Introduction
Gastric cancer is the fourth most commonly occurring
malignancy and the second leading cause of cancer-associated
mortality worldwide (1). Almost
two-thirds of gastric cancer cases occur in developing countries,
with an incidence of ~42% in China alone (2). Despite improvements in therapy in the
past decades, this type of cancer remains highly lethal due to its
aggressive metastatic behavior and the fact that it is often
diagnosed at an advanced stage (3).
An improved understanding of the disease-causing mechanism and the
identification of specific biomarkers for gastric cancer
progression are urgently required for the prediction and
improvement of clinical outcomes.
Human genome studies have identified a large number
of non-coding RNAs (ncRNAs) that are differentially-expressed in
varying organs and tissue types (4–7). Such
developments have been equaled through discoveries made by
analyzing the role of ncRNAs in human diseases, particularly
cancer, which has corroborated the importance of their cellular
functions (8,9). Preliminary results have indicated that
ncRNAs, particularly long ncRNAs (lncRNA), exhibit key roles in
tumorigenesis (8), and that
lncRNA-mediated biology is focal to the progression of cancer
(8,10–13). Those
lncRNAs associated with cancer are often aberrantly expressed and
affect cancer progression through different mechanisms (14,15).
Therefore, a better understanding of the expression and function of
lncRNAs may lead to the identification of novel biomarkers and
therapeutic targets for the treatment of cancer.
The present primary investigation of lncRNAs that
may be involved in gastric cancer progression led to the
identification of several noteworthy candidates. One of these was
MLLT4 antisense RNA 1 (MLLT4-AS1), which is also known as
chromosome 6 open reading frame 124 (C6orf124), dJ431P23.3 or
HGC6.4. This gene is located in chromosome
6:167,823,876-167,826,709, and 3 transcripts (splice variants) have
been identified, namely MLLT4-AS1-001 (2,238 bp), MLLT4-AS1-002
(311 bp) and MLLT4-AS1-003 (182 bp) (www.ensembl.org). It is unknown whether this gene is
associated with cancer. In the present study, the expression level
of MLLT4-AS1 was examined in gastric cancer tissues and the
potential correlation between its expression level and the
clinicopathological features of gastric cancer patients was
evaluated. These findings indicated that decreased expression of
MLLT4-AS1 is associated with a poor prognosis in gastric
cancer.
Materials and methods
Sample preparation
A total of 103 human primary gastric cancer samples
and paired adjacent non-cancerous tissue samples were collected
after obtaining informed consent from patients who underwent D2
radical resection between January 2007 and December 2008 in
Shanghai Songjiang Hospital Affiliated to Nanjing Medical
University (Shanghai, China). Of these, 5 tissue samples were
randomly selected for human lncRNA microarray analysis and the
remaining 98 were used for quantitative polymerase chain reaction
(qPCR) analysis. The study was approved by the Ethics Committee of
the Shanghai Songjiang Hospital Affiliated to Nanjing Medical
University. All subjects provided informed written consent at the
time of surgery for donation of their tissue for this study.
Specimens were obtained immediately after surgical resection and
stored at −80°C for further analysis. Lymph nodes (LNs) with or
without metastasis were also harvested during gastrectomy. The 98
samples analyzed by qPCR were obtained from 51 men and 47 women,
with a median age of 57 years (range, 31–83 years). Tumor stage was
defined according to the American Joint Committee on
Cancer/International Union against Cancer Tumor-Node-Metastasis
(TNM) classification system (seventh edition) (16). Clinical data, including date of birth,
gender, date of surgery, serum carcinoembryonic antigen (CEA)
level, Helicobacter pylori status, tumor size, tumor location and
other content of histopathological reports, were extracted from the
computerized clinical database.
RNA preparation
RNA preparation. Briefly, gastric cancer and paired
adjacent non-cancerous tissues were homogenized in TRIzol reagent
(1 ml per 50–100 mg tissue; Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). After sample homogenization, total RNA was
extracted following the manufacturer's instructions. The
concentration and quality of total RNA from each sample were
measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.),
and RNA integrity was assessed by 1.5% agarose-formaldehyde gel
electrophoresis.
lncRNA and mRNA microarray
The Human lncRNA 4*180K array was manufactured by
Agilent Technologies, Inc. (Santa Clara, CA, USA). Each array
represented all long transcripts, including protein coding mRNAs
and lncRNAs in the human genome. More than 41,053 lncRNAs were
collected. Each transcript was represented by 1–5 probes to improve
statistical confidence.
Microarray analysis
For microarray analysis, RNA purity and integrity
was analyzed by Agilent Bioanalyzer 2100 (Agilent Technologies,
Inc.). Qualified total RNA was further purified by RNeasy mini kit
(Qiagen, Hilden, Germany) and RNase-free DNase set (Qiagen). Total
RNA was then amplified and labeled by a Low Input Quick Amp
Labeling kit, One-Color (Agilent), following the manufacturer's
instructions. Labeled cRNA were purified by RNeasy mini kit
(Qiagen). Each Slide was hybridized with 600 ng Cy3-labeled cRNA
using a Gene Expression Hybridization kit (Agilent Technologies,
Inc.) in a Hybridization Oven (Agilent Technologies, Inc.),
according to the manufacturer's instructions. After 17 h of
hybridization, the slides were washed in staining dishes (Thermo
Fisher Scientific, Inc.) with Gene Expression Wash Buffer kit
(Agilent Technologies, Inc.), following the manufacturer's
instructions. Slides were scanned by Agilent Microarray Scanner
(Agilent) with default settings as follows: Dye channel, green;
scan resolution, 3 µm; 20 bit. Data were extracted with Feature
Extraction software 10.7 (Agilent Technologies, Inc.). Raw data
were normalized by Quantile algorithm, Gene Spring Software 11.0
(Agilent).
Reverse transcription (RT)-qPCR
The mRNA from gastric cancer samples and paired
adjacent non-cancerous tissues was analyzed by reverse
transcription using M-MLV Reverse Transcriptase (Takara
Biotechnology, Co., Ltd., Dalian, China). The cDNA template was
amplified by RT-qPCR using the SYBR® Premix Dimmer Eraser kit
(Takara Biotechnology, Co., Ltd.). Primer sequences used for
MLLT4-AS1 amplifications were as follows: Forward,
5′-TGCTGTGCGGTGTTCCTCTC-3′ and reverse,
5′-CGAAGAATTGGCAGATAACGATGT-3′. Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) was used as an internal control (forward,
5′-ACCCACTCCTCCACCTTTGAC-3′ and reverse,
5′-TGTTGCTGTAGCCAAATTCGTT-3′), and MLLT4-AS1 values were normalized
to GAPDH. RT-qPCR was performed with the ABI7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The cycling conditions
were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for
15 sec and 60°C for 45 sec. All experiments were repeated 3 times.
The relative expression fold-change of the mRNA was calculated
using the 2−ΔΔCq method (17).
Statistical analysis
Comparisons of continuous data between the two
groups were performed with the independent t-test or paired t-test,
whereas categorical data were analyzed using the χ2 test. Overall
survival was analyzed by the Kaplan-Meier method, and the
differences between groups were estimated by the log-rank test.
Independent prognostic indicators were assessed by multivariate
analysis using Cox's proportional hazards regression model. All
statistical analyses were performed using SPSS for Windows v.16.0
(SPSS, Inc., Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad
Software, La Jolla, CA, USA). P<0.05 was considered to indicate
a statistically significant difference.
Results
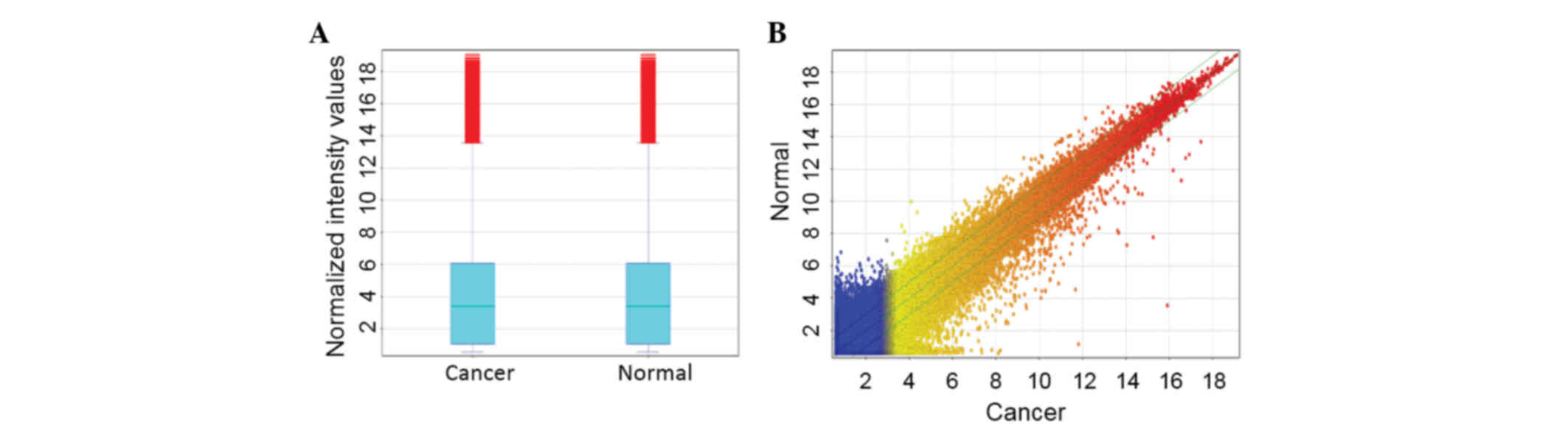
lncRNAs are aberrantly expressed in
gastric cancer compared with adjacent non-cancerous tissues
To investigate the potential biological functions of
lncRNAs in gastric cancer, the lncRNA expression profiles in human
gastric cancer were examined using microarray analysis. The lncRNA
expression profiling data revealed 41,053 lncRNAs expressed in
gastric cancer (Fig. 1); of these,
1,102 lncRNAs showed different expression profiles (fold-change,
≥2.0 or ≤0.5; P<0.01) between the gastric cancer and adjacent
non-cancerous tissues. Among these, 448 lncRNAs were upregulated
and 654 were downregulated in the gastric cancer tissues compared
with the adjacent non-cancerous tissues. MLLT4-AS1 was
significantly downregulated (fold-change, 0.48).
MLLT4-AS1 is downregulated in human
gastric carcinoma tissues
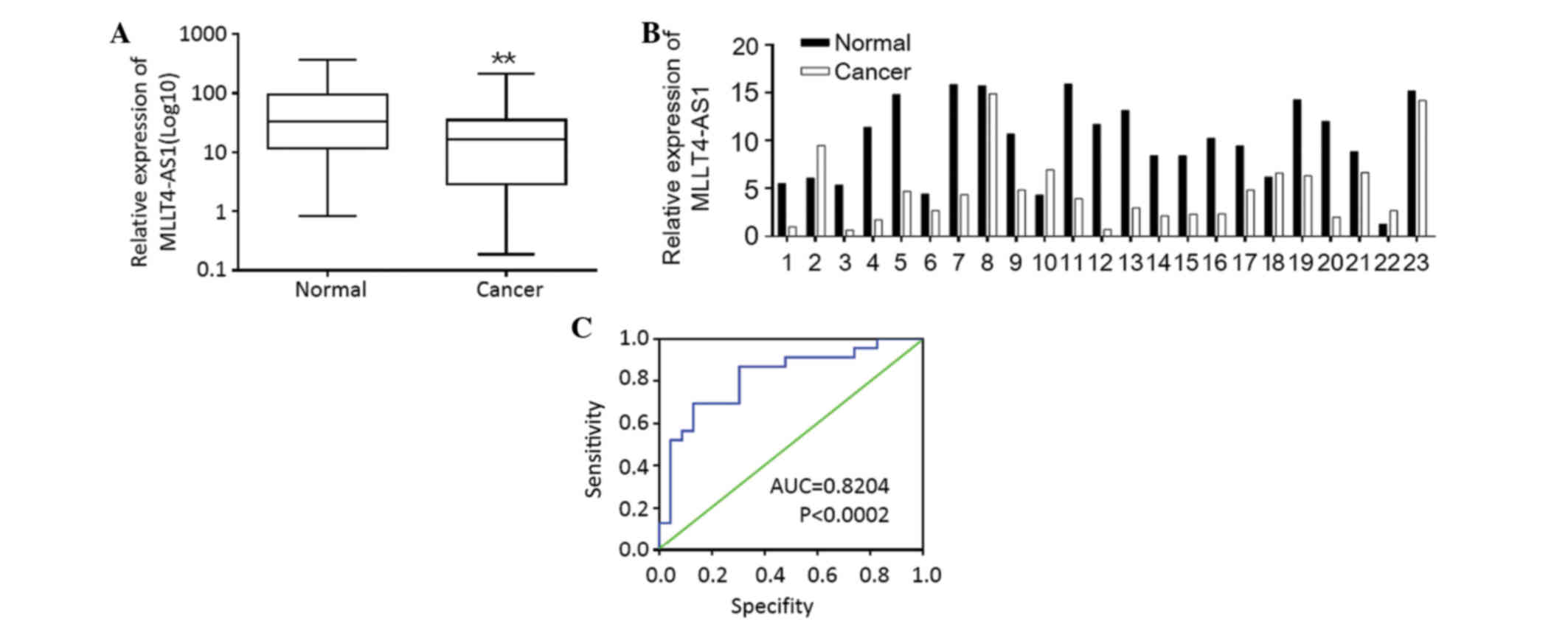
The expression of MLLT4-AS1, which was identified as
a significantly downregulated lncRNA in gastric cancer, was further
examined in 98 pairs of human gastric cancer and adjacent
non-cancerous tissues using qPCR. Downregulation of MLLT4-AS1 was
detected in 77/98 (78.6%) gastric cancer samples compared with
their non-tumorous counterparts (P=0.006; Fig. 2A), indicating that MLLT4-AS1 was
frequently downregulated in gastric cancer.
Next, the association between MLLT4-AS1 expression
and various clinicopathological parameters was evaluated. Low
MLLT4-AS1 expression was positively correlated with advanced TNM
stage (P=0.007) and LN metastasis (P=0.008). No significant
correlation was observed between MLLT4-AS1 expression and gender,
age, location of tumor, size of tumor, liver metastasis, Lauren's
classification or serum CEA levels (Table
I).
 | Table I.Association between MLLT4-AS1
expression and clinicopathological features. |
Table I.
Association between MLLT4-AS1
expression and clinicopathological features.
|
|
| MLLT4-AS1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
variable | n | Low | High | χ2 | P-value |
|---|
| All cases | 98 | 77 | 21 |
|
|
| Age, years |
|
|
| 0.766 | 0.381 |
| ≤50 | 36 | 30 | 6 |
|
|
|
>50 | 62 | 47 | 15 |
|
|
| Gender |
|
|
| 0.001 | 0.972 |
| Male | 51 | 40 | 11 |
|
|
|
Female | 47 | 37 | 10 |
|
|
| HP |
|
|
| 1.787 | 0.181 |
|
Positive | 50 | 42 | 8 |
|
|
|
Negative | 48 | 35 | 13 |
|
|
| Size of tumor,
cm |
|
|
| 1.265 | 0.261 |
| <5
(small) | 32 | 23 | 9 |
|
|
| ≥5
(large) | 66 | 54 | 12 |
|
|
| Location of
tumor |
|
|
| 1.874 | 0.392 |
|
Cardia | 22 | 15 | 7 |
|
|
|
Body | 25 | 20 | 5 |
|
|
|
Antrum | 51 | 42 | 9 |
|
|
| Depth of tumor
invasion |
|
|
| 0.466 | 0.495 |
|
T1-T2 | 39 | 32 | 7 |
|
|
|
T3-T4 | 59 | 45 | 14 |
|
|
| Lymph node
metastasis |
|
|
| 7.052 | 0.008 |
|
Present | 75 | 64 | 11 |
|
|
|
Absent | 23 | 13 | 10 |
|
|
| Liver
metastasis |
|
|
| 0.429 | 0.513 |
|
Absent | 69 | 53 | 16 |
|
|
|
Present | 29 | 24 | 5 |
|
|
| Invasion of
contiguous organs |
|
|
| 3.655 | 0.056 |
|
Yes | 26 | 17 | 9 |
|
|
| No | 72 | 60 | 12 |
|
|
| Vessel
invasion |
|
|
| 0.839 | 0.360 |
|
Negative | 52 | 39 | 13 |
|
|
|
Positive | 46 | 38 | 8 |
|
|
| Stage |
|
|
| 7.289 | 0.007 |
| I,
II | 32 | 20 | 12 |
|
|
| III,
IV | 66 | 57 | 9 |
|
|
| Lauren's
classification |
|
|
| 0.705 | 0.401 |
|
Diffuse | 30 | 22 | 8 |
|
|
|
Intestinal | 68 | 55 | 13 |
|
|
| Grade of
differentiation |
|
|
| 1.767 | 0.184 |
| Well
and moderate | 39 | 28 | 11 |
|
|
| Poor
and undifferentiated | 59 | 49 | 10 |
|
|
| Pre-operative
chemotherapy |
|
|
| 1.445 | 0.229 |
|
Yes | 44 | 37 | 7 |
|
|
| No | 54 | 40 | 14 |
|
|
| Serum CEA value,
µg/l |
|
|
| 0.105 | 0.746 |
|
<5 | 59 | 47 | 12 |
|
|
| ≥5 | 39 | 30 | 9 |
|
|
Downregulation of MLLT4-AS1 is
associated with LN metastasis
LN metastasis is one of the most important
prognostic factors in patients with gastric cancer. To further
investigate the role of MLLT4-AS1 in LN metastasis, MLLT4-AS1
expression was compared between 23 paired LN specimens using
RT-qPCR. Each paired LN specimen consisted of one LN with
metastasis and one without metastasis, derived from the same
patient. Overall, 19/23 pairs of LNs (82.6%) showed lower MLLT4-AS1
expression in the metastatic LNs than in their matched
non-metastatic counterparts (P=0.017; Fig. 2B).
In addition, the study investigated whether
MLLT4-AS1 expression status in the primary tumor could predict the
presence of LN metastasis. Calculation of predictive values by
receiver operating curve analysis showed that the area under the
curve was 0.8204 (Fig. 2C).
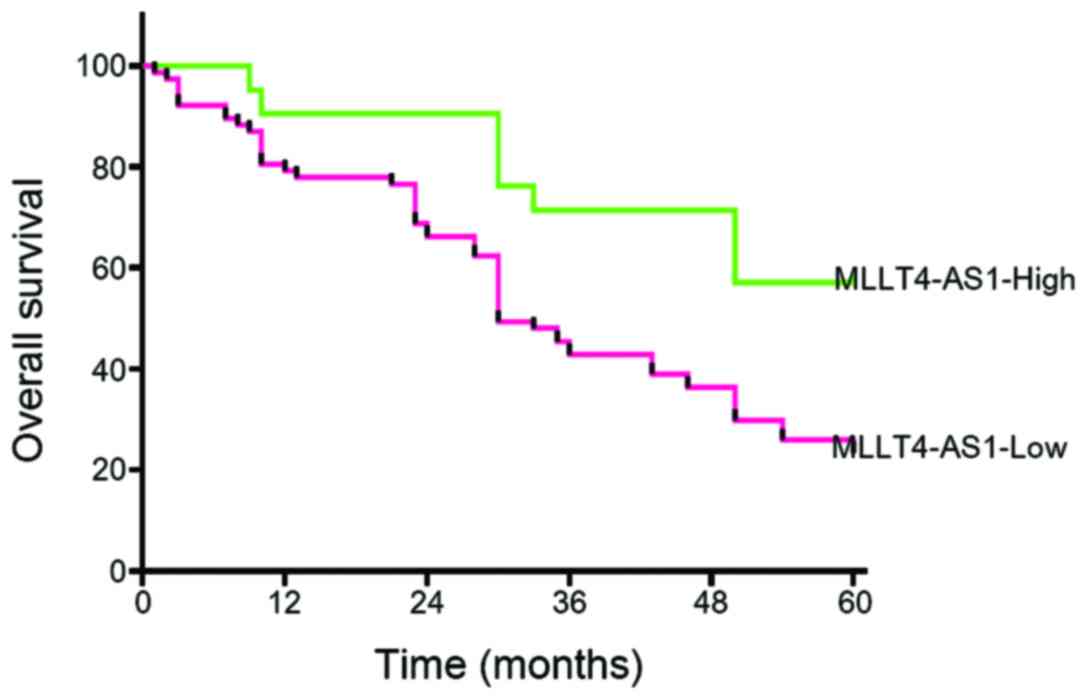
MLLT4-AS1 expression and clinical
outcomes
The 1-, 3- and 5-year cumulative survival rates for
patients with high MLLT4-AS1 expression were 90, 71 and 57%
respectively, whereas the corresponding values for patients with
low MLLT4-AS1 expression were 78, 43 and 23%, respectively. These
results indicated that gastric cancer patients with low MLLT4-AS1
expression had a poorer prognosis than those with high MLLT4-AS1
expression (P<0.05; Fig. 3).
Potential prognostic factors of 98 cases gastric cancer patients
were analyzed by the Cox's proportional hazards regression model to
investigate the association between patient survival and several
clinicopathological parameters (Table
II). The results indicated that MLLT4-AS1 expression was an
independent prognostic factor for patients with gastric cancer
[Hazard ratio (HR), 13.136; 95% CI, 5.065–34.068; P<0.001], in
addition to the TNM stage (HR, 6.489; 95% CI, 2.932–14.360;
P<0.001) and LN metastasis (HR, 4.330; 95% CI, 1.572–11.930;
P=0.005)(Table II).
 | Table II.Univariate and multivariate analyses
of factors associated with overall survival. |
Table II.
Univariate and multivariate analyses
of factors associated with overall survival.
|
|
| Multivariate |
|---|
|
|
|
|
|---|
| Clinicopathological
variable | Univariate
P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age: ≤50 vs. >50
years | 0.301 | 0.914 | 0.349–2.393 | 0.855 |
| Gender: Male vs.
female | 0.342 | 1.303 | 0.708–2.393 | 0.396 |
| HP: Positive vs.
negative | 0.280 | 0.824 | 0.459–1.480 | 0.518 |
| Size: <5 vs. 5
cm | 0.262 | 0.962 | 0.542–1.707 | 0.893 |
| Location: Cardia
vs. body vs. antrum | 0.324 | 1.164 | 0.935–1.449 | 0.173 |
| Invasion depth:
T1-T2 vs. T3-T4 | 0.550 | 0.824 | 0.457–1.488 | 0.522 |
| LNM: N0 vs. N1 vs.
N2 vs. N3a vs. N3b | <0.001 | 4.330 | 1.572–11.930 | 0.005 |
| Liver metastasis:
Yes vs. no | 0.254 | 1.192 | 0.633–2.245 | 0.586 |
| MLLT4-AS1: High vs.
low | <0.001 | 13.136 | 5.065–34.068 | <0.001 |
| Invasion of
contiguous organs: Yes vs. no | 0.869 | 0.684 | 0.356–1.314 | 0.254 |
| Microvessel
invasion: Yes vs. no | 0.823 | 1.156 | 0.676–1.977 | 0.596 |
| Stage: I, II vs.
III, IV | <0.001 | 6.489 | 2.932–14.360 | <0.001 |
| Lauren's
classification: Diffuse vs. intestinal | 0.618 | 0.724 | 0.371–1.416 | 0.724 |
| Grade of
differentiation: Well and moderate vs. poor | 0.650 | 0.960 | 0.534–1.725 | 0.892 |
| Preoperative
chemotherapy: Yes vs. no | 0.030 | 1.100 | 0.613–1.974 | 0.750 |
| CEA: 5 vs. >5
µg/ml | 0.797 | 0.660 | 0.376–1.158 | 0.147 |
Discussion
The present study showed for the first time that the
lncRNA MLLT4-AS1 is downregulated in gastric cancer tissues. The
downregulation of MLL4-AS1 expression was significantly associated
with histological grade, LN metastasis, distant metastasis and a
shorter disease-free interval. These data suggested that MLLT4-AS1
functions as a tumor suppressor gene and that downregulation of
MLLT4-AS1 is a potential predictor of a poor disease prognosis.
Two issues remain to be addressed. Firstly, the
mechanism by which MLLT4-AS1 is silenced in gastric cancer. In
cancer cells, tumor suppressive genes are usually silenced by
genetic (18) and epigenetic
(19) alterations. Two main pathways
are involved in the process of genetic alteration. One pathway is
the hypermutability pathway, in which repair gene inactivation
results in an increased mutation rate, affecting a number of
different genes (20) and leading to
deregulated cancer cell proliferation. In the second pathway, the
chromosomal instability pathway, gross chromosomal alterations
result in aneuploidy of cancer cells and lead to tumor suppressor
gene inactivation and oncogene activation (21). Studies have reported that chromosome 6
is a target of chromosome instability that is associated with
gastric cancer development. Deletions of the long arm of chromosome
6 have been observed in 26–45% of primary gastric carcinomas
(22–26). Two regions on chromosome 6 undergo
heterozygous loss in primary gastric carcinomas; the region between
6q16.3 and 6q23 is lost in 50% of informative cases, whereas the
region between 6q26 and 6q27 is lost in 37% of informative cases
(27). MLLT4-AS1 is located in 6q27
(www.ensembl.org), which indicates that the
silencing of MLLT4-AS1 in gastric cancer may result from the
heterozygous loss of regions on chromosome 6. However, the
possibility that epigenetic alterations may also play a role cannot
be excluded.
The second issue to be addresses is the mechanism
linking MLLT4-AS1 loss to enhanced gastric cancer metastasis. To
date, the majority of well-characterized lncRNAs have exhibited a
functional role in gene expression regulation, and normally in
transcriptional rather than post-transcriptional regulation. This
may occur through the targeting of genomically local
(cis-regulation) or genomically distant (trans-regulation) genes
(28). Typically, antisense lncRNAs
regulate gene transcription by suppressing the expression of their
sense counterparts (29). The
counterpart of MLLT4-AS1 is MLLT4, which encodes afadin/AF6, an
actin-binding protein that regulates cell-cell adhesions. Previous
studies have revealed an association between afadin/AF6 and cancer
(30–32). For instance, loss of afadin/AF6
expression, which is associated with adverse prognosis and
increased risk of metastatic relapse in breast cancer, induces cell
migration, invasiveness, and tumor growth (33). Nevertheless, in future studies, it
would be of interest to investigate whether the role of MLLT4-AS1
in gastric cancer metastasis involves the regulation of the
expression of its sense counterpart.
In summary, the present study showed that the lncRNA
MLLT4-AS1 was downregulated in gastric cancer. Decreased expression
of MLLT4-AS1 was associated with LN metastasis and a poor prognosis
in patients with gastric cancer. These data suggest that MLLT4-AS1
is a potential biomarker for the diagnosis of gastric cancer.
Acknowledgements
This study was supported by the Medical Leading
Project of Songjiang Commission of Health and Family Planning
(grant no. 2011LX07), the Youth Scientific Research Fund of
Shanghai Municipal Commission of Health and Family Planning (grant
no. 20144Y0162), the Key Medical Specialties Fund of Shanghai
(grant no. ZK2012A38), the Science and Technology Development Fund
of Nanjing Medical University (grant no. 2016NJMU161) and the
Shanghai Municipal Natural Science Foundation (grant no.
16ZR1432000).
References
|
1
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: Defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hartgrink HH, Jansen EP, van Grieken NC
and van de Velde CJ: Gastric cancer. Lancet. 374:477–490. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mattick JS: Challenging the dogma: The
hidden layer of non-protein-coding RNAs in complex organisms.
Bioessays. 25:930–939. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mattick JS: RNA regulation: A new
genetics? Nat Rev Genet. 5:316–323. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Szymanski M, Barciszewska MZ, Erdmann VA
and Barciszewski J: A new frontier for molecular medicine:
Noncoding RNAs. Biochim Biophys Acta. 1756:65–75. 2005.PubMed/NCBI
|
|
7
|
Prasanth KV and Spector DL: Eukaryotic
regulatory RNAs: An answer to the ‘genome complexity’ conundrum.
Genes Deve. 21:11–42. 2007. View Article : Google Scholar
|
|
8
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pauli A, Rinn JL and Schier AF: Non-coding
RNAs as regulators of embryogenesis. Nat Rev Genet. 12:136–149.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales D Rivea, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:pp. 11667–11672. 2009;
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spitale RC, Tsai MC and Chang HY: RNA
templating the epigenome: Long noncoding RNAs as molecular
scaffolds. Epigenetics. 6:539–543. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Costa FF: Non-coding RNAs: Meet thy
masters. Bioessays. 32:599–608. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Popov N and Gil J: Epigenetic regulation
of the INK4b-ARF-INK4a locus: In sickness and in health.
Epigenetics. 5:685–690. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA, Liu CG, Ferracin M, Hyslop T,
Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et
al: Ultraconserved regions encoding ncRNAs are altered in human
leukemias and carcinomas. Cancer Cell. 12:215–229. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sobin LH and Compton CC: TNM seventh
edition: What's new, what's changed: Communication from the
International Union Against Cancer and the American Joint Committee
on Cancer. Cancer. 116:5336–5339. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak and Schmittgen: Analysis of relative
gene expression data using real-time quantitative PCR and the
2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nault JC and Zucman-Rossi J: Genetics of
hepatobiliary carcinogenesis. Semin Liver Dis. 31:173–187. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanwal R and Gupta S: Epigenetic
modifications in cancer. Clin Genet. 81:303–311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oliveira C, Seruca R, Seixas M and
Sobrinho-Simões M: The clinicopathological features of gastric
carcinomas with microsatellite instability may be mediated by
mutations of different ‘target genes’: A study of the TGF beta RII
IGFII R and BAX genes. Am J Pathol. 153:1211–1219. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dos Santos NR and Van Kessel AG:
Chromosomal abnormalities: Detection and implications for cancer
development. Anticancer Res. 19:4697–4714. 1999.PubMed/NCBI
|
|
22
|
Ochi H, Douglass HO Jr and Sandberg AA:
Cytogenetic studies in primary gastric cancer. Cancer Genet
Cytogenet. 22:295–307. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seruca R, Castedo S, Correia C, Gomes P,
Carneiro F, Soares P, de Jong B and Sobrinho-Simões M: Cytogenetic
findings in eleven gastric carcinomas. Cancer Genet Cytogenet.
68:42–48. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Panani AD, Ferti A, Malliaros S and Raptis
S: Cytogenetic study of 11 gastric adenocarcinomas. Cancer Genet
Cytogenet. 81:169–172. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seruca R, Constancia M, Dossantos N, David
L, Queimado L, Carvalho F and Carneiro F: Allele loss in human
gastric carcinomas-relation to tumor progression and
differentiation. Int J Oncol. 7:1159–1166. 1995.PubMed/NCBI
|
|
26
|
Gleeson CM, Sloan JM, McGuigan JA, Ritchie
AJ, Weber JL and Russell SE: Allelotype analysis of adenocarcinoma
of the gastric cardia. Br J Cancer. 76:1455–1465. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Queimado L, Seruca R, Costa-Pereira A and
Castedo S: Identification of two distinct regions of deletion at 6q
in gastric carcinoma. Genes Chromosomes Cancer. 14:28–34. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Morris KV and Vogt PK: Long antisense
non-coding RNAs and their role in transcription and oncogenesis.
Cell Cycle. 9:2544–2547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu Y, Chang R, Peng Z, Wang Y, Ji W, Guo
J, Song L, Dai C, Wei W, Wu Y, et al: Loss of polarity protein AF6
promotes pancreatic cancer metastasis by inducing Snail expression.
Nat Commun. 6:71842015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun TT, Wang Y, Cheng H, Xiao HZ, Xiang
JJ, Zhang JT, Yu SB, Martin TA, Ye L, Tsang LL, et al: Disrupted
interaction between CFTR and AF-6/afadin aggravates malignant
phenotypes of colon cancer. Biochim Biophys Acta. 1843:618–628.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamamoto T, Mori T, Sawada M, Matsushima
H, Ito F, Akiyama M and Kitawaki J: Loss of AF-6/afadin induces
cell invasion, suppresses the formation of glandular structures and
might be a predictive marker of resistance to chemotherapy in
endometrial cancer. BMC Cancer. 15:2752015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fournier G, Cabaud O, Josselin E, Chaix A,
Adélaïde J, Isnardon D, Restouin A, Castellano R, Dubreuil P,
Chaffanet M, et al: Loss of AF6/afadin, a marker of poor outcome in
breast cancer, induces cell migration, invasiveness and tumor
growth. Oncogene. 30:3862–3874. 2011. View Article : Google Scholar : PubMed/NCBI
|