyses were performed
using SPSS for Windows (version 13.0; SPSS, Inc., Chicago, IL,
USA).
Results
Patient characteristics
Of the 40 patients enrolled in the present study,
the median follow-up duration was 69.5 months (range, 7.4–76.3
months). During the follow-up period, 12 (30.0%) of patients
relapsed and 9 (22.5%) succumbed to colorectal cancer. The
five-year DFS and OS rates were 76.3% and 81.6%, respectively.
Other clinical characteristics are summarized in Table I.
Expression levels of telomerase
activity and hTERT
Of the 40 tumor tissues, 32 (80%) expressed
telomerase activity and 32 (80%) expressed hTERT. Of the 32
patients with telomerase activity, 26 (81%) expressed hTERT. In the
8 patients without telomerase activity, 6 (75%) expressed hTERT. A
total of 2 patients did not express telomerase activity or hTERT
(Table II).
 | Table II.Comparison of telomerase activity and
hTERT expression levels in colon cancer tissues (n=40). |
Table II.
Comparison of telomerase activity and
hTERT expression levels in colon cancer tissues (n=40).
| Telomerase
activity | hTERT
expression | Patient no.
(%) |
|---|
| Positive | Expressed | 26 (65%) |
| Positive | Non-expressed | 6
(15%) |
| Negative | Expressed | 6
(15%) |
| Negative | Non-expressed | 2 (5%) |
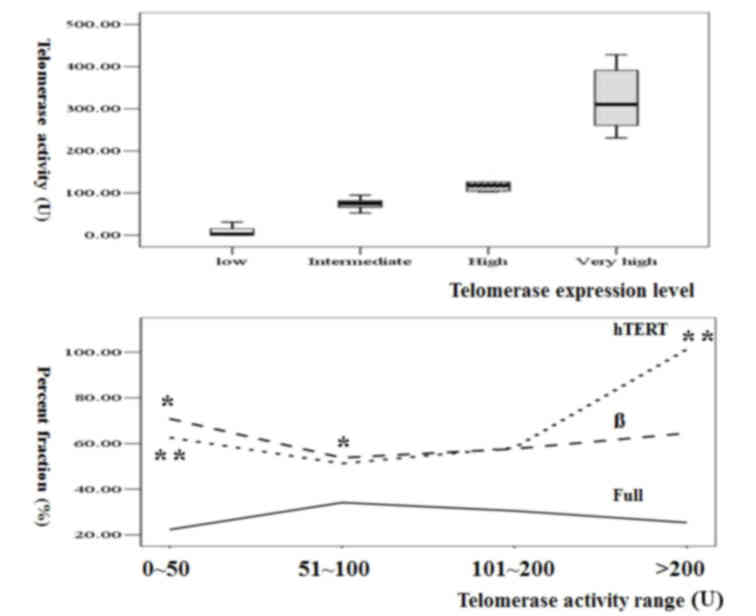
In the lower range of telomerase activity (0–100 U),
the percentage of β variant decreased with the increment in
telomerase activity (P=0.048), whereas hTERT did not significantly
differ. In the higher range of telomerase activity (≥100 U), total
hTERT level revealed a trend toward increment (P=0.06) without any
difference in the percentage of each spliced type (Table III; Fig.
1). These results suggested that a change of β variant
percentage was the main mechanism during the early phase and that
hTERT increment was the main mechanism of the late phase of
telomerase activity increment in colorectal cancer.
 | Table III.Comparison of telomerase activity and
percent fraction of hTERT variants. |
Table III.
Comparison of telomerase activity and
percent fraction of hTERT variants.
| Telomerase activity
range and mean ± SD (U) | Full (%) | α (%) | β (%) | αβ (%) | hTERT (U) |
|---|
| 0–50 (8±11) |
22±13 | 2±2 |
71±18a |
5±4 |
63±47b |
| 51–100 (75±14) |
34±21 | 4±4 |
53±23a |
9±7 | 51±79 |
| 101–200
(122±21) | 31±6 | 3±3 | 58±7 | 10±8 | 58±36 |
| >200
(323±76) |
25±15 | 4±3 |
65±19 |
6±5 | 101±55b |
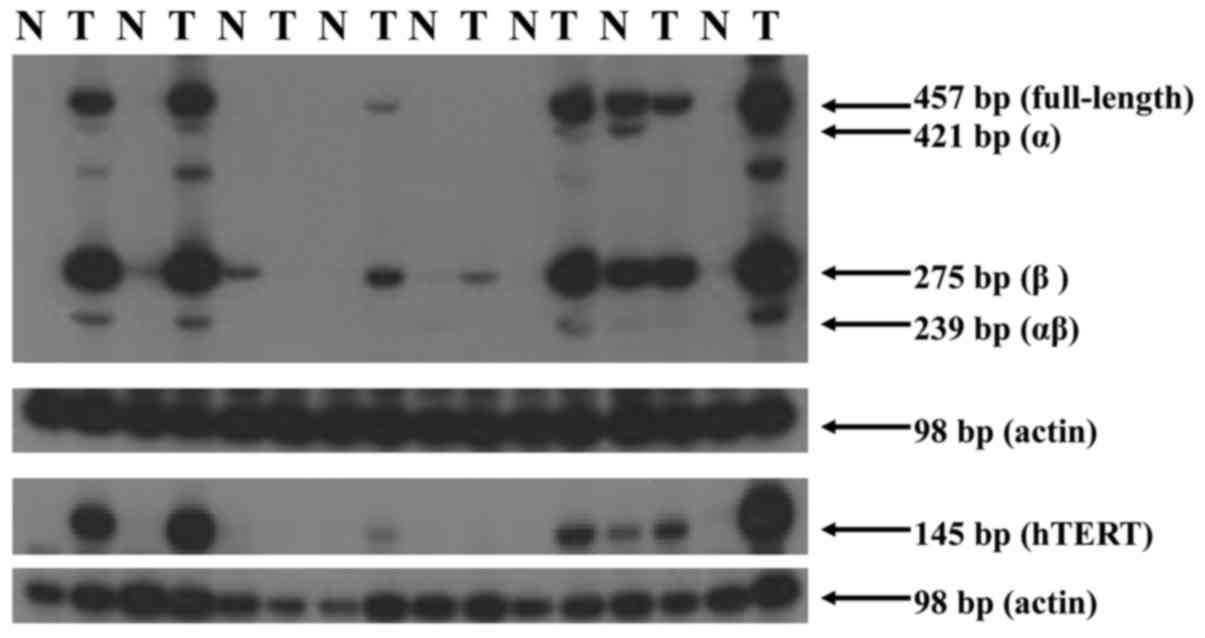
Expression levels of hTERT splicing
variants
Among the 30 paired normal tissues, 1 patient (3.3%)
expressed a low level of telomerase activity (36 U) and 1 patient
expressed hTERT (20 U). A total of 4 patients (13.3%) expressed
alternatively spliced variants [1 full-length (87 U), α (25 U), β
(120 U) variants; 1 full-length (1 U) and β (1 U) variants; 2 β
variant alone (12, 21 U)]. In cancer tissues with telomerase
activity, 17 (53%) of patients expressed all 4 types of variants
(full length, α, β and α/β). A total of 7 patients (22%) expressed
full-length, β and α/β variants. A total of 3 (9%) patients
expressed full length and β variants (Fig. 2). A total of 3 patients expressed none
of the hTERT types, even with telomerase activity expression. Of
the 8 patients without telomerase activity, 2 did not express any
hTERT splicing type. A total of 2 patients expressed all 4 types
(Table IV).
 | Table IV.Expression level of hTERT splicing
variants based on telomerase activity. |
Table IV.
Expression level of hTERT splicing
variants based on telomerase activity.
| Group | Patient number | Telomerase activity
(U) | hTERT expression
level (U) |
|---|
| Telomerase activity
(+) (n=32) |
|
Full/α/β/αβ | 17 | 146±130 | 92±62a,b |
|
Full/β/αβ | 7 | 111±133 | 55±45a |
|
Full/β | 3 | 144±149 | 13±14b |
|
Full/α/β | 1 | 95 | 1 |
| β | 1 | 21 | 0 |
|
None | 3 | 210±336 | 0 |
| Telomerase activity
(−) (n=8) |
|
Full/α/β/αβ | 2 |
|
|
|
Full/α/αβ | 2 |
|
|
|
β/αβ | 1 |
|
|
| β | 1 |
|
|
|
None | 2 |
|
|
Comparison of telomerase activity
based on hTERT splicing variant expression level
There was no difference in telomerase activity in
each group when telomerase activity based on the expression levels
of splicing types was compared; however, hTERT expression levels
were lower in 2 groups (full-length, β and α/β; P=0.09; and
full-length, β; P<0.0008) compared with the group with all 4
variant types expressed (Table
IV).
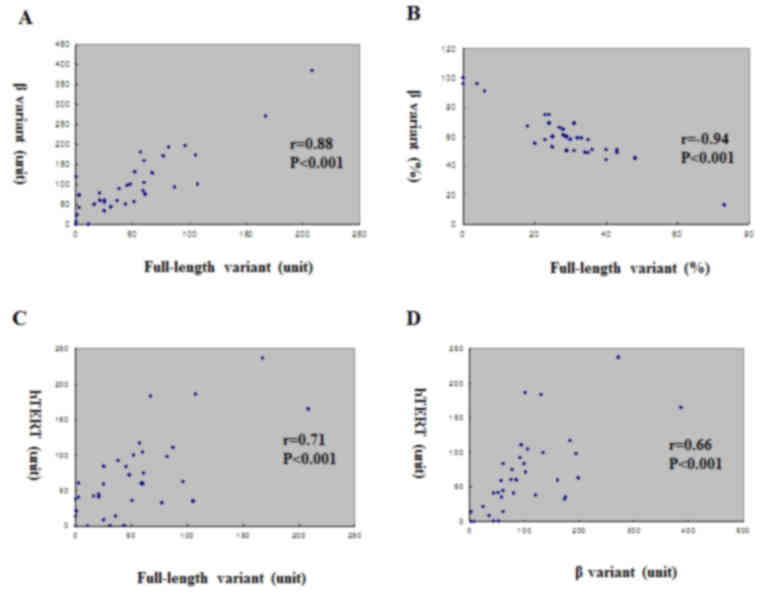
There was no correlation between telomerase activity
and expression level of each variant. However, among the variants,
there was a positive correlation between full-length variant level
and β variant level (r=0.88, P<0.001; Fig. 3). Conversely, there was a negative
correlation in the percentage between the full-length variant and β
variant (r=−0.939, P<0.001; Fig.
3). Expression fractions of full-length and β variants were
positively correlated with hTERT expression level (r=0.71,
P<0.001 and r=0.66, P<0.001, respectively; Fig. 3). Even if total levels of full length
and β variants increased with the increment of hTERT expression
level, the main factor for the hTERT increment was full-length
variant increment, which resulted in a negative correlation between
hTERT level and percent fraction of β variant. This finding
suggested that full-length variant was the main subtype of splicing
during the increment of hTERT in colorectal cancer.
Comparison of clinical parameters with
splicing type pattern
The present study compared the hazard ratio for
survival with clinical factors [age, sex, tumor-node-metastasis
(TNM) stage (20)] and molecular
markers (telomerase activity, percent fraction of alternative
splicing types, hTERT). TNM stage was demonstrated to be the
strongest prognostic factor by multivariate analysis, and percent
fraction of the full-length variant was an independent prognostic
factor for survival (Table V).
 | Table V.Multivariate analysis of hazard ratio
for overall survival in patients with colorectal cancer. |
Table V.
Multivariate analysis of hazard ratio
for overall survival in patients with colorectal cancer.
| Variable | P-value | Hazard ratio | 95% CI |
|---|
| Age | 0.231 |
0.757 | 0.480–1.194 |
| Sex | 0.525 |
5.471 |
0.029–1033.948 |
| Telomerase
activity | 0.596 |
0.994 | 0.973–1.016 |
| α/β ratio (%) | 0.215 |
1.209 | 0.895–1.633 |
| TNM stage |
0.027a | 24.125 |
1.429–407.290 |
| Full length
(%) |
0.037b |
1.294 | 1.016–1.647 |
| hTERT | 0.903 |
0.998 | 0.968–1.029 |
Discussion
The discrepancy between telomerase activity and the
hTERT mRNA expression levels may be due to the difficulty of
quantification of either telomerase activity or hTERT expression
level, constituents of intratumoral lymphocytes or leukocytes or
the presence of heterogeneity in tumor tissues. Finally,
post-transcriptional modification of telomerase activity by
alternative splicing may result in truncated and potentially
dysfunctional protein products.
The association of telomerase activity with the
presence of the full-length variant was significant (3). However, the presence of a full-length
hTERT variant was not sufficient to allow telomerase activity when
an abundance of hTERT-spliced variants were concomitantly present
(11). In melanoma, the β-spliced
form was a negative regulator of telomerase activity (21). In the present study, 4 (12.5%) of the
32 patients with telomerase activity lacked the full length
variant, and of the 8 patients without detectable telomerase
activity, 4 (50%) lacked the full-length variant and 4 (50%) lacked
the β variant. These cases suggested the presence of an additional
regulatory mechanism, for example post-translational modification
by phosphorylation or truncation outside the reverse transcriptase
motif of hTERT.
Various splice variants, depending on the tissue
type, continue to be synthesized when the full-length variant is
not (22). Two potential mechanisms
underlying the decrement of telomerase activity with
differentiation have been previously suggested (22,23). These
are posttranslational alterations in hTERT with stable total
transcription and splicing patterns, and minimal change in total
transcription with a decrement of the full-length variant
predominantly remaining in β-spliced form (12). In a previous in vitro
transcription and translation study, the β-splice variant was
revealed to not reconstitute an active telomerase enzyme (3). It is possible that protein products of
α- or β-spliced transcripts are negative inhibitors of the
formation of active telomerase enzymes. In the patients
investigated in the present study, when there was a low increment
of telomerase activity, the initial decrement of the β variant
fraction was demonstrated without any specific increment of hTERT.
However, in patients with high telomerase activity, total hTERT
level was increased with stable maintenance of each variant
fraction. Various regulatory mechanisms may be involved based on
different levels of telomerase activity in colorectal cancer,
including shifting of β variant fractions and total increments of
hTERT.
In general, the band density of the full-length
variant was equal to or higher than that of alternatively spliced
variants. Transcriptional activation of hTERT is not necessarily
the rate-limiting step in the generation of functional telomerase.
The present study observed decreased hTERT levels without any
changes in telomerase activity in patients without the α-splice
variant compared with patients with all 4 variants. Although the α
variant did not completely abolish telomerase activity, the
resultant reduction in activity was sufficient to reverse the
immortal phenotype (9). Clones
expressing the α variant underwent apoptosis and clones expressing
the β variant or α/β variant revealed no sign of apoptosis and
continued to proliferate (12).
The hTERT level was positively correlated with
expression levels of the full-length variant and the β variant in
the patients included in the present study. With the increment of
total hTERT, the full-length variant and β variant expression
levels increased with positive correlations. However, when the
percent fraction of each variant and hTERT expression level were
compared, the full-length variant was positively correlated and the
β variant was negatively correlated. Therefore, in patients with
hTERT increment, the increment rate of the full-length variant was
higher than the increment rate of the β variant. As a result, even
if the total amount of the β variant was increased, the total
fraction was relatively low compared with the fraction of the
full-length variant. These relative changes in the fraction and
total amount of each spliced variant may be the regulatory
mechanism underlying hTERT, and thus telomerase activity in
colorectal cancer. Of the transcriptional regulators of hTERT,
nuclear factor of activated T cells (24), SET and MYND domain-containing protein
3, a histone methyltransferase (25),
Mutant-S homolog 2, heterogeneous nuclear ribonuclear protein
(hnRNP) D, hnRNP K and grainyhead-like 2 (26) have been observed in cancer cells, and
mad1 was identified in myelodysplastic syndrome (27), which may be targets for
anti-telomerase or anti-hTERT treatment in cancer.
The cyclopentenone prostaglandin 15-deoxy-delta
12,14-prostaglandin J2 demonstrated anti-neoplastic activity by
decreasing hTERT expression level and suppressing c-Myc mRNA
expression levels (28). A low dose
of chelidonine reduced telomerase activity via downregulation of
hTERT expression, suggesting that hTERT may be a potential target
molecule in cancer (29). In the
patients enrolled in the present study, there was no significant
difference in clinical outcome between patients with
telomerase-positive and negative colorectal cancer. However, the
percent fraction of the full-length variant was a prognostic
factor, as was American Joint Committee on Cancer (TNM) stage. When
developing an anti-telomerase drug treatment, 80% of patients with
colorectal cancer may be a potential target population, using hTERT
variant as a patient selection biomarker (30).
In conclusion, telomerase activity changes and
alternative splicing of full-length and β variants of hTERT in
colorectal cancer were demonstrated. Clinical trials with an
anti-telomerase compound using alternative splicing variants as
patient selection biomarkers are warranted for the validation of
the clinical significance of hTERT variants in colorectal
cancer.
Acknowledgements
The present study was supported by the Korea Health
21 R&D Project, Ministry of Health & Welfare, Republic of
Korea (grant no. 0405-BC01-0604-0002).
References
|
1
|
Liu K, Schoonmaker MM, Levine BL, June CH,
Hodes RJ and Weng NP: Constitutive and regulated expression of
telomerase reverse transcriptase (hTERT) in human lymphocytes. Proc
Natl Acad Sci USA. 96:pp. 5147–5152. 1999; View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramakrishnan S, Eppenberger U, Mueller H,
Shinkai Y and Narayanan R: Expression profile of the putative
catalytic subunit of the telomerase gene. Cancer Res. 58:622–625.
1998.PubMed/NCBI
|
|
3
|
Krams M, Claviez A, Heidorn K, Krupp G,
Parwaresch R, Harms D and Rudolph P: Regulation of telomerase
activity by alternate splicing of human telomerase reverse
transcriptase mRNA in a subset of neuroblastomas. Am J Pathol.
159:1925–1932. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Villa R, Porta CD, Folini M, Daidone MG
and Zaffaroni N: Possible regulation of telomerase activity by
transcription and alternatively splicing of telomerase reverse
transcriptase in human melanoma. J Invest Dermatol. 116:867–873.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Killian A, Bowtell DD, Abud HE, Hime GR,
Venter DJ, Keese PK, Duncan EL, Reddel RR and Jefferson RA:
Isolation of a candidate human telomerase catalytic subunit gene,
which reveals complex splicing patterns in different cell types.
Hum Mo Genet. 6:2011–2019. 1997. View Article : Google Scholar
|
|
6
|
Ulaner GA, Hu JF, Vu TH, Giudice LC and
Hoffman AR: Telomerase activity in human development is regulated
by human telomerase reverse transcriptase (hTERT) transcription and
by alternate splicing of hTERT transcripts. Cancer Res. 58:1–4172.
1998.PubMed/NCBI
|
|
7
|
Ulaner GA, Hu JF, Vu TH, Giudice LC and
Hoffman AR: Tissue-specific alternate splicing of human telomerase
reverse transcriptase (hTERT) influences telomere lengths during
human development. Int J Cancer. 91:644–649. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brenner CA, Wolny YM, Adler RR and Cohen
J: Alternative splicing of the telomerase catalytic subunit in
human oocytes and embryos. Mol Human Repro. 5:845–850. 1999.
View Article : Google Scholar
|
|
9
|
Colgin LM, Wilkinso C, Englezou A, Kilian
A, Robinson MO and Reddel RR: The hTERTalpha splice variant is a
dominant negative inhibitor of telomerase activity. Neoplasia.
2:426–432. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weinrich SL, Pruzan R, Ma L, Ouellette M,
Tesmer VM, Holt SE, Bodnar AG, Lichtsteiner S, Kim NW, Trager JB,
et al: Reconstitution of human telomerase with the template RNA
component hTR and the catalytic protein subunit hTRT. Nat Genet.
17:498–502. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rha SY, Jeung HC, Yang WI, Kim JJ, Oh TJ,
An SW and Chung HC: Alteration of hTERT full-length variant
expression level showed different gene expression profiles and
genomic copy number changes in breast cancer. Oncol Rep.
15:749–755. 2006.PubMed/NCBI
|
|
12
|
Yi X, Shay JW and Wright WE: Quantitation
of telomerase components and hTERT mRNA splicing patterns in
immortal human cells. Nucleic Acid Res. 29:4818–4825. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hara T, Noma T, Yamashiro Y, Naito K and
Nakazawa A: Quantitative analysis of telomerae activity and
telomerase reverse transcriptase expression in renal cell
carcinoma. Urol Res. 29:1–6. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zaffaroni N, Porta C Della, Villa R, Botti
C, Buglioni S, Mottolese M and Daidone M Grazia: Transcription and
alternative splicing of telomerase reverse transcriptase in benign
and malignant breast tumours and in adjacent mammary glandular
tissues: Implications for telomerase activity. J Pathol. 198:37–46.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yokoyama Y, Wan X, Takahashi Y, Shinohara
A and Tamaya T: Alternatively spliced variant deleting exon 7 and 8
of the human telomerase reverse transcriptase gene is dominantly
expressed in the uterus. Mol Hum Repro. 7:853–857. 2001. View Article : Google Scholar
|
|
16
|
Wright WE, Shay JW and Piatyszek MA:
Modifications of a telomeric repeat amplification protocol (TRAP)
result in increased reliability, linearity and sensitivity. Nucleic
Acid Res. 23:3794–3795. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park KH, Rha SY, Kim CH, Kim TS, Yoo NC,
Kim JH, Roh JK, Noh SH, Min JS, Lee KS, et al: Telomerase activity
and telomere lengths in various cell lines: Changes of telomerase
activity can be another method for chemosensitivity evaluation. Int
J Oncol. 13:489–495. 1998.PubMed/NCBI
|
|
18
|
Rha SY, Park KH, Kim TS, Yoo NC, Yang WI,
Roh JK, Min JS, Lee KS, Kim BS, Choi JH, et al: Changes of
telomerase and telomere lengths in paired normal and cancer tissues
of breast. Int J Oncol. 15:839–845. 1999.PubMed/NCBI
|
|
19
|
Yeo M, Rha SY, Jeung HC, Hu SX, Yang SH,
Kim YS, An SW and Chung HC: Attenuation of telomerase activity by
hammerhead ribozyme targeting human telomerase RNA induces growth
retardation and apoptosis in human breast tumor cells. Int J
Cancer. 114:484–489. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumors. 7th. New York: Wiley;
pp. 200921
|
|
21
|
Lincz LF, Mudge LM, Scorgie FE, Sakoff JA,
Hamilton CS and Seldon M: Quantitation of hTERT splice variants in
melanoma by SYBR green real-time polymerase chain reaction
indicates a negative regulatory role for the beta deletion variant.
Neoplasia. 10:1131–1137. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jalink M, Ge Z, Liu C, Björkholm M, Gruber
A and Xu D: Human normal T lymphocutes and lymphoid cell lines do
express alternative splicing variants of human telomerase reverse
transcriptase (hTERT) mRNA. Biochem Biophys Res Commun.
353:999–1003. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shervington A and Patel A: Differential
hTERT mRNA processing between young and older glioma patients. FEBS
Lett. 582:1707–1710. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chebel A, Rouault JP, Urbanowicz I,
Baseggio L, Chien WW, Salles G and Ffrench W: Transcriptional
activation of HTERT, the human telomerase reverse transcriptase, by
nuclear factor of activated T cells. J Biol Chem. 284:35725–35734.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu C, Fang X, Ge Z, Jalink M, Kyo S,
Björkholm M, Gruber A, Sjöberg J and Xu D: The telomerase reverse
transcriptase (hTERT) gene is a direct target of the histone
methyltransferase SMYD3. Cancer Res. 67:2626–2631. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kang X, Chen W, Kim RH, Kang MK and Park
NH: Regulation of the hTERT promoter activity by MSH2, the hnRNPs K
and D, and GRHL2 in human oral squamous cell carcinoma cells.
Oncogene. 28:565–574. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Briatore F, Barrera G, Pizzimenti S,
Toaldo C, Casa CD, Laurora S, Pettazzoni P, Dianzani MU and Ferrero
D: Increase of telomerase activity and hTERT expression in
myelodysplastic syndromes. Cancer Biol and Ther. 8:883–889. 2009.
View Article : Google Scholar
|
|
28
|
Moriai M, Tsuji N, Kobayashi D,
Kuribayashi K and Watanabe N: Down-regulation of hTERT expression
plays an important role in 15-deoxy-Delta 12,14-prostaglandin
J2-indiced apoptosis in cancer cells. Int J Oncol. 34:1363–1372.
2009.PubMed/NCBI
|
|
29
|
Noureini SK and Wink M: Transcriptional
down regulation of hTERT and senescence induction in HepG2 cells by
chelidonine. World J Gastroenterol. 15:3603–3610. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu XH, Zhang JS, Zhang N and Zhang YD:
Combination of telomerase antisense oligonucleotides simultaneously
targeting hTR and hTERT produces synergism of inhibition of
telomerase activity and growth in human colon cancer cell line.
World J Gastroenterol. 11:785–790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oken MM, Creech RH, Tormey DC, Horton J,
Davis TE, McFadden ET and Carbone PP: Toxicity and response
criteria of the Eastern Cooperative Oncology Group. Am J Clin
Oncol. 5:649–655. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Astler VB and Coller FA: The prognostic
significance of direct extension of carcinoma of the colon and
rectum. Ann Surg. 139:846–852. 2003. View Article : Google Scholar
|