Introduction
Inhibitor of growth (ING) is a family of tumor
suppressor genes, and the well-known members of this family are
inhibitor of growth family member 1 (ING1), ING2, ING3, ING4 and
ING5 (1,2). The ING4 gene is localized at chromosome
12p13, consists of 8 exons and encodes a 29-kDa nuclear protein;
its expression is ubiquitous in multiple different human tissues
(3–5).
The biological functions of ING4 have been extensively
investigated, with a previous study demonstrating that ING4 protein
promoted tumor protein p53 activity through direct interaction with
it (6). As a consequence of the
interaction between ING4 and p53, tumor protein p21 expression was
upregulated, leading to cell cycle arrest (7,8). In
addition, ING4 altered apoptosis, contact inhibition and DNA repair
(9,10). ING4 has been considered as an
important tumor suppressor gene, whose expression was significantly
downregulated in a number of malignant tumors, including breast
cancer, glioma and lung cancer (8,11,12). However, the expression of ING4 has not
been investigated in renal cell carcinoma. Approximately 15 million
people worldwide are diagnosed with renal cell carcinoma annually
(13), of which 75% of cases are
clear cell renal carcinoma (CCRC) (14). The molecular mechanism of renal cell
carcinoma has been extensively investigated, but there is no
targeted therapy owing to a lack of targets. As ING4 was
downregulated, or even mutated, in multiple cancer types (8,15,16), In the present study, ING4 was inferred
to be associated with multiple cancer types, potentially making it
an ideal target for cancer therapy. In the present study, the level
of ING4 mRNA and protein was probed in renal cancer tissue
specimens in 125 patients with CCRC, and the adjacent normal tissue
from 40 patients were used as a control.
Materials and methods
Patients and tissue specimens
Tumor tissue specimens were harvested from 125
patients with CCRC who underwent nephrectomy between October 2007
and September 2009 in the Department of Urology Surgery, The First,
Second, and Fourth Affiliated Hospitals of Harbin Medical
University (Heilongjiang, China). The 125 patients consisted of 79
males and 46 females with a median age of 52.5 years (range, 31–74
years). The average tumor diameter was 7.1 (range, 2.9–11.3 cm); 40
specimens of adjacent normal tissue were collected along with
resection of the kidney to serve as controls. Neoplasm staging was
performed according to the tumor-node-metastasis (TNM)
classification of renal cell carcinoma (17). A total of 32 patients had stage I
disease, 39 had stage II, 49 had stage III and 5 had stage IV. The
nuclear grade of CCRC was as follows: 23 cases at grade I, 51 cases
at grade II, 42 cases at grade III and 9 cases at grade IV. Tissue
samples were divided into two types; one was stored at −80°C
following snap-freezing in liquid nitrogen and the other was fixed
in 4% neutral formalin and embedded in paraffin. Informed written
consent was obtained from the patients prior to tissue sample
collection. The research protocol was approved by the Ethics
Committee of Harbin Medical University.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
cDNA was reverse-transcribed from mRNA using the Reverse
Transcription kit (Takara Biotechnology Co., Ltd., Dalian, China).
cDNA samples were stored at −20°C prior to use. A One-Step
SYBR-Green I-based qPCR kit (PerkinElmer, Inc., Waltham, MA, USA)
was used according to the manufacturer's protocol to detect ING4
mRNA levels in 40 CCRC tissue samples and corresponding normal
renal tissues. The primer pairs for qPCR were as follows: ING4
forward, 5′-TCGTGCTCGTTCCAAAGG-3′ and ING4 reverse,
5′-GGCAATAGGTGGGTTCGTT-3′; β-actin forward,
5′-CCCAGCACAATGAAGATCAAGATCAT-3′ and β-actin reverse,
5′-ATCTGCTGGAAGGTGGACAGCGA-3′. The PCR program was as follows:
Initial denaturation at 95°C for 10 sec, followed by 40 cycles of
95°C for 5 sec and 53°C for 30 sec. There were three replicates of
each PCR reaction. Results were quantified using the
2ΔΔCq method (18)
following normalization to β-actin.
Western blotting
Western blotting was performed to detect the protein
level of ING4 in the renal tissues obtained from the 40 patients.
Tissue samples of ~50 mg were minced and ground in liquid nitrogen.
The proteins were extracted using a Protein Extraction kit (Pierce;
Thermo Fisher Scientific, Inc.), and the protein concentration was
determined using the Bradford assay. A total of 100 µg protein was
separated by SDS-PAGE and transferred onto nitrocellulose
membranes. The membranes were blocked with 5% skimmed milk powder
in PBS and subsequently incubated with rabbit anti-human ING4
polyclonal antibody (1:300; cat no. 40-7700; Invitrogen; Thermo
Fisher Scientific, Inc.). Following washing, the membranes were
incubated with horseradish peroxidase-conjugated goat anti-rabbit
antibody (1:10,000; cat no. ZB-5301; Beijing Zhongshan
Biotechnology Jinqiao Co., Ltd., Beijing, China). An enhanced
chemiluminescence detection system (Shanghai Jiapeng Technology
Co., Ltd., Shanghai, China) was used to visualize the protein
bands. β-actin (1:1,000; cat no. ab8226; Abcam, Cambridge, MA, USA)
was used as an internal control. All experiments were repeated
three times independently, and the level of ING4 protein expression
was quantified using Scion imaging software 4.0 (Scion Corporation,
Frederick, MD, USA).
Immunohistochemistry (IHC)
analysis
IHC analysis was performed using a two-step
immunohistochemical staining protocol as previously described
(19). Briefly, 4-µm-thick sections
were blocked with 5% bovine serum albumin in PBS prior to
incubation overnight at 4°C with rabbit anti-human ING4 polyclonal
antibody (1:100; cat no. 40-7700; Invitrogen; Thermo Fisher
Scientific, Inc.). Following three washes with PBS containing
Tween-20, the sections were then incubated with goat anti-rabbit
secondary antibody (1:100; cat no. ZB-5301; Beijing Zhongshan
Biotechnology Jinqiao Co., Ltd.) for 30 min at 37°C. The tissues
were visualized using 3,3′-diaminobenzidine tetrahydrochloride in
water. Sections were counterstained using hematoxylin and sealed
with neutral gum. Tissue sections incubated in PBS were used as
negative controls and normal lung tissue, which was ING4-positive,
was used as positive control. Cell membrane, cytoplasm and nucleus
that were positively stained appeared as yellow-brown granules. The
IHC results were quantified according to the number of positive
cells and expressed as follows: 0%, negative; <25%, +; 25–50%,
++; 51–75%, +++; >75%, ++++.
Statistical analysis
The densitometric analysis of western blotting and
PCR results was performed using ImageJ software version 1.48
(National Institutes of Health, Bethesda, MD, USA). Comparison was
performed using an unpaired Student's t-test. A non-parametric test
was used to analyze the results from IHC staining and Spearman's
rank correlation was used to analyze tissue positive for the
expression of ING4. P<0.05 was considered to indicate a
statistically significant difference.
Results
Decrease in ING4 mRNA in CCRC
tissues
Initially, the level of ING4 mRNA expression was
quantified in 40 paired tumor and adjacent normal tissues using
RT-qPCR, with the data indicating that ING4 mRNA was downregulated
in 100% (40/40) of CCRC tissues when compared with that in adjacent
normal renal tissues (0.4869±0.0448 in CCRC vs. 0.7303±0.0434 in
normal tissue, P<0.0001; Fig. 1).
The level of ING4 mRNA was positively associated with the nuclear
grade of renal cancer (rs=−0.94076; P<0.0001), the
clinical stage of CCRC (rs=−0.92400; P<0.0001) and
lymphatic metastasis (rs=−0.78291; P<0.0001);
however, no association with patient sex or age, or the size of
tumor was identified (P>0.05).
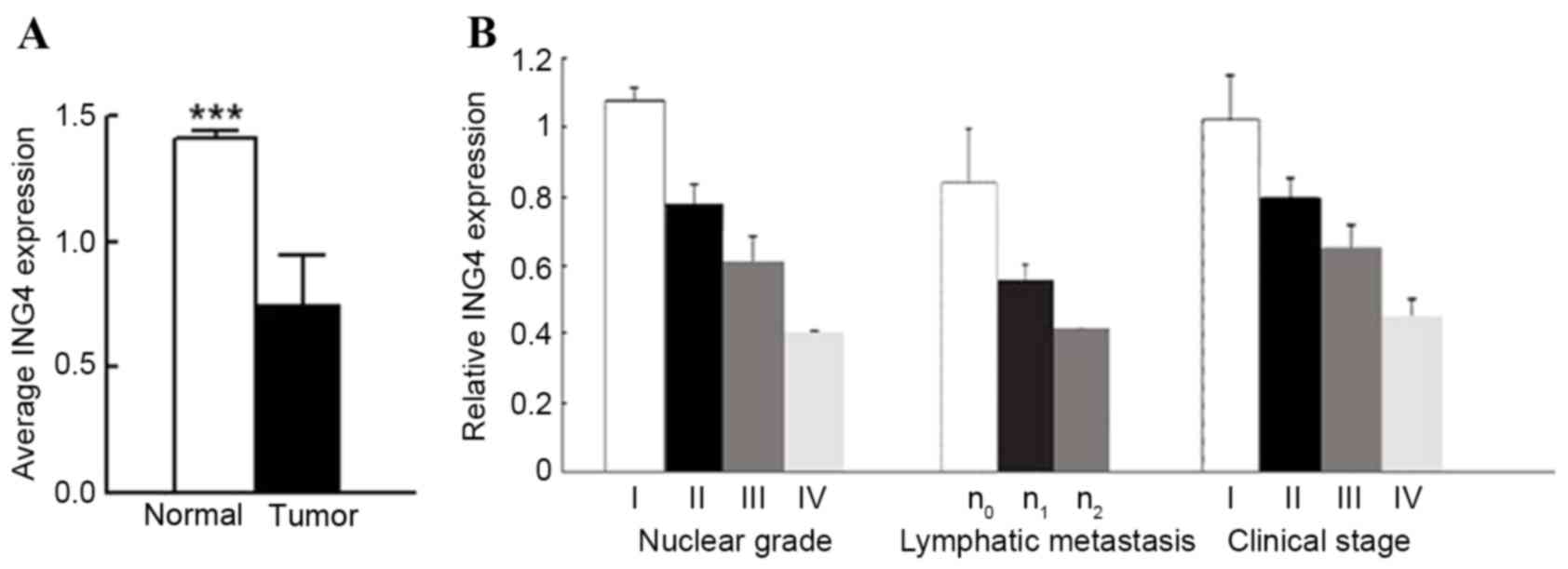 | Figure 1.ING4 mRNA is significantly decreased
in CCRC tissues when compared with normal renal tissues. (A) A
total of 40 paired tumor and adjacent normal renal tissues were
analyzed for ING4 mRNA level by reverse transcription-quantitative
polymerase chain reaction. β-actin was used as an internal control.
(B) Analysis of relative mRNA level (compared with normal renal
tissue) according to Fuhrman nuclear grade (24) (grade I, n=7; grade II, n=16; grade 3,
n=13; grade 4, n=4), lymphatic metastasis (n0, n=29;
n1, n=7; n2, n=4), tumor clinical stage
(stage I, n=9; stage II, n=12; stage III, n=14; stage IV, n=5).
***P<0.001 by t-test. CCRC, clear cell renal carcinoma; ING4,
inhibitor of growth family member 4. |
Decrease in ING4 protein expression in
CCRC tissues
ING4 protein expression was evaluated by western
blot analysis in the same 40 CCRC tissues and adjacent normal renal
tissues. The expression of ING4 protein level in CCRC tissues was
significantly decreased compared with in adjacent normal renal
tissues (0.4127±0.0723 vs. 0.7488±0.0572; P<0.0001; Fig. 2). The expression level of ING4 protein
correlated with the nuclear grade of renal cancer
(rs=−0.94655; P<0.0001), the clinical stage of CCRC
(rs=−0.90465, P<0.0001) and lymphatic metastasis
(rs=−0.60608; P<0.0001); however, no association
between this expression and sex, age or tumor volume was identified
(P>0.05).
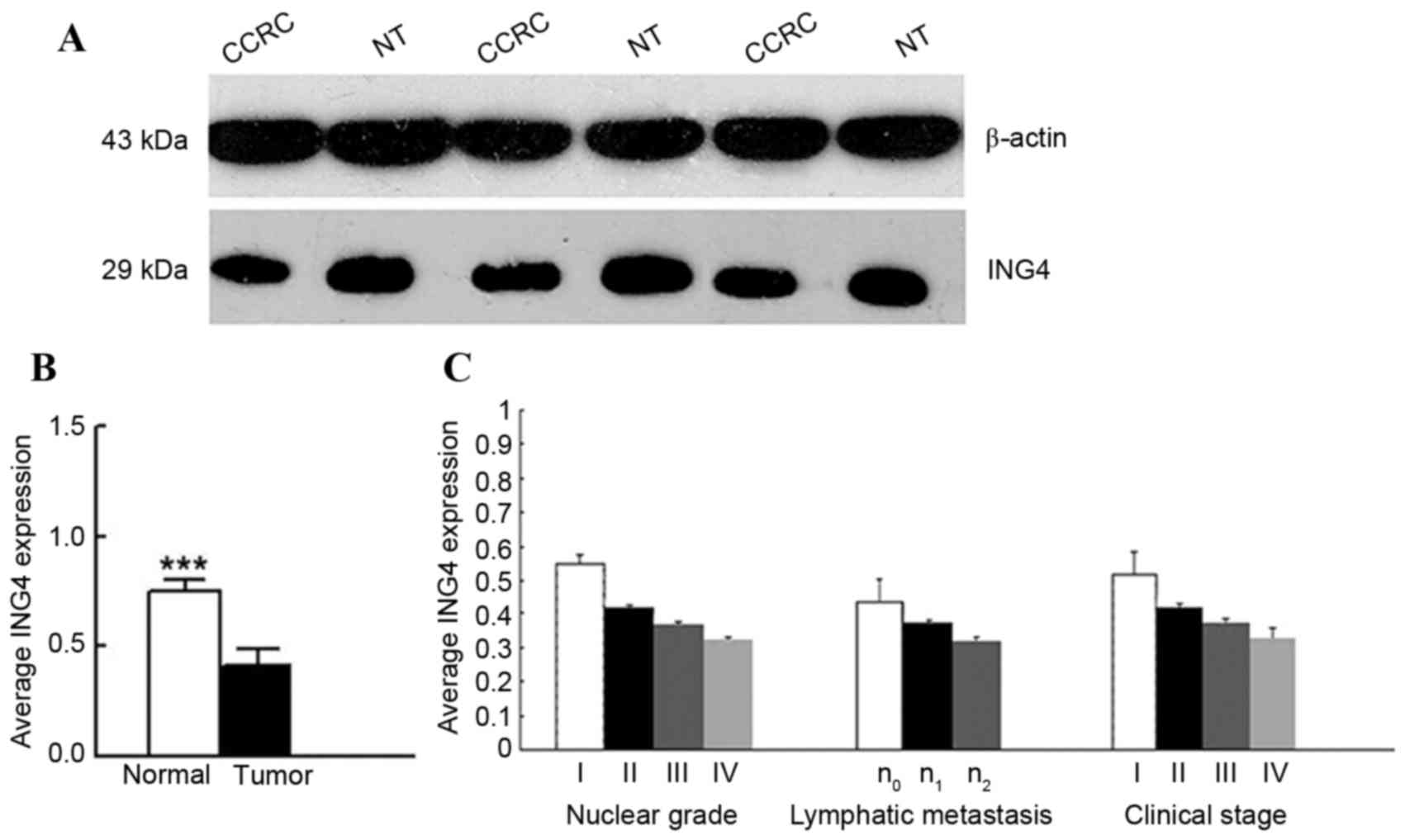 | Figure 2.ING4 expression is significantly
decreased in CCRC tissues when compared with normal renal tissues.
(A) A total of 40 paired tumor and adjacent normal renal tissues
were analyzed for ING4 expression using western blotting. Decreased
ING4 expression was observed in CCRC tissues compared with adjacent
normal renal tissues. β-actin was used as internal control. (B)
Average ING4 expression in 40 paired tumor and normal renal tissues
samples. (C) Analysis of the expression level of ING4 protein
(compared with normal renal tissue) according to tumor nuclear
grade (grade I, n=7; grade II, n=16; grade III, n=13; grade IV,
n=4), lymphatic metastasis (n0, n=29; n1,
n=7; n2, n=4), tumor clinical stage (stage I, n=9; stage
II, n=12; stage III, n=14; stage IV n=5). ***P<0.001 by t-test.
CCRC, clear cell renal carcinoma; ING4, inhibitor of growth family
member 4; NT, normal tissue. |
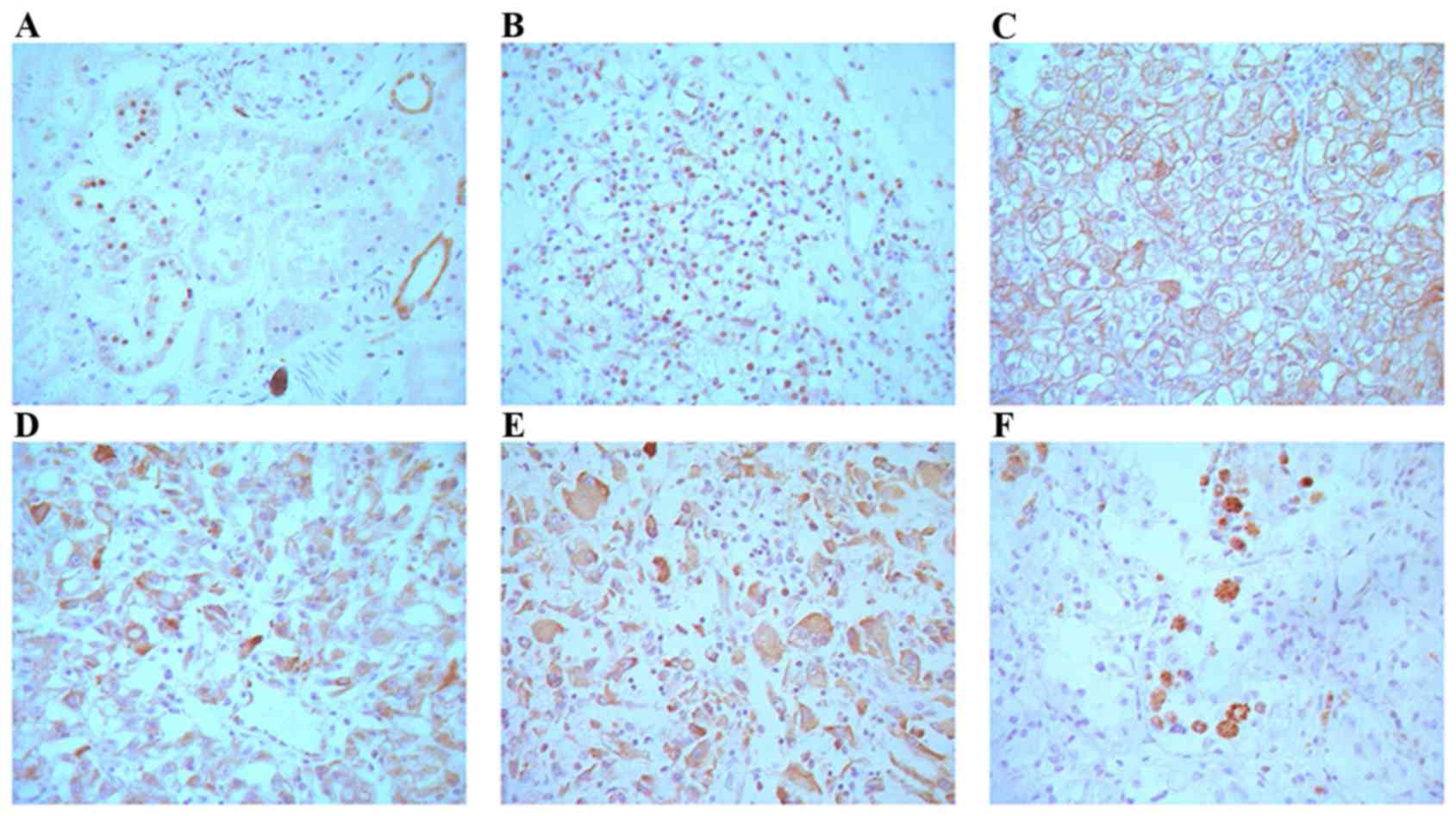
Localization of ING4 in normal and
CCRC tissue
All 40 normal renal tissue samples were positive for
ING4 (100%) in the renal tubular epithelial cell nuclei. ING4
protein expression was detected in all 125 CCRC samples. Of the 62
cases, ING4 was expressed in the nuclei of 11 cases, whereas in the
remaining 51 cases, ING4 was mainly expressed in both cell membrane
and cytoplasm (82.3%, 51/62 cases) (Fig.
3B-F). The spatial expression of ING4 and the nuclear grade of
CCRC is presented in Table I. The
results indicated that ING4 protein expression was inversely
associated with the nuclear grade of CCRC, and the rate of ING4
expression was increased with a low nuclear grade of CCRC
(P<0.0001). No association was identified between ING4
expression and the sex, age, tumor size and clinical stage of the
patients (P>0.05; Table II).
 | Table I.ING4 protein in different nuclear
grade renal cell carcinoma tissues. |
Table I.
ING4 protein in different nuclear
grade renal cell carcinoma tissues.
|
|
|
|
ING4-positivea |
|---|
|
|
|
|
|
|---|
| Nuclear grade | Total cases, n |
ING4-negativea | + | ++ | +++ |
|---|
| I | 34 | 7 | 6 | 10 | 11 |
| II | 39 | 15 | 9 | 7 | 8 |
| III | 29 | 21 | 6 | 1 | 1 |
| IV | 23 | 20 | 2 | 1 | 0 |
 | Table II.ING4 protein expression in patients
with clear cell renal carcinoma of the association between the
clinical and pathological features. |
Table II.
ING4 protein expression in patients
with clear cell renal carcinoma of the association between the
clinical and pathological features.
| Clinical
indicators | Patients, n | ING4-positive
patients, n | ING4-positive
expression, % | P-value |
|---|
| Sex |
|
|
|
|
| Male | 79 | 40 | 50.63 | 0.7621 |
|
Female | 46 | 22 | 47.83 |
|
| Age, years |
|
|
|
|
| ≤50 | 41 | 19 | 46.34 | 0.6107 |
|
>50 | 84 | 43 | 51.19 |
|
| Tumor size, cm |
|
|
|
|
| ≤7 | 51 | 23 | 45.09 | 0.0991 |
|
>7 | 74 | 39 | 52.70 |
|
| Staging grade |
|
|
|
|
| II | 27 | 12 | 44.44 | 0.8463 |
| II | 14 | 6 | 42.86 |
|
| III | 53 | 28 | 52.83 |
|
| IV | 31 | 16 | 51.61 |
|
Discussion
ING is a family of tumor suppressor genes that
includes ING1, ING2, ING3, ING4 and ING5. These genes participate
in a number of cellular events, including the cell cycle, apoptosis
and DNA repair. Overexpression of ING4 disrupted the cell cycle
distribution of the cells, decreasing the number of cells in S
phase (7). As a tumor suppressor
gene, ING4 expression can suppress the growth of gliomas, breast
tumors and squamous cell carcinomas of the head and neck (4,5); however,
its roles in CCRC remain unclear. In the present study, 125
clinical CCRC specimens underwent western blot analysis, RT-qPCR
and IHC to probe ING4 expression in CCRC. The results of these
analyses revealed that ING4 expression was significantly decreased
in CCRC tissues compared with in the normal renal tissues at the
mRNA and protein levels. ING4 was expressed in 100% of normal renal
tissue samples (40/40 cases), but its expression was detectable
only in 49.6% of CCRC samples (62/125). Garkavtsev et al
(3) identified that the expression of
ING4 mRNA in 50 glioma samples was significantly decreased compared
with that in 5 adjacent brain tissues. Previous studies indicated
that ING4 was downregulated in prostate cancer cell lines,
melanoma, and in cancer of the stomach, liver, breast and lung
(15,16,20–22) and in
76% of the head and neck squamous cell carcinoma samples assessed
(4). Taken together, the results of
the present study indicated that a decrease in ING4 expression was
associated with a variety of tumors, including CCRC.
Besides the dysregulation of ING4 expression,
deletions and missense mutations in ING4 transcripts were detected
in several tumor cell lines (4).
Hybridization-mediated deletion of the ING4 locus was identified in
between 10 and 20% of breast cancer cell lines and primary breast
tumors, and in 66% of head and neck squamous carcinomas (4,5). When
deletions and missense mutations occurred in ING4 transcripts, the
expression of ING4 decreased, leading to cell cycle
dysregulation.
The results of the present study demonstrated that
ING4 expression was not significantly associated with sex, age or
tumor volume (P>0.05); however, ING4 expression was identified
to be significantly associated with nuclear grade, clinical stage
and lymphatic metastasis. ING4 expression decreased as the nuclear
grade level increased (P=0.0038). These results were confirmed by
RT-qPCR, western blot analysis and IHC. Therefore, downregulation
of ING4 was associated with the progression and metastasis of CCRC.
ING4 may cooperate with other genes to inhibit tumor cell growth in
human renal cancer and may serve as a prognostic indicator for
renal cancer.
In normal renal tissue, ING4 is expressed in the
renal tubular epithelial cell nuclei. Of the 125 CCRC patient
samples, 62 were positive for ING4 expression. In 51/62
ING4-positive samples (82.3%), ING4 expression was observed outside
nuclei in sites including the cell membrane and cytoplasm. The
presence of membrane-bound ING4 has not yet been reported in other
malignancies, indicating that the genetic changes of ING4 in the
development of renal cell carcinoma may be distinct from those in
other types of tumor. The nuclear localization signal (NLS) domain
of ING4 is necessary for its interaction with p53 (19). Alternative splice variants of ING4,
ING4_V2, ING4_V3 and ING4_V4 localized to the cytoplasm and lacked
classical ING4 functions owing to a lack of NLS domain (23), suggesting that alternative splicing
events may be one of the reasons for the cytoplasmic localization
of ING4 in renal cancer.
Renal cell carcinoma does not typically respond to
chemotherapy or radiation therapy, but responds well to
immunotherapy (24). Therefore, it is
important to develop novel therapeutic approaches for the treatment
of CCRC. On the basis of the conclusions drawn from studies of
other tumors, ING4 may serve as a valuable marker for CCRC,
particularly the membrane-bound ING4 splicing variants, which may
be targets for future immunotherapy strategies.
In conclusion, the results of the present study
demonstrated that ING4 expression was downregulated in CCRC, and
its expression was identified for the first time, to the best of
our knowledge, to be associated with the advancement of clinical
stage and nuclear grade level. Considering its roles in other types
of tumor, ING4 may be a novel target for the diagnosis and therapy
of CCRC.
Acknowledgements
The present study was partly supported by the
Research Project of Heilongjiang Health Department (grant no.
2009-107), China.
Glossary
Abbreviations
Abbreviations:
|
ING
|
inhibitor of growth
|
|
ING4
|
inhibitor of growth family member
4
|
|
CCRC
|
clear cell renal carcinoma
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
NLS
|
nuclear localization signal
|
References
|
1
|
Garkavtsev I, Kazarov A, Gudkov A and
Riabowol K: Suppression of the novel growth inhibitor p33ING1
promotes neoplastic transformation. Nat Genetics. 14:415–420. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garkavtsev I and Riabowol K: Extension of
the replicative life span of human diploid fibroblasts by
inhibition of the p33ING1 candidate tumor suppressor. Mol Cell
Biol. 17:2014–2019. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garkavtsev I, Kozin SV, Chernova O, Xu L,
Winkler F, Brown E, Barnett GH and Jain RK: The candidate tumour
suppressor protein ING4 regulates brain tumour growth and
angiogenesis. Nature. 428:328–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gunduz M, Nagatsuka H, Demircan K, Gunduz
E, Cengiz B, Ouchida M, Tsujigiwa H, Yamachika E, Fukushima K,
Beder L, et al: Frequent deletion and down-regulation of ING4, a
candidate tumor suppressor gene at 12p13, in head and neck squamous
cell carcinomas. Gene. 356:109–117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim S, Chin K, Gray JW and Bishop JM: A
screen for genes that suppress loss of contact inhibition:
Identification of ING4 as a candidate tumor suppressor gene in
human cancer. Proc Natl Acad Sci USA. 101:pp. 16251–16256. 2004;
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo Y, Meng X, Wang Q, Wang Y and Shang H:
The ING4 binding with p53 and induced p53 acetylation were
attenuated by human papillomavirus 16 E6. PLoS One. 8:e714532013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Masayuki S, Makoto N, Pedeux RM,
Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y,
Appella E, Yokota J and Harris CC: p29ING4 and p28ING5 bind to p53
and p300, and enhance p53 activity. Cancer Res. 63:2373–2378.
2003.PubMed/NCBI
|
|
8
|
Liu E, Wu J, Cao W, Zhang J, Liu W, Jiang
X and Zhang X: Curcumin induces G2/M cell cycle arrest in a
p53-dependent manner and upregulates ING4 expression in human
glioma. J Neurooncol. 85:263–270. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Russell M, Berardi P, Wei G and Riabowol
K: Grow-ING, Age-ING and Die-ING: ING proteins link cancer,
senescence and apoptosis. Exp Cell Res. 312:951–961. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagashima M, Shiseki M, Pedeux RM, Okamura
S, Kitahama-Shiseki M, Miura K, Yokota J and Harris CC: A novel
PHD-finger motif protein, p47ING3, modulates p53-mediated
transcription, cell cycle control, and apoptosis. Oncogene.
22:343–350. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tapia C, Zlobec I, Schneider S, Kilic E,
Güth U, Bubendorf L and Kim S: Deletion of the inhibitor of growth
4 (ING4) tumor suppressor gene is prevalent in human epidermal
growth factor 2 (HER2)-positive breast cancer. Hum Pathol.
42:983–990. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang QS, Li M, Zhang LY, Jin Y, Tong DD,
Yu Y, Bai J, Huang Q, Liu FL, Liu A, et al: Down-regulation of ING4
is associated with initiation and progression of lung cancer.
Histopathology. 57:271–281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Organization WH, Ferlay J, Pisani P and
Parkin DM: Globocan 2000: Cancer incidence, mortality and
prevalence worldwide. Bray Freddie. 2001.
|
|
14
|
Zbar B, Klausner R and Linehan WM:
Studying cancer families to identify kidney cancer genes. Annu Rev
Med. 54:217–233. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang F, Luo LB, Tao YM, Wu F and Yang L:
Decreased expression of inhibitor of growth 4 correlated with poor
prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomarkers
Prev. 18:409–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cai L, Li X, Zheng S, Wang Y, Li H, Yang J
and Sun J: Inhibitor of growth 4 is involved in melanomagenesis and
induces growth suppression and apoptosis in melanoma cell line M14.
Melanoma Res. 19:1–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao W, Yang L, Wang L, Zuo W, Yuan S, Yu
J, Yu Q, Hu X, Wang S, Liu N, et al: Primary clear cell carcinoma
of nasal cavity: Report of six cases and review of literature. Int
J Clin Exp Med. 7:5469–5476. 2014.PubMed/NCBI
|
|
20
|
Zhang X, Wang KS, Wang ZQ, Xu LS, Wang QW,
Chen F, Wei DZ and Han ZG: Nuclear localization signal of ING4
plays a key role in its binding to p53. Biochem Biophys Res Commun.
331:1032–1038. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li M, Jin Y, Sun WJ, Yu Y, Bai J, Tong DD,
Qi JP, Du JR, Geng JS, Huang Q, et al: Reduced expression and novel
splice variants of ING4 in human gastric adenocarcinoma. J Pathol.
219:87–95. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yang HC, Sheng WH, Xie YF, Miao JC, Wei WX
and Yang JC: In vitro and in vivo inhibitory effect of Ad-ING4 gene
on proliferation of human prostate cancer PC-3 cells. Ai Zheng.
28:1149–1157. 2009.(In Chinese). PubMed/NCBI
|
|
23
|
Raho G, Miranda C, Tamborini E, Pierotti
MA and Greco A: Detection of novel mRNA splice variants of human
ING4 tumor suppressor gene. Oncogene. 26:5247–5257. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bleumer I, Oosterwijk E, De Mulder P and
Mulders PF: Immunotherapy for renal cell carcinoma. Eur Urol.
44:65–75. 2003. View Article : Google Scholar : PubMed/NCBI
|