Introduction
Lung cancer is the leading cause of global
cancer-related death. According to the recent statistics by
national cancer institute, there are approximately 224,390 new
cases reported annually and 158,080 will die from it (1). Although the morbidity due to lung cancer
has gradually been decreasing world-wide since the 1990s, it is
persistently rising in China (2,3). Non-small
cell lung cancer (NSCLC) accounts for 85–90% of all lung cancer
diagnoses (4). For the majority of
these patients, current treatments do not cure the disease, and the
prognosis remains poor (5). However,
results from lots of Phase III trials had been demonstrated that
better prognosis could still be achieved if molecular targeted
therapies (e.g., based on EGFR, ALK) rather than standard
chemotherapies were adopted (6–12).
Therefore, the discovery of new prognostic markers and potential
drug targets as well as a better understanding of molecular
mechanisms for lung tumorigenesis are very essential.
The tripartite motif (TRIM) family proteins,
comprising over 60 members, are evolutionarily conserved proteins
that share a RBCC motif, which consist of a common N-terminal
Really Interesting New Gene (RING) finger domain, one or two B-box
motifs and a coiled-coil region (13,14). These
proteins are involved in a plethora of cellular processes such as
cell proliferation, migration, apoptosis, cell cycle regulation,
differentiation and development (15–18).
Recently, several groups reported that TRIM proteins including
TRIM11, TRIM28, TRIM29, TRIM44 and others seemed to act as
oncogenes in lung cancer (19–22),
indicating significant roles of the TRIM family in lung
tumorigenesis. TRIM59, a novel TRIM family member, is characterized
by the presence of a RING finger domain, a B-box 2 domain, two
coiled-coil domains and a transmenbrane domain in its structure
(17), implicated in a wide range of
biological processes in lung cancer and other multiple tumors. It
may be used as a novel multiple tumor biomarker in
immunohistochemical detection for early tumorigenesis (23). Upregulation of the TRIM59 gene
promotes gastric carcinogenesis via facilitating the p53
ubiquitination and degradation (24),
and increases the proliferation, migration and invasion in human
osteosarcoma cells (25). Knockdown
of TRIM59 inhibits the cellular proliferation and migration in
cervical cancer (26). Additionally,
TRIM59 may act as a proto-oncogene that affects both the
Ras/Braf/MEK/ERK signaling pathway and the SV40 Tag/pRB/p53 pathway
in prostate cancer models (27).
Furthermore, a recent study of TRIM59 in lung cancer showed that it
promotes the proliferation and migration of NSCLC cells by
affecting the expression of cell cycle proteins (17). However, the molecular mechanisms of
this protein are still poorly defined, and to date, no reports have
evaluated the prognostic value of TRIM59 in lung cancer.
In this study, a screening approach was performed on
microarray datasets from Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo) to
explore possible targets in the TRIM family whose expressions were
significantly altered in lung cancer. Interestingly, TRIM59 was
screened out by this approach. Then, the clinical relevance, the
prognostic value and the functional mechanisms of TRIM59 in NSCLC
were further examined by using bioinformatics approaches in order
to elucidate the possibility of this protein being used as a
biomarker in NSCLC patients. Our results showed that TRIM59 was a
novel prognostic biomarker modulating oncogenic mammalian target of
rapamycin (MTOR) and eukaryotic initiation factor 4E (EIF4E)
signaling in NSCLC. It might be served as an independent predictor
for prognosis and a potential therapeutic target for NSCLC.
Materials and methods
The expression profile datasets
Gene expression datasets used for statistical
analysis were acquired from the National Center for Biotechnology
Information GEO database with the accession codes GSE19804
(28), GSE30219 (29), GSE31210 (30,31),
GSE32863 (32), GSE37745 (33) and GSE43580 (34). The TCGA data of lung cancer was
available on cBioPortal (www.cbioportal.org).
Screening of the TRIM family which was
overexpressed in NSCLC dataset
The screening was performed in GSE19804 which
contained both the lung tumor samples and the matched adjacent
normal lung samples. Two criteria were followed during the
selection of the probes for screening: i) the probe specificity
corresponding to a single gene of the TRIM family; ii) for multiple
probes corresponding to a same gene, one with the maximum
expression value in tumor samples would be chosen. The average
value of log2(Tumor/Normal) was calculated for each
selected probe and listed in the rank order. The identified
overexpressed genes had relative high log2(Tumor/Normal)
values in GSE19084 while being selected.
Gene set enrichment analysis
(GSEA)
GSEA was performed using the GSEA program provided
by the Broad Institute (http://www.broadinstitute.org/gsea/index.jsp). In all
datasets, the samples were divided into 2 groups according to their
TRIM59 expression levels (top 50%: High vs. bottom 50%: Low). GSEA
was carried out to compare the 2 groups within each indicated
geneset and to examine the relative enrichment of the genes in a
specific group. The genesets were downloaded from the Molecular
Signatures Database. Significantly genesets were confirmed with
nominal P-value <0.05 and false discovery rates (FDR) <0.25
after performing 1,000 permutations (35). Cytoscape and Enrichment Map were used
for visualization of the GSEA results.
Protein-protein interaction (PPI)
network construction
STRING 10.0 software (http://string-db.org/) is a web-based database for
providing comprehensive interactions information for the already
known or predicted proteins (36).
Complex cellular functions of TRIM59 are formed by tightly
interacted with other protein partners. To explore the interactions
between TRIM59 and some oncogenic proteins, we mapped them onto
STRING database, selected the interactions pertaining to Homo
sapiens and grew a PPI network with combined score >0.4.
Functional enrichments of the network were analyzed and displayed
on the webpage.
Statistical analysis
In this study, the analyses were performed using
GraphPad Prism 5.0 and SPSS 19.0 software. Unpaired comparisons
were assessed by a two-tailed t-test. Matched-sample comparisons
were performed by a paired t-test. Multigroup analyses were carried
out by ANOVA analysis. Associations between TRIM59 expression and
clinicopathologic parameters were assessed by χ2 test.
The Pearson correlation was used to analyze the strength of the
association between expression levels of TRIM59 and the survival or
recurrence statuses in NSCLC patients. Survival analysis was
performed by using Kaplan-Meier method and differences were tested
by a Log-rank test. It was also obtained using the Kaplan Meier
Plotter (K-M Plotter) website for lung cancer (v2015) (http://kmplot.com/analysis/index.php?p=service&cancer=lung)
(37) with auto select best cut-off
for splitting groups. Univariate and Multivariate Cox proportional
hazards regression models were performed to identify the
independent factors with a significant impact on patient survival.
The hazard ratios (HR) and 95% confidence intervals (95% CI) of the
prognostic factors were calculated. All P-values were two-sided,
and a significant difference was defined as P<0.05.
Results
Upregulation of TRIM59 expression in
NSCLC
Genes of the TRIM family that aberrantly expressed
in GSE19804 of NSCLC were firstly screened. Among the 59 probes
selected from GPL570 platform for study, 27 genes were elevated
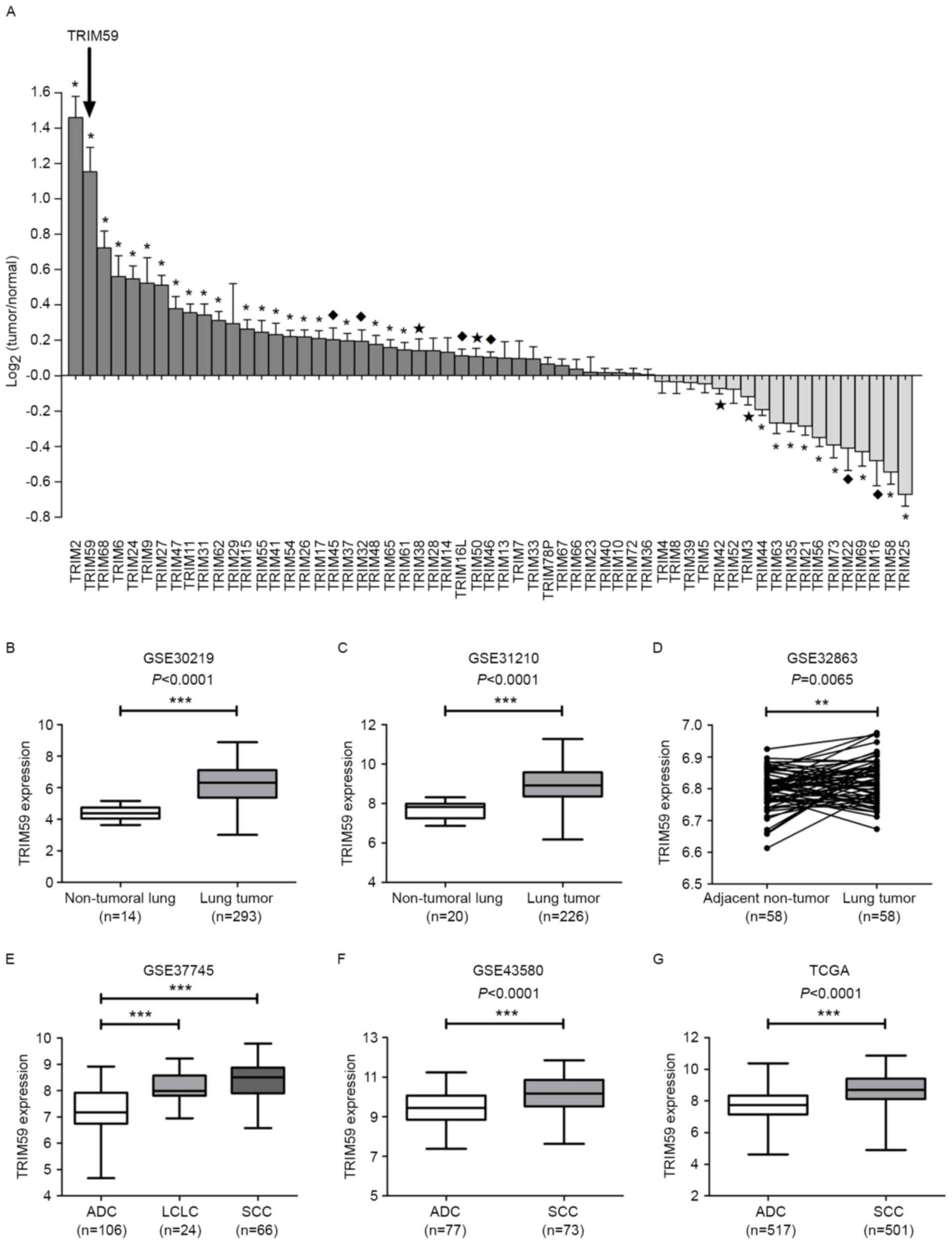
while 13 were decreased in lung tumors (Table I). TRIM59 was defined to be
upregulated with positive and relatively high
log2(Tumor/Normal) value (P<0.0001) (Fig. 1A). Statistical analysis was also
performed to confirm the overexpression of TRIM59 in other two
datasets (GSE30219 and GSE31210, all P<0.0001, Fig. 1B-C) and another paired-sample dataset
(GSE32863, P=0.0065, Fig. 1D). The
subtypes of NSCLC are adenocarcinoma (ADC), squamous cell carcinoma
(SCC) and large cell lung carcinoma (LCLC) (38). TRIM59 expression was notably enhanced
in LCLC and SCC compared to ADC in GSE37745 (all P<0.0001,
Fig. 1E). Similar results were also
obtained from GSE43580 and TCGA data, which verified the
enhancement of TRIM59 expression in SCC (all P<0.0001, Fig. 1F-G).
 | Table I.Comparison of the TRIM family genes
expression within lung tumor and the adjacent normal lung in
GSE19804. |
Table I.
Comparison of the TRIM family genes
expression within lung tumor and the adjacent normal lung in
GSE19804.
|
|
| Log-2 mRNA signal
intensity (mean ± SEM) |
|
|---|
|
|
|
|
|
|---|
| Gene symbol | ID | Tumor | Normal | P-value |
|---|
| TRIM2 | 202341_s_at |
9.71±0.12 |
8.25±0.08 | <0.0001 |
| TRIM59 | 227801_at |
6.13±0.14 |
4.98±0.06 | <0.0001 |
| TRIM68 | 219405_at |
8.86±0.09 |
8.14±0.07 | <0.0001 |
| TRIM6 | 223599_at |
7.06±0.12 |
6.50±0.07 | <0.0001 |
| TRIM24 | 213301_x_at |
8.96±0.08 |
8.41±0.05 | <0.0001 |
| TRIM9 | 209859_at |
5.13±0.15 |
4.61±0.04 | 0.0006 |
| TRIM27 | 212118_at |
9.48±0.05 |
8.97±0.03 | <0.0001 |
| TRIM47 | 225868_at |
8.82±0.07 |
8.44±0.07 | <0.0001 |
| TRIM11 | 226566_at |
7.83±0.04 |
7.48±0.04 | <0.0001 |
| TRIM31 | 215444_s_at |
5.40±0.06 |
5.06±0.02 | <0.0001 |
| TRIM62 | 219272_at |
7.21±0.05 |
6.90±0.05 | <0.0001 |
| TRIM29 | 202504_at |
7.85±0.21 |
7.55±0.09 | 0.1942 |
| TRIM15 | 210177_at |
6.84±0.06 |
6.58±0.02 | <0.0001 |
| TRIM55 | 232721_at |
4.46±0.07 |
4.21±0.02 | 0.0005 |
| TRIM41 | 226445_s_at |
8.71±0.06 |
8.48±0.04 | 0.0006 |
| TRIM54 | 233669_s_at |
5.29±0.03 |
5.07±0.02 | <0.0001 |
| TRIM26 | 202702_at |
8.81±0.05 |
8.59±0.04 | <0.0001 |
| TRIM17 | 220279_at |
6.34±0.04 |
6.13±0.03 | <0.0001 |
| TRIM45 | 219923_at |
6.34±0.07 |
6.14±0.03 | 0.0041 |
| TRIM37 | 213009_s_at |
9.07±0.05 |
8.88±0.03 | <0.0001 |
| TRIM32 | 203846_at |
7.63±0.07 |
7.43±0.04 | 0.0046 |
| TRIM48 | 220534_at |
4.35±0.05 |
4.18±0.02 | 0.0009 |
| TRIM65 | 235081_x_at |
7.28±0.05 |
7.12±0.04 | 0.0009 |
| TRIM61 | 240342_at |
6.62±0.04 |
6.47±0.02 | 0.0007 |
| TRIM38 | 203568_s_at |
8.94±0.07 |
8.80±0.05 | 0.0349 |
| TRIM28 | 200990_at | 10.48±0.07 | 10.34±0.07 | 0.0500 |
| TRIM14 | 203148_s_at |
9.08±0.06 |
8.95±0.06 | 0.1116 |
| TRIM16L | 1559682_at |
7.11±0.04 |
7.00±0.03 | 0.0045 |
| TRIM50 | 1556554_at |
5.88±0.04 |
5.77±0.03 | 0.0238 |
| TRIM46 | 220909_at |
5.51±0.03 |
5.41±0.02 | 0.0014 |
| TRIM13 | 229943_at |
8.52±0.11 |
8.42±0.11 | 0.3087 |
| TRIM7 | 223694_at |
6.00±0.10 |
5.91±0.03 | 0.3279 |
| TRIM33 | 210266_s_at |
9.70±0.08 |
9.61±0.06 | 0.1793 |
| TRIM78P | 232464_at |
5.65±0.03 |
5.58±0.03 | 0.0710 |
| TRIM67 | 233357_at |
3.83±0.02 |
3.82±0.02 | 0.5171 |
| TRIM66 | 229466_at |
6.21±0.06 |
6.17±0.05 | 0.4846 |
| TRIM23 | 204732_s_at |
6.63±0.09 |
6.62±0.08 | 0.8399 |
| TRIM40 | 1553079_at |
4.55±0.02 |
4.53±0.02 | 0.5134 |
| TRIM10 | 210579_s_at |
5.36±0.02 |
5.34±0.02 | 0.3981 |
| TRIM72 | 1554803_s_at |
5.75±0.02 |
5.74±0.03 | 0.6289 |
| TRIM36 | 1565812_at |
5.76±0.04 |
5.76±0.03 | 0.8415 |
| TRIM4 | 223384_s_at |
9.07±0.07 |
9.10±0.05 | 0.6288 |
| TRIM8 | 223132_s_at |
9.83±0.08 |
9.86±0.05 | 0.6242 |
| TRIM39 | 222732_at |
8.20±0.04 |
8.24±0.03 | 0.2768 |
| TRIM5 | 210705_s_at |
7.83±0.06 |
7.88±0.05 | 0.3822 |
| TRIM42 | 1566851_at |
4.97±0.02 |
5.04±0.03 | 0.0163 |
| TRIM52 | 221897_at |
7.22±0.09 |
7.30±0.06 | 0.3504 |
| TRIM3 | 213885_at |
6.23±0.04 |
6.34±0.04 | 0.0165 |
| TRIM44 | 217759_at | 10.18±0.04 | 10.37±0.03 | <0.0001 |
| TRIM63 | 236972_at |
4.95±0.04 |
5.21±0.07 | <0.0001 |
| TRIM35 | 227102_at |
7.31±0.04 |
7.58±0.04 | <0.0001 |
| TRIM21 | 204804_at |
8.04±0.05 |
8.33±0.04 | <0.0001 |
| TRIM56 | 231876_at |
8.74±0.06 |
9.09±0.05 | <0.0001 |
| TRIM73 | 1554250_s_at |
6.01±0.08 |
6.40±0.07 | <0.0001 |
| TRIM22 | 213293_s_at | 10.58±0.12 | 10.99±0.06 | 0.0022 |
| TRIM69 | 1568592_at |
8.72±0.07 |
9.14±0.05 | <0.0001 |
| TRIM16 | 204341_at |
7.09±0.14 |
7.57±0.09 | 0.0012 |
| TRIM58 | 215047_at |
3.73±0.04 |
4.27±0.06 | <0.0001 |
| TRIM25 | 224806_at |
9.04±0.10 |
9.72±0.08 | <0.0001 |
TRIM59 expression was associated with
various clinicopathological characteristics in NSCLC
In order to confirm the correlation between TRIM59
expression and the clinicopathological parameters of NSCLC,
GSE30219 and GSE31210, which had a large cohort of samples and
corresponding clinical information, were chosen and analyzed
statistically. As shown in Tables II
and III, TRIM59 expression was
closely associated with gender (all P<0.05), N stage (P=0.037),
smoking status (P=0.011), and pathological stage (P<0.001).
These results strongly indicated that TRIM59 expression was closely
associated with gender, smoking habits and tumor development.
 | Table II.Correlations of TRIM59 with
clinicopathological features of NSCLC in GSE30219. |
Table II.
Correlations of TRIM59 with
clinicopathological features of NSCLC in GSE30219.
|
|
| TRIM59
expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | No. of
patients | High | Low | χ2
value | P-value |
|---|
| Age at surgery
(years) |
|
|
| 0.028 | 0.866 |
|
≤50 | 41 | 21 | 20 |
|
|
|
>50 | 251 | 126 | 125 |
|
|
| Gender |
|
|
| 6.253 | 0.012 |
|
Male | 250 | 133 | 117 |
|
|
|
Female | 43 | 14 | 29 |
|
|
| T stage |
|
|
| 3.355 | 0.187 |
| T1 | 166 | 78 | 88 |
|
|
| T2 | 121 | 67 | 54 |
|
|
|
T3-4 | 52 | 32 | 20 |
|
|
| N stage |
|
|
| 4.332 | 0.037 |
|
Positive | 95 | 56 | 39 |
|
|
|
Negative | 198 | 91 | 107 |
|
|
| Metastasis |
|
|
| 0.872 | 0.351 |
|
Yes | 11 | 4 | 7 |
|
|
| No | 282 | 143 | 139 |
|
|
 | Table III.Correlations of TRIM59 with
clinicopathological features of NSCLC in GSE31210. |
Table III.
Correlations of TRIM59 with
clinicopathological features of NSCLC in GSE31210.
|
|
| TRIM59
expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | No. of
patients | High | Low | χ2
value | P-value |
|---|
| Age (years) |
|
|
| 2.061 | 0.151 |
|
≤50 | 27 | 17 | 10 |
|
|
|
>50 | 199 | 96 | 103 |
|
|
| Gender |
|
|
| 4.002 | 0.045 |
|
Male | 105 | 60 | 45 |
|
|
|
Female | 121 | 53 | 68 |
|
|
| Smoking status |
|
|
| 6.391 | 0.011 |
|
Ever-smoker | 111 | 65 | 46 |
|
|
|
Never-smoker | 115 | 48 | 67 |
|
|
| Pathological
stage |
|
|
| 26.812 | <0.001 |
| I | 168 | 67 | 101 |
|
|
| II | 58 | 46 | 12 |
|
|
| EGFR/KRAS/ALK |
|
|
| 2.103 | 0.147 |
| alteration
status |
|
|
|
|
|
|
Mutation | 158 | 74 | 84 |
|
|
|
Triple-negative | 68 | 39 | 29 |
|
|
| c-MYC
expression |
|
|
| 0.516 | 0.472 |
|
High | 17 | 10 | 7 |
|
|
|
Low | 207 | 103 | 104 |
|
|
TRIM59 expression showed a negative
correlation with survival time in NSCLC
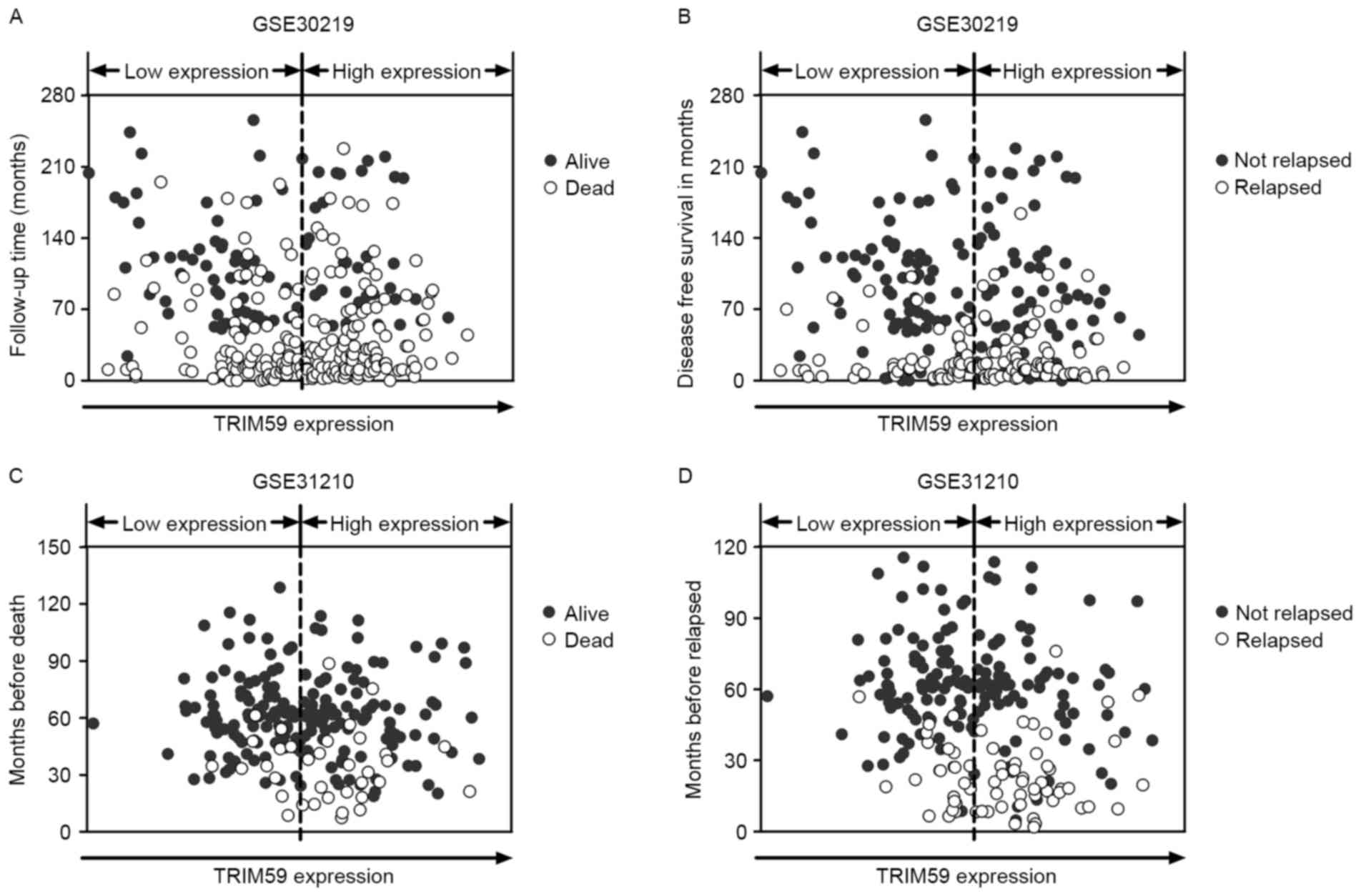
The distribution of the survival and recurrence
statuses was analyzed independently in GSE30219 and GSE31210. For
each dataset, samples were arranged according to TRIM59 expression
and patients' survival time. As shown in Fig. 2A and C, more deaths were noted for
NSCLC patients with higher TRIM59 expression than for those with
lower TRIM59 expression in both datasets. In addition, more
recurrent cases were observed in higher TRIM59 expression groups in
both datasets (Fig. 2B and D).
The correlations between TRIM59 expression and the
survival or recurrence statuses of NSCLC patients were further
examined. In consistence with the results described above, TRIM59
expression showed a negative correlation with overall survival (OS)
time (R=−0.155, P=0.008), disease-free survival (DFS) time
(R=−0.168, P=0.005) and relapse-free survival (RFS) time (R=−0.200,
P=0.002). These results indicated that higher expression level of
TRIM59 was related to shorter survival time in NSCLC patients.
High expression of TRIM59 led to poor
prognosis in NSCLC
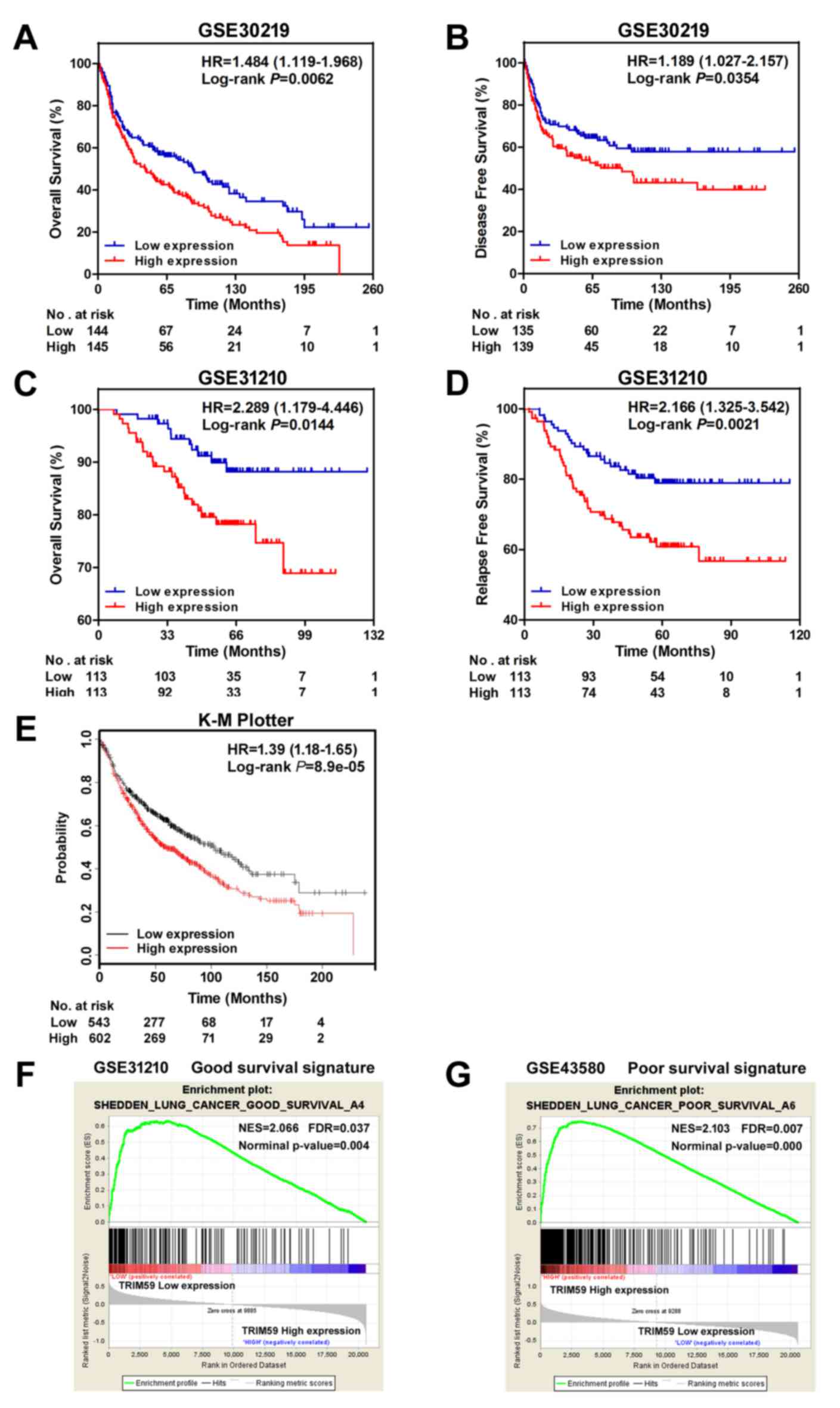
The survival curves for lung cancer patients were
further analyzed in GSE30219 and GSE31210. Ranked on the expression
level of TRIM59, top 50% samples were counted as the
high-expression group, and the remaining 50% were as the
low-expression group. The Kaplan-Meier analysis and Log-rank test
were used to compare the survival of patients in the 2 groups. As
shown in Fig. 3A and B, low
expression of TRIM59 was found to be correlated to better OS in
GSE30219 (HR=1.484, 95% CI 1.119–1.968, P=0.0062) and GSE31210
(HR=2.289, 95% CI 1.179–4.446, P=0.0144). In addition, better DFS
rate (HR=1.189, 95% CI 1.027–2.157, P=0.0354) (Fig. 3C) and RFS rate (HR=2.166, 95% CI
1.325–3.542, P=0.0021) (Fig. 3D) were
presented in low-expression group. Survival analysis was also
performed through online software, the K-M Plotter (www.kmplot.com), with auto selected best cut-off. The
survival curves plotted for all lung cancer patients from
multi-datasets (all datasets provided by the website) revealed the
similar results (Fig. 3E).
To further corroborate the prognostic role of
TRIM59, we performed a GSEA using NSCLC datasets with the
‘c2.cgp.v5.2.’ genesets from MSigDB database. It was observed that
genes in a good survival signature were enriched in the group with
low TRIM59 expression (Fig. 3F),
while a group of poor survival signature genes were enriched in the
subset of high TRIM59 expression (Fig.
3G).
Additionally, the relevance of TRIM59 expression
with other prognostic features of NSCLC patients was also
determined by using the K-M Plotter software. As shown in Table IV, high expression of TRIM59 acted as
a risk factor in SCC (but not in ADC) patients, pathological stage
I cases, T1-2, N0-1 and M0 stages, both male and female, and the
surgical margins negative cases.
 | Table IV.Correlation of TRIM59 expression with
prognostic factors of NSCLC patients in the K-M Plotter
software. |
Table IV.
Correlation of TRIM59 expression with
prognostic factors of NSCLC patients in the K-M Plotter
software.
| Prognostic
factors | Cases | HR | 95% CI of HR | P-value |
|---|
| Histological
type |
|
|
|
|
|
ADC | 673 | 1.27 | 1–1.61 | 0.052 |
|
SCC | 271 | 1.52 | 1.03–2.22 | 0.032 |
| Pathological
stage |
|
|
|
|
| I | 449 | 2.03 | 1.49–2.77 | 4.9e-06 |
| II | 161 | 0.66 | 0.41–1.06 | 0.083 |
|
III | 44 | 1.92 | 0.89–4.15 | 0.091 |
| T stage |
|
|
|
|
| T1 | 224 | 2.78 | 1.65–4.7 | 6.5e-05 |
| T2 | 190 | 1.95 | 1.21–3.15 | 0.005 |
| N stage |
|
|
|
|
| N0 | 324 | 1.96 | 1.36–2.83 | 0.00023 |
| N1 | 102 | 2.63 | 1.58–4.39 | 0.00011 |
| N2 | 32 | 0.52 | 0.22–1.23 | 0.13 |
| M stage |
|
|
|
|
| M0 | 462 | 1.86 | 1.39–2.49 | 1.9e-05 |
| Gender |
|
|
|
|
|
Male | 659 | 1.48 | 1.19–1.83 | 0.00032 |
|
Female | 375 | 1.55 | 1.1–2.17 | 0.011 |
| Smoking
history |
|
|
|
|
|
Ever-smoker | 300 | 0.8 | 0.53–1.19 | 0.27 |
|
Never-smoker | 141 | 1.86 | 0.74–4.7 | 0.18 |
| Surgery
success |
|
|
|
|
| Only surgical
margins negative | 204 | 2.24 | 1.05–4.79 | 0.033 |
Effect of TRIM59 expression on
survival by Cox regression analysis
Furthermore, a significant association between
TRIM59 expression signature and OS in the univariable Cox
regression model was observed. As is shown in Table V, the HR values of TRIM59 expression
signature of the high-expression group vs. the low-expression group
for OS in the two datasets were 1.437 (95% CI 1.086–1.901, P=0.011
for GSE30219) and 2.373 (95% CI 1.162–4.847, P=0.018 for GSE31210).
Gender (HR=1.698, 95% CI 1.080–2.670, P=0.022), T stage (HR=4.128,
95% CI 2.547–6.689, P<0.001), N stage (HR=4.613, 95% CI
2.385–8.920, P<0.001), metastasis (HR=2.555, 95% CI 1.300–5.020,
P=0.007), and pathological stage (HR=4.232, 95% CI 2.175–8.236,
P=0.001) were also contribute factors to shorter OS of patients.
Multivariate Cox proportional hazards model analysis from GSE30219
indicated that TRIM59 expression signature (high vs. low, HR=1.369,
95% CI 1.012–1.851, P=0.042) was an independent prognostic factor
in tumor tissues as compared with age, gender, T stage, N stage and
metastasis.
 | Table V.Univariable and multivariable Cox
regression analysis of TRIM59 expression signature and OS of NSCLC
patients in GSE30219. |
Table V.
Univariable and multivariable Cox
regression analysis of TRIM59 expression signature and OS of NSCLC
patients in GSE30219.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value |
|---|
| GSE30219 |
|
|
|
|
|
|
|
Age | 1.038 | 1.024–1.052 | <0.001 | 1.039 | 1.024–1.054 | <0.001 |
|
Gender | 1.698 | 1.080–2.670 | 0.022 | 1.492 | 0.931–2.391 | 0.096 |
| T
stage | 4.128 | 2.547–6.689 | <0.001 | 2.339 | 1.225–4.468 | 0.010 |
| N
stage | 4.613 | 2.385–8.920 | <0.001 | 2.988 | 1.379–6.474 | 0.006 |
|
Metastasis | 2.555 | 1.300–5.020 | 0.007 | 1.646 | 0.700–3.871 | 0.254 |
| TRIM59
expression | 1.437 | 1.086–1.901 | 0.011 | 1.369 | 1.012–1.851 | 0.042 |
| GSE31210 |
|
|
|
|
|
|
|
Age | 1.025 | 0.977–1.075 | 0.306 | 1.034 | 0.986–1.085 | 0.170 |
|
Gender | 1.519 | 0.780–2.955 | 0.219 | 0.951 | 0.379–2.386 | 0.914 |
| Smoking
status | 1.637 | 0.837–3.201 | 0.150 | 1.331 | 0.528–3.357 | 0.545 |
|
Pathological stage | 4.232 | 2.175–8.236 | <0.001 | 3.489 | 1.685–7.225 | 0.001 |
|
EGFR/KRAS/ALK | 0.457 | 0.235–0.890 | 0.021 | 0.548 | 0.277–1.085 | 0.084 |
|
alteration status |
|
|
|
|
|
|
| c-MYC
expression | 0.696 | 0.167–2.900 | 0.618 | 0.793 | 0.186–3.370 | 0.753 |
| TRIM59
expression | 2.373 | 1.162–4.847 | 0.018 | 1.505 | 0.696–3.252 | 0.299 |
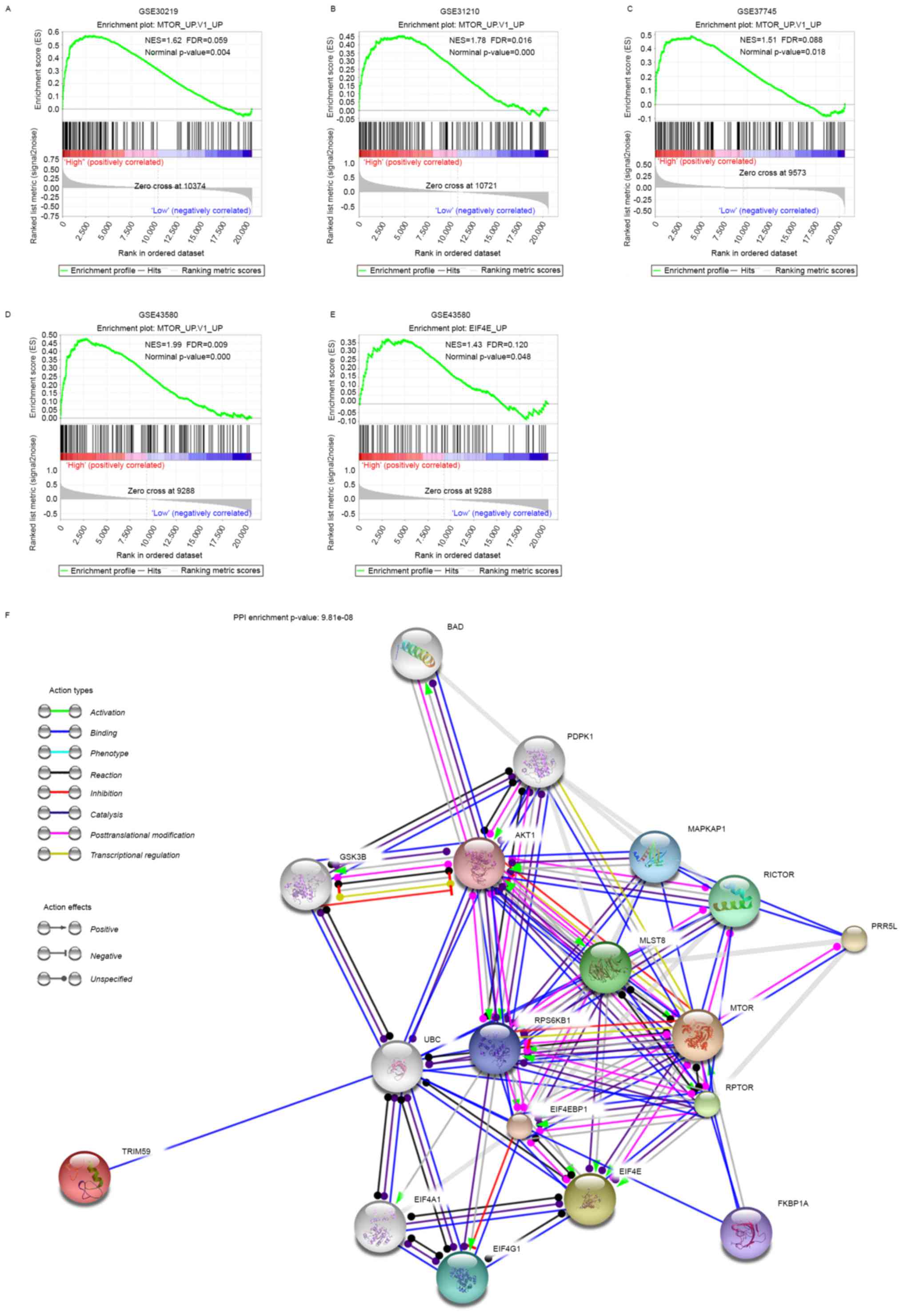
Overexpression of TRIM59 promoted MTOR
and EIF4E signaling
After screening the association of TRIM59 expression
with the oncogenic signatures within the ‘c6.all.v5.2’ genesets
from MSigDB database, the results revealed that high expression of
TRIM59 was associated with the activation of MTOR signaling in the
four datasets (Fig. 4A-D) and
activation of EIF4E signaling which has been considered as a
down-stream effector of MTOR (Fig.
4E), suggesting the involvement of TRIM59 in MTOR pathways. To
better understand the association of TRIM59 with MTOR and EIF4E, we
mapped TRIM59, MTOR and EIF4E onto STRING database to build a PPI
network (Fig. 4F). By using the ‘+
more proteins’ option, additional 15 predicted functional partners
were allowed into the network. As is shown in Fig. 4F, UBC (ubiquitin C) acted as a bridge
to connect TRIM59 with MTOR, EIF4E and other predicted functional
partners. Functional enrichments of the PPI network by STRING
software, using KEGG Pathway and GO analysis filter, revealed that
they were involved in a wide variety of processes and pathways. 9
proteins (PDPK1, AKT1, RICTOR, MLST8, RPS6KB1, MTOR, RPTOR,
EIF4EBP1 and EIF4E) were enriched in MTOR signaling pathway
(FDR=9.96e-17). 4 proteins (BAD, AKT1, GSK3B and MTOR) were
enriched in Pathways in cancer (FDR=0.00228). 3 proteins (BAD,
PDPK1 and AKT1) were enriched in Non-small cell lung cancer
(FDR=0.000305). 10 proteins (BAD, PDPK1, AKT1, GSK3B, MLST8, MTOR,
RPS6KB1, RPTOR, EIF4EBP1, EIF4E) were enriched in PI3K-Akt
signaling pathway (FDR=4.66e-12).
In addition, GO enrichment analysis showed that the
proteins in the network were enriched in 10 cell cycle processes. 9
proteins (BAD, AKT1, UBC, RPS6KB1, MLST8, MTOR, RPTOR, EIF4EBP1 and
EIF4E) were enriched in regulation of cell cycle (FDR=3.37e-06). 9
proteins (GSK3B, AKT1, UBC, RPS6KB1, MLST8, MTOR, RPTOR, EIF4EBP1
and EIF4E) were enriched in cell cycle process (FDR=6.88e-06) and
cell cycle (FDR=4.56e-05). 5 proteins (AKT1, UBC, RPS6KB1, EIF4EBP1
and EIF4E) were enriched in G1/S transition of mitotic cell cycle
(FDR=4.57e-05), positive regulation of cell cycle (FDR=0.000478),
and mitotic cell cycle (FDR=0.021). 3 proteins (RPS6KB1, EIF4EBP1
and EIF4E) were enriched in positive regulation of mitotic cell
cycle (FDR=0.00779). 3 proteins (MLST8, MTOR and RPTOR) were
enriched in cell cycle arrest (FDR=0.013). 4 proteins (MLST8, UBC,
MTOR and RPTOR) were enriched in negative regulation of cell cycle
(FDR=0.0218). 4 proteins (UBC, RPS6KB1, EIF4EBP1 and EIF4E) were
enriched in regulation of mitotic cell cycle (FDR=0.0233).
Discussion
In the last decade, much attention has been garnered
in focusing on the role of TRIM proteins in innate immunity and
antiviral defense (39–41). Recently, their biological functions in
tumor biology have become an attractive research area. A number of
TRIM proteins have been revealed to play a crucial role in
proliferation, migration and invasion of lung cancer (19–22).
However, no reports have evaluated the prognostic value of TRIM59
in lung cancer. In this study, we tried to explore a possible
target in the TRIM family and elucidated its possibility being used
as a biomarker in NSCLC patients. By taking the advantage of the
availability of online expression profile datasets, bioinformatics
analysis is a well-established method which has been widely
utilized to help researchers find out potential biomarkers.
Therefore, bioinformatics analysis was also adopted in our study
and a microarray dataset of matched-samples was screened on the
focus of the TRIM family. As a result, TRIM59 was identified as an
aberrantly overexpressed gene in lung cancer tissues. Among the
major histological subtypes of NSCLC, our results showed that
TRIM59 expression was notably enhanced in LCLC and SCC compared to
ADC. We also revealed that the positive expression of TRIM59 was
significantly associated with gender, smoking habits, and the
unfavorable conditions on the depth of tumor N stage and
pathological stage, which suggested that TRIM59 might play an
important role in the development and progression of NSCLC.
Nowadays, surgery and chemotherapy are considered as
the first choice of NSCLC treatments, but even for the early stage
patients, the therapeutic effect of these treatments remains
unsatisfactory (42). The results
above indicated that TRIM59 acted as a risk factor even in the
surgical margins negative cases and was negatively correlated with
clinical outcome. It is worthy to note that TRIM59 could represent
as a potential independent prognostic marker in NSCLC patients,
which might help doctors make optimal clinical decisions and
individualized treatment strategies in order to provide better
prognosis. Considering that distinct histological subtypes might
make a significant contribution to selecting appropriate treatment
programs (42), the prognostic values
of TRIM59 in ADC and SCC were further evaluated, using the K-M
Plotter online software with auto selected best cut-off. High
expression of TRIM59 was found to act as a risk factor in SCC
patients but not in ADC patients, which implied that TRIM59 might
act as a prognostic marker in different histological subtypes of
NSCLC.
GSEA showed the enrichment of MTOR signaling and its
down-stream signaling genes within high TRIM59 expression of lung
cancer. MTOR is a key component of PI3K/AKT/MTOR pathway, a
potential candidate served as an effectively therapeutic target of
cancers (43–45). Abnormal MTOR activity may result in
tumorigenesis, aberrant proliferation, metastasis, and chemotherapy
resistance (46–48). MTOR contains two independent
functional complexes, MTOR complex 1 (MTORC1) and MTOR complex 2
(MTORC2) (49). In general, MTORC1
regulates cell autonomous growth by controlling nutrient
availability and growth factors, whereas MTORC2 mediates cell
proliferation and survival by regulating cell surface area
(45,49). Dysregulation of upstream signals, such
as PI3K/AKT mutation (50),
Phosphatase and tensin homolog (PTEN) mutation (51), Tuberous sclerosis complex (TSC) loss
of function (52), and RAS mutation
(53), often results in the
alteration of MTOR, which has been demonstrated to contribute to a
poor prognosis in serious cancers including NSCLC, breast cancer,
gastric cancer and esophageal squamous cell carcinoma (54–57). The
activation of MTOR is mediated by Ser2448 phosphorylation through
the PI3K/AKT/MTOR pathway, and then it activates a potent oncogene,
EIF4E (58). The oncogenic ability of
EIF4E is formed by activating translation and being phosphorylated,
which leads to tumor formation primarily by suppressing apoptosis
(59). Furthermore, elevated levels
of EIF4E on the one hand induce cellular proliferation, invasion
and acquired drug resistance, and on the other hand enhance
translation of many malignancy-related proteins, thus may present
negative effects on survival of NSCLC patients (60–62).
The complicated cellular processes of TRIM59 related
to cancer are undertaken by closely connected to proteins which
have oncogenic signatures. The PPI network constructed from STRING
database vividly delineated the functional interactions of TRIM59
with other proteins through UBC binding. Because of the presence of
RING finger domain, many TRIM proteins can act as E3 ubiquitin
ligases and partake in the ubiquitin-proteasome system (63). That might be one of the reasons why
TRIM59 could be tied to UBC. The TRIM family of E3 ubiquitin
ligases is necessary for regulation of many key and diverse
processes in various malignancies, such as TRIM4 which sensitizes
the tumor cells to hydrogen peroxide induced cell death (64), TRIM32 which negatively regulates tumor
suppressor p53 to promote tumorigenesis (65), TRIM11 which may promote cell motility
and invasiveness through AKT pathway in lung cancer (19) and TRIM25 which acts as an oncogene in
gastric cancer and exerts its function through TGF-β pathway
(66).
In particular, several other tumor related molecules
were also observed in the PPI network, such as AKT1, BAD and
EIF4EBP1 (EIF4E binding protein 1). AKT1, one of AKT kinase family
members, is the predominant isoform responsible for cell
proliferation and survival (67). AKT
has been reported to transduce antiapoptotic signals by
inactivating BAD (68) and mediate
cell growth through MTOR, which activates p70 ribosomal protein S6
kinase 1 and inhibits EIF4EBP1 (69).
EIF4EBP1, a critical regulator of MTOR downstream signaling, may be
associated with drug resistance in human tumors (70). Phosphorylation of EIF4EBP1 results in
release of EIF4E, which enhances the oncogenic protein synthesis
and correlates with the poor prognosis in lung cancer (71,72).
In addition, a recent study of TRIM59 in NSCLC
showed that TRIM59 knocking down arrests NSCLC cell cycle in G2
phase and decreases the expression of cell cycle proteins (17). Consistent with this paper, functional
enrichments of the PPI network by STRING software showed that the
proteins selected by STRING Database for network construction were
enriched in 10 cell cycle processes. These results not only
implicated the involvement of TRIM59 in cell cycle process but also
revealed how it would affect this process.
In conclusion, we have demonstrated the clinical
relevance, the prognostic value and the functional mechanisms of
TRIM59 in NSCLC. TRIM59 was frequently elevated in NSCLC,
associated with various unfavorable conditions of
clinicopathological characteristics. TRIM59 was negatively
correlated with clinical outcome and represented as a potential
independent prognostic marker in NSCLC patients. GSEA and PPI
network construction revealed that TRIM59 was associated with
oncogenic MTOR and EIF4E signaling through UBC binding. Taken
together, high expression of TRIM59 could serve as a valuable
independent predictor for the poor prognosis of NSCLC patients, and
aberrant TRIM59 expression might be a novel biomarker for molecular
targeted therapies against the disease.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
TRIM59
|
Tripartite motif 59
|
|
LCLC
|
large cell lung carcinoma
|
|
SCC
|
squamous cell carcinoma
|
|
ADC
|
adenocarcinoma
|
|
GEO
|
gene expression omnibus
|
|
GSEA
|
gene set enrichment analysis
|
|
MTOR
|
mammalian target of rapamycin
|
|
EIF4E
|
eukaryotic initiation factor 4E
|
|
PPI
|
Protein-protein interaction
|
|
K-M Plotter
|
Kaplan Meier plotter
|
|
HR
|
hazard ratios
|
|
CI
|
confidence intervals
|
|
FDR
|
false discovery rates
|
|
NES
|
normal enrichment score
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
RFS
|
relapse-free survival
|
|
PTEN
|
phosphatase and tensin homolog
|
|
TSC
|
tuberous sclerosis complex
|
|
MTORC1
|
MTOR complex 1
|
|
MTORC2
|
MTOR complex 2
|
|
UBC
|
ubiquitin C
|
|
EIF4EBP1
|
EIF4E binding protein 1
|
References
|
1
|
Surveillance epidemiology and end results
SEER stat fact sheets: Lung and bronchus cancer. 2016.http://seer.cancer.gov/statfacts/html/lungb.htmlNovember
17–2016
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zago G, Muller M, van den Heuvel M and
Baas P: New targeted treatments for non-small-cell lung cancer-role
of nivolumab. Biologics. 10:103–117. 2016.PubMed/NCBI
|
|
5
|
Non-small cell lung cancer treatment
(PDQ®)-patient version. 2016, https://www.cancer.gov/types/lung/patient/non-small-cell-lung-treatment-pdqNovember
17–2016
|
|
6
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sequist LV, Yang JC, Yamamoto N, O'Byrne
K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, et al:
Phase III study of afatinib or cisplatin plus pemetrexed in
patients with metastatic lung adenocarcinoma with EGFR mutations. J
Clin Oncol. 31:3327–3334. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang
Y, Li W, Hou M, Shi JH, Lee KY, et al: Afatinib versus cisplatin
plus gemcitabine for first-line treatment of Asian patients with
advanced non-small-cell lung cancer harbouring EGFR mutations
(LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet
Oncol. 15:213–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó
L, Ahn MJ, De Pas T, Besse B, Solomon BJ, Blackhall F, et al:
Crizotinib versus chemotherapy in advanced ALK-positive lung
cancer. N Engl J Med. 368:2385–2394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa
K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al:
First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shaw AT, Kim DW, Mehra R, Tan DS, Felip E,
Chow LQ, Camidge DR, Vansteenkiste J, Sharma S, De Pas T, et al:
Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J
Med. 370:1189–1197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meroni G and Diez-Roux G: TRIM/RBCC, a
novel class of ‘single protein RING finger’ E3 ubiquitin ligases.
Bioessays. 27:1147–1157. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ikeda K and Inoue S: TRIM proteins as RING
finger E3 ubiquitin ligases. Adv Exp Med Biol. 770:27–37. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamada Y, Takayama KI, Fujimura T,
Ashikari D, Obinata D, Takahashi S, Ikeda K, Kakutani S, Urano T,
Fukuhara H, Homma Y, et al: A novel prognostic factor TRIM44
promotes cell proliferation and migration, and inhibits apoptosis
in testicular germ cell tumor. Cancer Sci. 180:32–41. 2017.
View Article : Google Scholar
|
|
16
|
Wang J, Zhu J, Dong M, Yu H, Dai X and Li
K: Knockdown of tripartite motif containing 24 by lentivirus
suppresses cell growth and induces apoptosis in human colorectal
cancer cells. Oncol Res. 22:39–45. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhan W, Han T, Zhang C, Xie C, Gan M, Deng
K, Fu M and Wang JB: TRIM59 promotes the proliferation and
migration of non-small cell lung cancer cells by upregulating cell
cycle related proteins. PLoS One. 10:e01425962015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bell JL, Malyukova A, Kavallaris M,
Marshall GM and Cheung BB: TRIM16 inhibits neuroblastoma cell
proliferation through cell cycle regulation and dynamic nuclear
localization. Cell Cycle. 12:889–898. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Shi W, Shi H, Lu S, Wang K, Sun C,
He J, Jin W, Lv X, Zou H and Shu Y: TRIM11 overexpression promotes
proliferation, migration and invasion of lung cancer cells. J Exp
Clin Cancer Res. 35:1002016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen L, Muñoz-Antonia T and Cress WD:
Trim28 contributes to EMT via regulation of E-cadherin and
N-cadherin in lung cancer cell lines. PLoS One. 9:e1010402014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song X, Fu C, Yang X, Sun D, Zhang X and
Zhang J: Tripartite motif-containing 29 as a novel biomarker in
non-small cell lung cancer. Oncol Lett. 10:2283–2288.
2015.PubMed/NCBI
|
|
22
|
Luo Q, Lin H, Ye X, Huang J, Lu S and Xu
L: Trim44 facilitates the migration and invasion of human lung
cancer cells via the NF-κB signaling pathway. Int J Clin Oncol.
20:508–517. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Khatamianfar V, Valiyeva F, Rennie PS, Lu
WY, Yang BB, Bauman GS, Moussa M and Xuan JW: TRIM59, a novel
multiple cancer biomarker for immunohistochemical detection of
tumorigenesis. BMJ Open. 2:pii:e0014102012. View Article : Google Scholar
|
|
24
|
Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH
and Gao WQ: TRIM59 is up-regulated in gastric tumors, promoting
ubiquitination and degradation of p53. Gastroenterology.
147:1043–1054. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang J, Xing D, Li Z, Shen J, Zhao H and
Li S: TRIM59 is upregulated and promotes cell proliferation and
migration in human osteosarcoma. Mol Med Rep. 13:5200–5206.
2016.PubMed/NCBI
|
|
26
|
Aierken G, Seyiti A, Alifu M and Kuerban
G: Knockdown of tripartrtite-59 (TRIM59) inhibits cellular
proliferation and migration in human cervical cancer cells. Oncol
Res. 25:381–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Valiyeva F, Jiang F, Elmaadawi A, Moussa
M, Yee SP, Raptis L, Izawa JI, Yang BB, Greenberg NM, Wang F and
Xuan JW: Characterization of the oncogenic activity of the novel
TRIM59 gene in mouse cancer models. Mol Cancer Ther. 10:1229–1240.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC,
Lin CW, Shih JY, Yang PC, Hsiao CK, Lai LC and Chuang EY:
Identification of a novel biomarker, SEMA5A, for non-small cell
lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers
Prev. 19:2590–2597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra662013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamauchi M, Yamaguchi R, Nakata A, Kohno
T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y,
et al: Epidermal growth factor receptor tyrosine kinase defines
critical prognostic genes of stage I lung adenocarcinoma. PLoS One.
7:e439232012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Botling J, Edlund K, Lohr M, Hellwig B,
Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, et
al: Biomarker discovery in non-small cell lung cancer: Integrating
gene expression profiling, meta-analysis, and tissue microarray
validation. Clin Cancer Res. 19:194–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tarca AL, Lauria M, Unger M, Bilal E, Boue
S, Dey K Kumar, Hoeng J, Koeppl H, Martin F, Meyer P, et al:
Strengths and limitations of microarray-based phenotype prediction:
Lessons learned from the IMPROVER diagnostic signature challenge.
Bioinformatics. 29:2892–2899. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:pp. 15545–15550. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Wang DC, Shi L, Zhu B, Min Z and
Jin J: Genome analyses identify the genetic modification of lung
cancer subtypes. Semin Cancer Biol. 42:20–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ozato K, Shin DM, Chang TH and Morse HC
III: TRIM family proteins and their emerging roles in innate
immunity. Nat Rev Immunol. 8:849–860. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McNab FW, Rajsbaum R, Stoye JP and O'Garra
A: Tripartite-motif proteins and innate immune regulation. Curr
Opin Immunol. 23:46–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chan E, Towers GJ and Qasim W: Gene
therapy strategies to exploit TRIM derived restriction factors
against HIV-1. Viruses. 6:243–263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Reck M, Popat S, Reinmuth N, De Ruysscher
D, Kerr KM and Peters S; ESMO Guidelines Working Group, :
Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical
practice guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 25 Suppl 3:iii27–iii39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zaytseva YY, Valentino JD, Gulhati P and
Evers BM: mTOR inhibitors in cancer therapy. Cancer Lett. 319:1–7.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheng H, Walls M, Baxi SM and Yin MJ:
Targeting the mTOR pathway in tumor malignancy. Curr Cancer Drug
Targets. 13:267–277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moschetta M, Reale A, Marasco C, Vacca A
and Carratù MR: Therapeutic targeting of the mTOR-signalling
pathway in cancer: Benefits and limitations. Br J Pharmacol.
171:3801–3813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen R, Duan J, Li L, Ma Q, Sun Q, Ma J,
Li C, Zhou X, Chen H, Jing Y, et al: mTOR promotes pituitary tumor
development through activation of PTTG1. Oncogene. 36:979–988.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mai H, Xu X, Mei G, Hong T, Huang J, Wang
T, Yan Z, Li Y, Liang Y, Li L, et al: The interplay between HPIP
and casein kinase 1α promotes renal cell carcinoma growth and
metastasis via activation of mTOR pathway. Oncogenesis. 5:e2602016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhuang H, Bai J, Chang JY, Yuan Z and Wang
P: MTOR inhibition reversed drug resistance after combination
radiation with erlotinib in lung adenocarcinoma. Oncotarget.
7:84688–84694. 2016.PubMed/NCBI
|
|
49
|
Eltschinger S and Loewith R: TOR complexes
and the maintenance of cellular homeostasis. Trends Cell Biol.
26:148–159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Willems L, Tamburini J, Chapuis N, Lacombe
C, Mayeux P and Bouscary D: PI3K and mTOR signaling pathways in
cancer: New data on targeted therapies. Curr Oncol Rep. 14:129–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tsujita Y, Mitsui-Sekinaka K, Imai K, Yeh
TW, Mitsuiki N, Asano T, Ohnishi H, Kato Z, Sekinaka Y, Zaha K, et
al: Phosphatase and tensin homolog (PTEN) mutation can cause
activated phosphatidylinositol 3-kinase δ syndrome-like
immunodeficiency. J Allergy Clin Immunol. 138:1672–1680.e10. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Habib SL, Al-Obaidi NY, Nowacki M, Pietkun
K, Zegarska B, Kloskowski T, Zegarski W, Drewa T, Medina EA, Zhao Z
and Liang S: Is mTOR inhibitor good enough for treatment all tumors
in TSC patients? J Cancer. 7:1621–1631. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Müller E, Bauer S, Stühmer T, Mottok A,
Scholz CJ, Steinbrunn T, Brünnert D, Brandl A, Schraud H, Kreßmann
S, et al: Pan-Raf co-operates with PI3K-dependent signalling and
critically contributes to myeloma cell survival independently of
mutated RAS. Leukemia. 31:922–933. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li L, Liu D, Qiu ZX, Zhao S, Zhang L and
Li WM: The prognostic role of mTOR and p-mTOR for survival in
non-small cell lung cancer: A systematic review and meta-analysis.
PLoS One. 10:e01167712015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Azim HA, Kassem L, Treilleux I, Wang Q, El
Enein MA, Anis SE and Bachelot T: Analysis of PI3K/mTOR pathway
biomarkers and their prognostic value in women with hormone
Receptor-Positive, HER2-Negative early breast cancer. Transl Oncol.
9:114–123. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xiao L, Wang YC, Li WS and Du Y: The role
of mTOR and phospho-p70S6K in pathogenesis and progression of
gastric carcinomas: An immunohistochemical study on tissue
microarray. J Exp Clin Cancer Res. 28:1522009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li S, Wang Z, Huang J, Cheng S, Du H, Che
G and Peng Y: Clinicopathological and prognostic significance of
mTOR and phosphorylated mTOR expression in patients with esophageal
squamous cell carcinoma: A systematic review and meta-analysis. BMC
Cancer. 16:8772016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shaw RJ and Cantley LC: Ras, PI(3)K and
mTOR signalling controls tumour cell growth. Nature. 441:424–430.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wendel HG, Silva RL, Malina A, Mills JR,
Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J,
Pelletier J and Lowe SW: Dissecting eIF4E action in tumorigenesis.
Genes Dev. 21:3232–3237. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li Y, Fan S, Koo J, Yue P, Chen ZG,
Owonikoko TK, Ramalingam SS, Khuri FR and Sun SY: Elevated
expression of eukaryotic translation initiation factor 4E is
associated with proliferation, invasion and acquired resistance to
erlotinib in lung cancer. Cancer Biol Ther. 13:272–280. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Thumma SC, Jacobson BA, Patel MR, Konicek
BW, Franklin MJ, Jay-Dixon J, Sadiq A, De A, Graff JR and Kratzke
RA: Antisense oligonucleotide targeting eukaryotic translation
initiation factor 4E reduces growth and enhances chemosensitivity
of non-small-cell lung cancer cells. Cancer Gene Ther. 22:396–401.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Khoury T, Alrawi S, Ramnath N, Li Q, Grimm
M, Black J and Tan D: Eukaryotic initiation factor-4E and cyclin D1
expression associated with patient survival in lung cancer. Clin
Lung Cancer. 10:58–66. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Li Y, Wu H, Wu W, Zhuo W, Liu W, Zhang Y,
Cheng M, Chen YG, Gao N, Yu H, et al: Structural insights into the
TRIM family of ubiquitin E3 ligases. Cell Res. 24:762–765. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Tomar D, Prajapati P, Lavie J, Singh K,
Lakshmi S, Bhatelia K, Roy M and Singh R, Bénard G and Singh R:
TRIM4; a novel mitochondrial interacting RING E3 ligase, sensitizes
the cells to hydrogen peroxide (H2O2) induced cell death. Free
Radic Biol Med. 89:1036–1048. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu J, Zhang C, Wang XL, Ly P, Belyi V,
Xu-Monette ZY, Young KH, Hu W and Feng Z: E3 ubiquitin ligase
TRIM32 negatively regulates tumor suppressor p53 to promote
tumorigenesis. Cell Death Differ. 21:1792–1804. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhu Z, Wang Y, Zhang C, Yu S, Zhu Q, Hou K
and Yan B: TRIM25 blockade by RNA interference inhibited migration
and invasion of gastric cancer cells through TGF-β signaling. Sci
Rep. 6:190702016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Koseoglu S, Lu Z, Kumar C, Kirschmeier P
and Zou J: AKT1, AKT2 and AKT3-dependent cell survival is cell
line-specific and knockdown of all three isoforms selectively
induces apoptosis in 20 human tumor cell lines. Cancer Biol Ther.
6:755–762. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu Y, Gao X, Deeb D, Zhang Y, Shaw J,
Valeriote FA and Gautam SC: Mycotoxin verrucarin A inhibits
proliferation and induces apoptosis in prostate cancer cells by
inhibiting prosurvival Akt/NF-κB/mTOR signaling. J Exp Ther Oncol.
11:251–260. 2016.PubMed/NCBI
|
|
69
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hsieh AC, Nguyen HG, Wen L, Edlind MP,
Carroll PR, Kim W and Ruggero D: Cell type-specific abundance of
4EBP1 primes prostate cancer sensitivity or resistance to PI3K
pathway inhibitors. Sci Signal. 8:ra1162015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lee HW, Lee EH, Lee JH, Kim JE, Kim SH,
Kim TG, Hwang SW and Kang KW: Prognostic significance of
phosphorylated 4E-binding protein 1 in non-small cell lung cancer.
Int J Clin Exp Pathol. 8:3955–3962. 2015.PubMed/NCBI
|
|
72
|
Roh MS, Lee JH, Kang KW, Nam HY, Jung SB,
Kim K, Lee EH, Park MI, Kim MS and Lee HW: Phosphorylated
4E-binding protein 1 expression is associated with poor prognosis
in small-cell lung cancer. Virchows Arch. 467:667–673. 2015.
View Article : Google Scholar : PubMed/NCBI
|