Introduction
Primary liver cancer is one of the most common
malignant tumors, and hepatocellular carcinoma (HCC) is the main
type (1). HCC is the result of a
complex pathological process involving multiple factors that has an
insidious onset, rapid progress and high mortality rates. The
5-year survival rate is <10% (2).
HCC mainly affects people between 40 and 50 years of age, but the
age of presentation has gradually been shifting to a younger
population; the incidence in males is higher than in females
(3). DNA damage can be caused by
various factors. The accumulation of DNA damage and errors
occurring during DNA repair can eventually lead to cell
carcinogenesis (4). DNA repair genes
include X-ray repair cross-complementing 1 (XRCC1) and xeroderma
pigmentosum complementary group D (XPD). Genetic polymorphisms in
those genes and the resulting specific interactions of their
products are a recognized basis for the occurrence of HCC. The
determination of genetic polymorphisms of those genes can be
helpful for treatment and prognosis of certain cancers (5,6). In this
study, XRCC1-194, XRCC1-280 and XPD-312 gene polymorphisms in liver
cancer patients were detected in order to provide a theoretical
basis for the treatment and prognosis of the disease.
Materials and methods
General information
A total of 172 patients with HCC in Qilu Hospital,
Shandong University, were enrolled in the study from January 2010
to September 2011. All the patients in the study had been diagnosed
with HCC by liver biopsy and imaging examination according to the
diagnostic criteria established by the American Association for the
Study of Liver Diseases (AASLD); the patients received radical
treatment and signed the informed consent form.
Patients with severe dysfunctions of the heart,
brain, lung, kidney or other organs and patients presenting mental
or neurological disorders or other diseases were excluded from the
study. There were a total of 121 males and 51 females. The age of
the patients ranged from 40 to 70 years with a mean age of
50.36±4.78 years. The patients were classified according to the
diameter of their tumor: There were 80 cases with diameters smaller
than 5 cm, 76 with diameters from 5 to 10 cm and 16 with diameters
larger than 10 cm. Also, according to the number of tumors present
in each patient, 145 cases had a single tumor and 27 cases
presented multiple tumors. In 79 cases the tumors were found in the
left lobe, in 69 cases the tumors were in the right lobe and in 24
cases in other positions. This study was approved by the Ethics
Committee of Qilu Hospital. Signed written informed consents were
obtained from the patients.
Epidemiological survey
The patient's demographic characteristics were
investigated and their medical records and physical examination
results were collected to establish a research database for
statistical analyses. Five-year follow-up was conducted and the
data collected was used in this study.
Sample collection
Peripheral blood samples (3–5 ml) were collected
after fasting for 8 h. The samples were transferred to blood
collection tubes containing ethylenediamine-tetraacetic acid. The
tubes were numbered and stored at −20°C.
XRCC1 and XPD gene detection
Blood genomic DNA extraction from the samples was
performed according to the instructions of DNA extraction kit
(Promega, Madison, WI, USA). The primers for PCR were synthesized
at Shanghai Boa Biotechnology (Shanghai, China) and the sequences
are listed in Table I. The PCR
amplification procedures followed the instructions in the One-Step
qRT-PCR kit (Invitrogen, Carlsbad, CA, USA). The protocol for the
amplification of the XRCC1-194 locus gene included an initial 95°C
for 5 min step, followed by 35 cycles of 94°C for 30 sec, 57°C for
30 sec and 72°C for 45 sec and then a final step of 72°C for 10
min. The protocol for the XRCC1-280 locus gene included an initial
step of 95°C for 5 min, followed by 35 cycles of 94°C for 30 sec,
62°C for 30 sec and 72°C for 45 sec, with a final step of 72°C for
7 min. PCR products were analyzed by agarose gel electrophoresis
(Six One Instrument Factory, Beijing, China). Briefly, a 2.0%
agarose gel was prepared, containing nucleic acid dye, the solid
gel was placed in a horizontal electrophoresis tank containing
electrophoresis buffer. A 5 µl PCR product sample was mixed with
loading buffer and the mixture was subjected to electrophoresis at
100 V for 30 min. The gel was then observed under an ultraviolet
transmittance analyzer and images were saved.
 | Table I.Primer sequences for XRCC1 and
XPD. |
Table I.
Primer sequences for XRCC1 and
XPD.
| Microsatellite
loci | Sequences | Length |
|---|
| XRCC1-194 | F:
5′-CTGACCTTGCGGGACCTTA-3′ | 455 bp |
|
| R:
5′-GTCGCTGGCTGTGACTATG-3′ |
|
| XRCC1-280 | F:
5′-CCCCAGTGGTGCTAACCTAA-3′ | 304 bp |
|
| R:
5′-CTACATGAGGTGCGTGCTGT-3′ |
|
| XPD-312 | F:
5′-CTGTTGGTGGGTGCCCGTATCTGTTGGTCT-3′ | 751 bp |
|
| R:
5′-TAATATCGGGGCTCACCCTGCAGCACTTCCT-3′ |
|
PCR products were sequenced using reagents from ABI
(Applied Biosystems, Grand Island, NY, USA). Briefly, PCR product
(1 µl) was mixed with 2 µl SAP MIX for digestion in a thermocycler
(Wenzhou Yongqiang medical equipment, Zhejiang, China). PCR
digestion products, sequencing reagents and sequencing primers were
mixed and the PCR amplification was performed. The sequencing
amplification product was mixed with a sodium acetate-ethanol
solution and centrifuged to remove the supernatant. A DNA
dissolving solution was added. The purified DNA was then subjected
to electrophoresis with a DNA sequencer.
Statistical analysis
Data were processed using SPSS 19.0 software (SPSS
Inc., Chicago, IL, USA). Countable data were expressed by number of
cases and the composition ratio. The relationship between different
genotypes and the clinical features of HCC was examined using the
χ2 test. Logistic regression analysis was used to
analyze the prognostic factors of HCC. The Kaplan-Meier survival
analysis was also performed. P-value <0.05 were considered to
indicate a statistically significant difference.
Results
Allele frequency and genotype
distribution of XRCC1 and XPD loci
The allele frequencies for CC, CT and TT genotypes
of the XRCC1-194 locus, AA and GG genotypes of the XRCC1-280 locus
and AA, GA and GG genotypes of the XPD-312 locus found in the
population of patients are listed in Table II.
 | Table II.Allele frequency and genotype
distribution for XRCC1 and XPD loci in HCC patients (n, %). |
Table II.
Allele frequency and genotype
distribution for XRCC1 and XPD loci in HCC patients (n, %).
| XRCC1-194 | Frequency | XRCC1-280 | Frequency | XPD-312 | Frequency |
|---|
| CC | 95
(55.23) | AA | 35 (20.35) | AA | 29
(16.86) |
| CT | 13 (7.56) | – | – | GA | 15 (8.72) |
| TT | 64
(37.21) | GG | 137 (79.65) | GG |
128 (74.42) |
Analysis of the relationship between
XRCC1-194, XRCC1-280 and XPD-312 gene polymorphisms and the
clinical characteristics of HCC
XRCC1-194, XRCC1-280 and XPD-312 gene polymorphisms
were significantly correlated with tumor number, tumor location and
tumor diameter (p<0.05), but were not correlated with capsule
invasion or the general type of cancer (p>0.05) (Tables III–V).
 | Table III.Relationship between XRCC1-194 gene
polymorphism and clinical characteristics of HCC. |
Table III.
Relationship between XRCC1-194 gene
polymorphism and clinical characteristics of HCC.
|
| XRCC1-194 (n, %) |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
features | CC | CT | TT | χ2 | P-value |
|---|
| General types |
|
|
| 0.653 | 0.957 |
| Giant
block type | 30 (54.55) | 4 (7.27) | 21 (38.18) |
|
|
| Nodular
type | 27 (54.00) | 3 (6.00) | 20 (40.00) |
|
|
| Diffuse
type | 38 (56.72) | 6 (8.96) | 23 (34.33) |
|
|
| No. of tumors |
|
|
| 8.621 | 0.013 |
| Single
tumor | 83 (57.24) | 7 (4.83) | 55 (37.93) |
|
|
| Multiple
tumors | 12 (44.44) | 6 (22.22) | 9 (33.33) |
|
|
| Liver tumor
location |
|
|
| 10.149 | 0.038 |
| Left
lobe | 42 (53.16) | 5 (6.33) | 32 (40.51) |
|
|
| Right
lobe | 38 (55.07) | 3 (4.35) | 28 (40.58) |
|
|
|
Other | 15 (62.50) | 5 (20.83) | 4 (16.67) |
|
|
| Tumor diameter |
|
|
| 10.432 | 0.034 |
| <5
cm | 42 (52.50) | 5 (6.25) | 33 (41.25) |
|
|
| 5-10
cm | 43 (56.58) | 4 (5.26) | 29 (38.16) |
|
|
| >10
cm | 10 (62.50) | 4 (25.00) | 2 (12.50) |
|
|
| Capsule invasion |
|
|
| 2.239 | 0.327 |
| Yes | 32 (53.33) | 7 (11.67) | 21 (35.00) |
|
|
| No | 63 (56.25) | 6 (5.36) | 43 (38.39) |
|
|
 | Table V.Relationship between XPD-312 gene
polymorphism and clinical characteristics of HCC. |
Table V.
Relationship between XPD-312 gene
polymorphism and clinical characteristics of HCC.
|
| XPD-312 (n, %) |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
features | AA | GA | GG | χ2 | P-value |
|---|
| General typing |
|
|
| 2.507 | 0.643 |
| Giant
block type | 10 (18.18) | 4 (7.27) | 41
(74.55) |
|
|
| Nodular
type | 11 (22.00) | 5
(10.00) | 34
(68.00) |
|
|
| Diffuse
type | 18 (11.94) | 6 (8.96) | 52
(79.10) |
|
|
| No. of tumors |
|
|
| 19.309 | <0.001 |
| Single
tumor | 19 (13.10) | 9 (6.21) | 117 (80.69) |
|
|
|
Multiple tumors | 10 (37.04) | 6
(22.22) | 11
(40.74) |
|
|
| Liver tumor
location |
|
|
| 11.831 | 0.019 |
| Left
lobe | 12 (15.19) | 5 (6.33) | 62
(78.48) |
|
|
| Right
lobe | 11 (15.94) | 4 (5.80) | 54
(78.26) |
|
|
|
Other | 6
(25.00) | 6
(25.00) | 12
(50.00) |
|
|
| Tumor diameter |
|
|
| 19.502 | <0.001 |
| <5
cm | 11 (13.75) | 4 (5.00) | 65
(81.25) |
|
|
| 5-10
cm | 13 (17.11) | 6 (7.89) | 57
(75.00) |
|
|
| >10
cm | 6
(37.50) | 5
(31.25) |
5 (31.25) |
|
|
| Capsule
invasion |
|
|
| 2.825 | 0.243 |
|
Yes | 11 (18.33) | 8
(13.33) | 41
(68.33) |
|
|
| No | 18 (16.07) | 7 (6.25) | 87
(77.68) |
|
|
Genotypes and survival of
patients
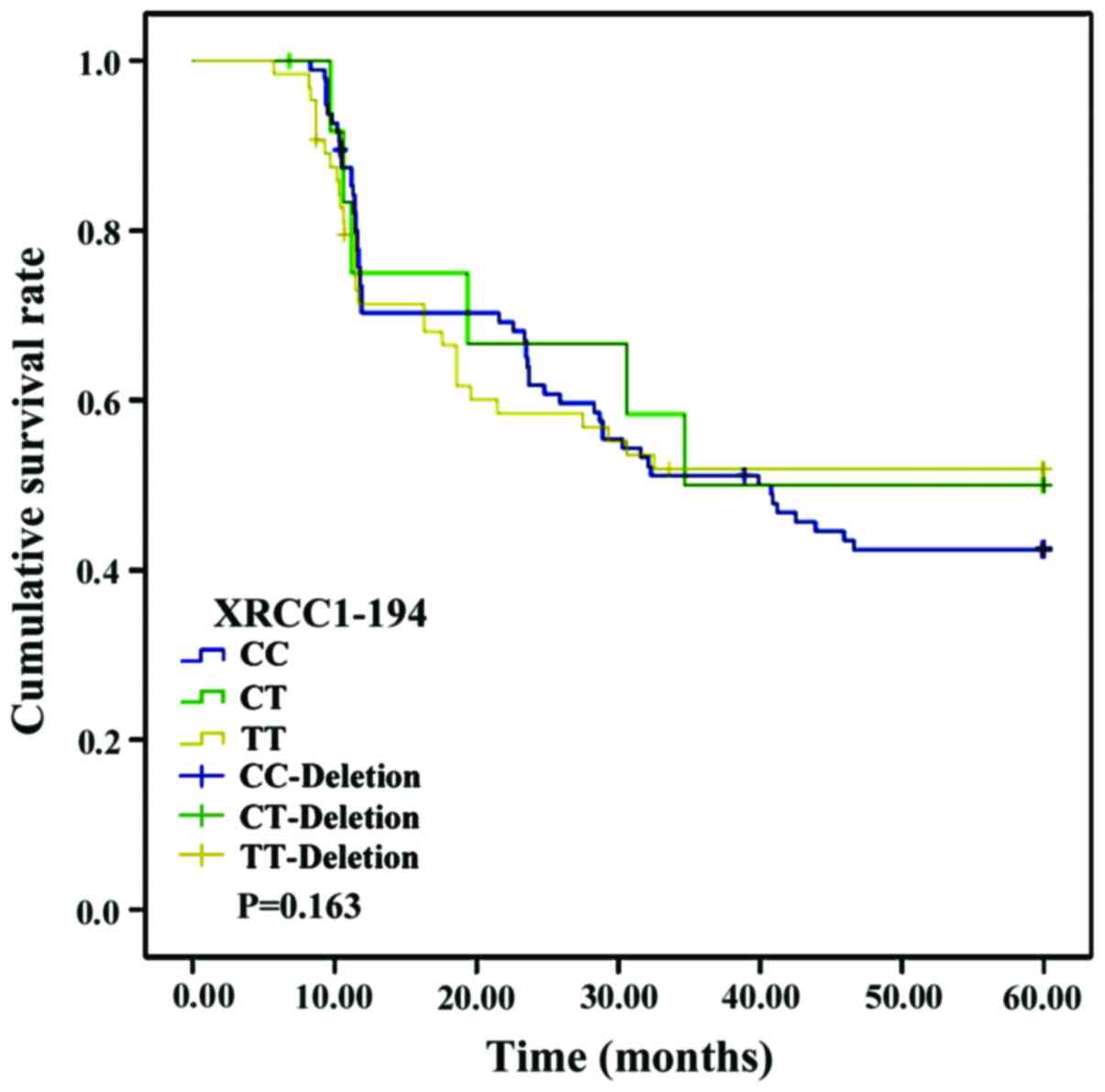
No significant differences were found in the
survival curves of patients with CC, CT or TT genotypes of the
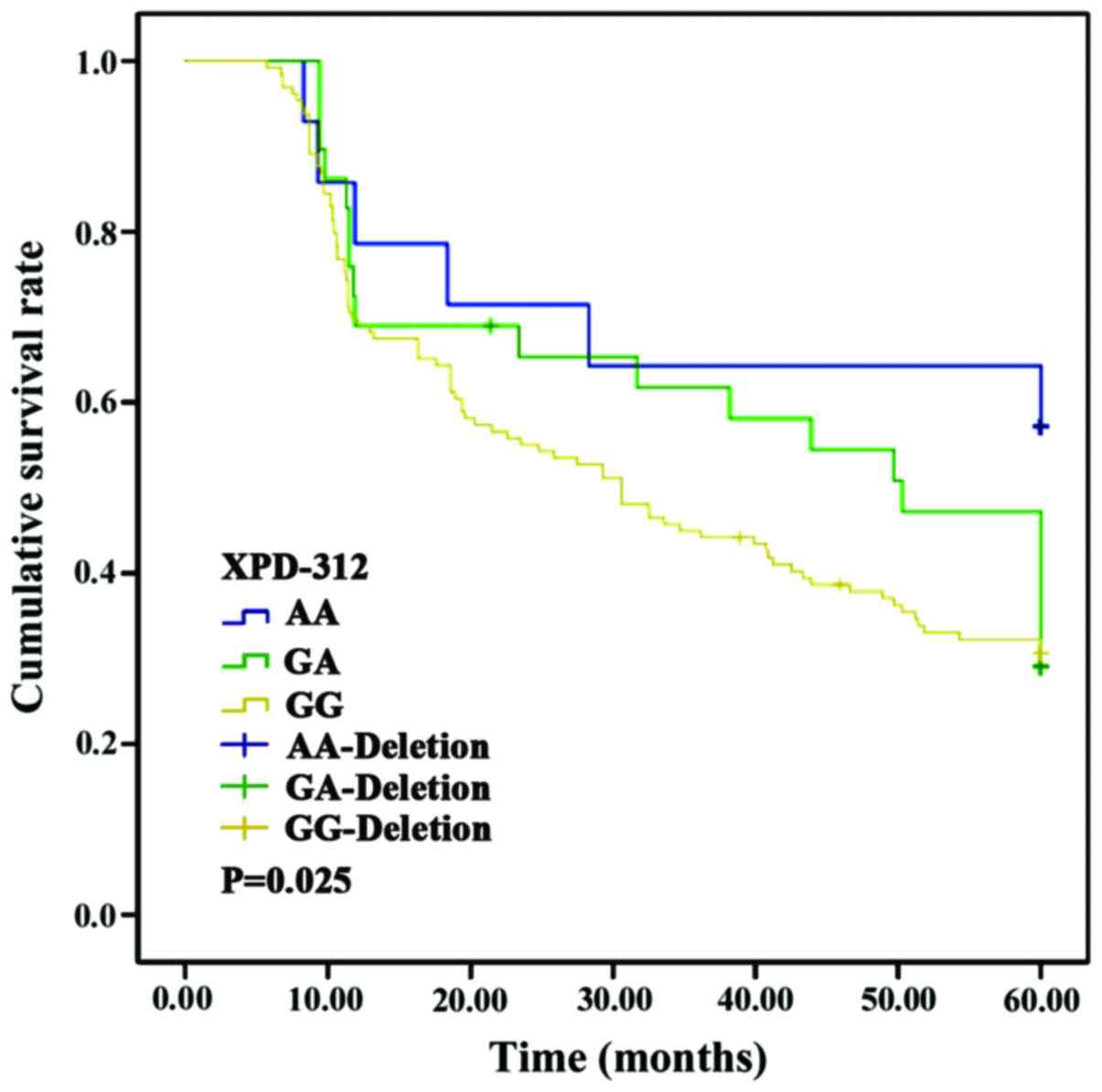
XRCC1-194 locus (p>0.05). Nevertheless, the survival curves of
patients with AA and GG genotypes at the XRCC1-280 locus and those
in patients with AA, GA and GG genotypes at the XPD-312 locus
differed significantly (p<0.05) (Figs.
1–3).
The influencing factors of HCC
prognosis
The risk of poor prognosis in patients with HCC was
set as a dependent variable; and factors such as age, sex,
educational level, living status and XRCC1-194, XRCC1-280 and
XPD-312 genotypes were set as independent variables. Logistic
regression analysis showed that age, sex, educational level, living
status and XRCC1-194 genotype (OR=0.945, p=0.734) were not
independent risk factors for a poor HCC prognosis, but XRCC1-280
(OR=1.323, p=0.006) and XPD-312 (OR=1.815, p=0.008) genotypes were
independent risk factors for poor HCC prognosis (p<0.05)
(Table VI).
 | Table VI.Logistic regression analysis of poor
prognosis factors for HCC. |
Table VI.
Logistic regression analysis of poor
prognosis factors for HCC.
| Factors | β | SE | Wald | OR | 95% CI | P-value |
|---|
| Age (years) |
0.331 | 0.512 | 3.783 | 0.631 | 0.375–0.952 | 0.214 |
| Sex |
0.437 | 0.517 | 6.372 | 0.215 | 0.104–0.779 | 0.309 |
| Educational
level | −0.467 | 0.673 | 5.321 | 0.013 | 0.456–0.854 | 0.153 |
| Living status | −0.635 | 0.814 | 6.425 | 0.217 | 0.196–0.542 | 0.217 |
| XRCC1-280 |
1.426 | 0.749 | 7.757 | 1.323 | 1.075–2.252 | 0.006 |
| XPD-312 |
1.433 | 0.517 | 4.524 | 1.815 | 1.103–3.347 | 0.008 |
| XRCC1-194 |
0.815 | 0.426 | 5.232 | 0.945 | 0.625–0.973 | 0.734 |
Discussion
Liver cancer often arises in hepatocytes or
intrahepatic bile duct cells, HCC and intrahepatic
cholangiocarcinoma are the most common types of liver cancer. HCC
accounts for more than 80% of liver cancers, and its incidence and
mortality rate both rank in the first three positions among all the
malignant tumors (3). The clinical
manifestations of HCC include flank pain, loss of appetite, weight
loss, ascites and jaundice, and even coma and systemic failure
(7,8).
The pathogenesis of HCC is complex; it involves a dynamic process
of long-term accumulation of the actions of multiple different
factors including the presence of genetic polymorphism and
environmental factors (9). At
present, treatment outcomes of HCC are not satisfactory. Radical
resection is the main treatment method, but the recurrence rate
after the procedure is still high (10). A better understanding of the genetic
underlying genetic mechanisms will aid in the development of
improved treatment strategies.
Genetic polymorphisms are usually classified into
one of three categories: Fragment length polymorphisms (FLPs),
repeated sequence polymorphisms (RSPs) and single nucleotide
polymorphisms (SNPs) (11). Genetic
polymorphisms may affect the protein expression, or the composition
of cytokine molecules leading to different clinical presentations,
and different prognoses for HCC (12). DNA repair systems play important roles
in the normal regulation of the cell cycle and maintenance of cell
integrity, and are essential body's defense mechanisms against
tumors. Altered DNA repair systems lead to the accumulation of gene
mutations in the human genome and can result in the development of
cancers (13). About 90% of mutations
in the human genome are caused by SNPs, and studies have confirmed
that gene mutations can lead to different repair capabilities that
can result in tumor malignancies (14). Ionizing radiation, ultraviolet light
and a variety of chemical factors can lead to DNA single and double
strand breaks, DNA protein cross-linking, base mismatch and other
genetic damage. The gene repair system can effectively repair these
DNA mistakes, maintaining the stability of the genetic information.
The known repair genes include hMSH2, XPD, XRCC1, hOGG1 and XRCC3
(15,16).
The length of the XRCC1 gene is ~32 kb. The XRCC1
gene contains 17 exons and 60 SNP loci; it participates in DNA
damage repair, cell cycle and DNA transcription regulation
(17). The product of XRCC1 can
combine with DNA ligase III, DNA polymerase β and polysaccharide
enzyme to form a complex, which repairs single-stranded DNA
mistakes. This complex can also act as a protein scaffold to
stablize the DNA structure (18). The
results of our study show that the XRCC1-194 alleles CC and TT were
mostly found in patients with a single tumor in the left lobe of
the liver and a tumor diameter smaller than 10 cm. On the other
hand, the XRCC1-280 GG allele was mostly found in patients
exhibiting a single left lobe tumor with a diameter smaller than 10
cm. Additionally, the XRCC1-194 and XRCC1-280 gene polymorphisms
were significantly correlated with tumor number, location and
diameter (p<0.05), indicating that the polymorphisms can be used
as markers predicting biological behavior of HCC.
The XPD gene, another DNA repair gene, contains 23
exons and 6 SNP sites. The main function of XPD is nucleotide
excision (19). However, XPD can also
participate in p53-mediated apoptosis and in transcription by being
part of the transcription factor II D (TFIID) (20). A single base mutation in XPD can cause
a change in the protein function, and lead to tumorigenesis
(21). The results of this study show
that XPD-312 AA and GG were mostly found in patients with single
tumors (<10 cm in diameter) in the left lobe of HCC patients.
The XPD-312 gene polymorphism was significantly correlated with
tumor number, location and diameter (p<0.05). This probably
means the XPD-312 SNP can alter the ability of its cognate protein
to participate in repair, ultimately promoting the progress of
HCC.
Due to the rapid growth of liver tumors, and their
vascular invasion, capsule infiltration and distant metastases, the
prognosis in HCC is extremely poor. Our study found no significant
differences in the survival curves between the patients with CC, CT
and TT genotypes of the XRCC1-194 locus (p>0.05). However, a
significant difference was found in the survival curve of patients
with AA and those with GG genotypes of the XRCC1-280 locus; and in
the patients with AA, GA and GG genotypes of the XPD-312 locus
(p<0.05). Logistic regression analysis showed that the OR of
RCC1-194 genotype is 0.945, the OR of XRCC1-280 is 1.323 and the OR
of XPD-312 is 1.815. Therefore, we think it is possible that
XRCC1-280 and XPD-312 may be involved in promoting the development
of HCC, shortening the time of recurrence and metastasis, and
negatively affecting the overall survival of patients. We are aware
of the small population size of our study and know that additional
studies are needed in order to confirm our findings. Nevertheless,
the evidence strongly suggests that XRCC1-280 and XPD-312 loci
should be useful as reference markers for predicting the evolution
of the disease in HCC patients.
References
|
1
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee SS, Shin HS, Kim HJ, Lee SJ, Lee HS,
Hyun KH, Kim YH, Kwon BW, Han JH, Choi H, et al: Analysis of
prognostic factors and 5-year survival rate in patients with
hepatocellular carcinoma: A single-center experience. Korean J
Hepatol. 18:48–55. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kew MC: Hepatocellular carcinoma:
Epidemiology and risk factors. J Hepatocell Carcinoma. 1:115–125.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seguí N, Mina LB, Lázaro C, Sanz-Pamplona
R, Pons T, Navarro M, Bellido F, López-Doriga A, Valdés-Mas R,
Pineda M, et al: Germline mutations in FAN1 cause hereditary
colorectal cancer by impairing DNA repair. Gastroenterology.
149:563–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Patrono C, Sterpone S, Testa A, Verna L,
Palma V, Gentile P and Cozzi R: Polymorphisms in X-ray repair
cross-complementing group 1 gene: Haplotypes, breast cancer risk
and individual radiosensitivity. Open Med J. 2:25–30. 2015.
View Article : Google Scholar
|
|
6
|
Tamura D, DiGiovanna JJ, Khan SG and
Kraemer KH: Living with xeroderma pigmentosum: Comprehensive
photoprotection for highly photosensitive patients. Photodermatol
Photoimmunol Photomed. 30:146–152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang YX, De Baere T, Idee JM and Ballet S:
Transcatheter embolization therapy in liver cancer: An update of
clinical evidences. Chin J Cancer Res. 27:96–121. 2015.PubMed/NCBI
|
|
8
|
Hung HH, Chao Y, Chiou YY, Li CP, Lee RC,
Huo TI, Huang YH, Chau GY, Su CW, Yeh YC, et al: A comparison of
clinical manifestations and prognoses between patients with
hepatocellular carcinoma and Child-Pugh scores of 5 or 6. Medicine
(Baltimore). 93:e3482014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu F, Wang J, Chang H, Lu J and Li H:
Relevance between HLA-DP gene rs2281388 polymorphism and
hepatocellular carcinoma risk. Int J Clin Exp Pathol. 8:7431–7435.
2015.PubMed/NCBI
|
|
10
|
Huang X, Zeng Y, Xing X, Zeng J, Gao Y,
Cai Z, Xu B, Liu X, Huang A and Liu J: Quantitative proteomics
analysis of early recurrence/metastasis of huge hepatocellular
carcinoma following radical resection. Proteome Sci. 12:222014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Petta S, Miele L, Bugianesi E, Cammà C,
Rosso C, Boccia S, Cabibi D, Di Marco V, Grimaudo S, Grieco A, et
al: Glucokinase regulatory protein gene polymorphism affects liver
fibrosis in non-alcoholic fatty liver disease. PLoS One.
9:e875232014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akhdar H, El Shamieh S, Musso O, Désert R,
Joumaa W, Guyader D, Aninat C, Corlu A and Morel F: The
rs3957357C>T SNP in GSTA1 is associated with a higher risk of
occurrence of hepatocellular carcinoma in european individuals.
PLoS One. 11:e01675432016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Duxin JP and Walter JC: What is the DNA
repair defect underlying Fanconi anemia? Curr Opin Cell Biol.
37:49–60. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bharati R, Jenkins MA, Lindor NM, Le
Marchand L, Gallinger S, Haile RW, Newcomb PA, Hopper JL and Win
AK: Does risk of endometrial cancer for women without a germline
mutation in a DNA mismatch repair gene depend on family history of
endometrial cancer or colorectal cancer? Gynecol Oncol.
133:287–292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Caldecott KW: DNA single-strand break
repair. Exp Cell Res. 329:2–8. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fahmideh M Adel, Schwartzbaum J, Frumento
P and Feychting M: Association between DNA repair gene
polymorphisms and risk of glioma: A systematic review and
meta-analysis. Neuro Oncol. 16:807–814. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang JY and Cai Y: X-ray repair
cross-complementing group 1 codon 399 polymorphism and lung cancer
risk: An updated meta-analysis. Tumour Biol. 35:411–418. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Ma QH, Huang ZY, Li SW, Liu F and
Tan S: P0163 X-ray repair cross-complementing group 1 (XRCC1)
genetic polymorphisms and cervical cancer risk: A systematic review
and meta-analysis. Eur J Cancer. 50:e552014. View Article : Google Scholar
|
|
19
|
Liu J, Fang H, Chi Z, Wu Z, Wei D, Mo D,
Niu K, Balajee AS, Hei TK, Nie L, et al: XPD localizes in
mitochondria and protects the mitochondrial genome from oxidative
DNA damage. Nucleic Acids Res. 43:5476–5488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bănescu C, Trifa AP, Demian S, Lazar E
Benedek, Dima D, Duicu C and Dobreanu M: Polymorphism of XRCC1,
XRCC3, and XPD genes and risk of chronic myeloid leukemia. BioMed
Res Int. 2014:2137902014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li P, Wang YD, Cheng J, Chen JC and Ha MW:
Association between polymorphisms of BAG-1 and XPD and chemotherapy
sensitivity in advanced non-small-cell lung cancer patients treated
with vinorelbine combined cisplatin regimen. Tumour Biol.
36:9465–9473. 2015. View Article : Google Scholar : PubMed/NCBI
|