Introduction
Renal cell carcinoma (RCC) is the most common
primary renal malignancy, and accounts for 2–3% of adult
malignancies (1). In China, the
incidence of RCC is ~540 cases per million individuals every year
(2). It has been shown that RCC has a
high mortality rate, and the 5-year survival rate of metastatic RCC
patients is <10% (3). RCC can be
divided into several subtypes according to the morphological and
microscopic features, and clear cell RCC (ccRCC) is the most
predominant subtype, which accounts for 75–80% of all RCCs
(4).
The tumor suppressor serine-threonine kinase 11
(STK11), also termed liver kinase B1 (LKB1), was
first identified as a germline-mutated gene in Peutz-Jeghers
Syndrome in 1996 (5). The product of
the STK11 gene is a 50-kDa serine-threonine kinase involved
in various biological functions, including cell polarity, cell
detachment and adhesion, cell structure and energy metabolism
(6). Germline mutations of the
STK11 gene are found in a variety of cancer types, including
lung cancer (7), hepatocellular
carcinoma (8) and breast cancer
(9). In addition, functional studies
showed that STK11 heterozygous knockout mice would develop
tumors in several organs (10,11).
Somatic mutations of the STK11 gene have also been
found in several tumors, including pancreatic cancer (12), biliary cancer (12), hepatocellular carcinoma (8) and testicular tumor (13); however, the frequency of mutations is
relatively rare, with a range of 0–6%. In lung cancer, a
geographically variable incidence was observed. Mutational
inactivation of the STK11 gene is frequently detected in
Caucasian, but not in Asian, lung cancer patients (14). As for RCC, a study by Avizienyte et
al (15) detected no somatic
mutations in the STK11 gene in 19 RCC specimens, whereas a
controversial result was observed by Yalniz et al (16), in which 51.6% of RCC patients were
found to have somatic mutations in the STK11 gene. Decreased
expression of STK11 has also been shown to occur in several
cancer types, such as non-small cell lung cancer (8), breast carcinoma (17) and ccRCC. Duivenvoorden et al
(18) showed that under-expression of
STK11 was a common event in all 10 examined ccRCC samples.
However, the mechanism of reduced expression of STK11 in
ccRCC remains to be elucidated.
Although inactivation of STK11 gene was found
in several cancers, its somatic mutations appear rare. This
indicates that the under-expression of the STK11 gene may be
also mediated by other mechanisms. In addition to mutation, the
expression of STK11 can also be regulated through epigenetic
modification, transcriptional regulation and post-translational
modification (19). Epigenetic
alterations that suppress the activity of tumor suppressor genes is
an alternative mechanism for tumor development and progression
(20). The methylation status of the
STK11 promoter has been investigated in colorectal cancer
(21), non-small cell lung cancers
(22), and breast, gastric,
pancreatic, thyroid, bladder and testicular carcinomas (23). These studies reported that frequency
of hypermethylation of the STK11 promoter in the described
tumors is low (0–13%) (21–23); however, this indicates that
STK11 promoter methylation contributes to the inactivation
of the STK11 gene and STK11-mediated functions.
At present, the methylation status of the
STK11 promoter in RCC cells remains unclear. In addition,
the role of the methylation status of the STK11 promoter in
the pathogenesis of RCC remains to be investigated. In order to
determine the possible inactivation of STK11 by epigenetic
mechanisms, the present study aimed to investigate the methylation
status of the STK11 promoter in ccRCC and its association
with tumor disease stage and survival of ccRCC patients. The
methylation status of the STK11 promoter in RCC was
determined by analysis of 42 cases of ccRCC Paraffin-embedded
specimens were assessed using methylation-specific polymerase chain
reaction (MSP) and the association with RCC progression was
analyzed.
Patients and methods
Patients
The present study was reviewed and approved by the
Institutional Review Board of the First Affiliated Hospital of Sun
Yat-sen University (Guangzhou, China). Paraffin-embedded tumor
specimens were obtained and prepared from 42 patients with ccRCC
(29 men and 13 women) admitted to the First Affiliated Hospital of
Sun Yat-sen University between February 1999 and August 2009.
Patients pathologically diagnosed with ccRCC were included, and
patients that did not comply with follow-up visits were excluded
from the study. Clinical data, including the tumor-node-metastasis
(TNM)/American Joint Committee on Cancer (AJCC) staging (9), hematological parameters and
post-operative follow-up were documented for further analyses.
Written informed consent was obtained from all patients prior to
inclusion in the present study.
MSP
A total of 42 paraffin-embedded tissues were
sectioned into 5 µm thick slices, then dried for 2 h at 60°C or
overnight at 37°C. The slices were immersed in 2X xylene for 15
min, and then dehydrated through a graded series of ethanol (70,
80, 90 and 95%; 5 min each). DNA from the tissue samples was
isolated using QIAamp DNA FFPE Tissue Kit (Qiagen, Inc., Valencia,
CA, USA), according to the manufacture's protocol. EZ DNA
Methylation kit (Zymo Research Corp., Irvine, CA, USA) was used for
bisulfite conversion to assess the DNA methylation status. The
bisulfite-modified DNA was then used as a template, together with
primers specific for methylated and unmethylated sequences for MSP.
Polymerase chain reaction (PCR) was performed with DNA polymerase
(Beijing Sunbiotech, Beijing, China) at a final volume of 25 µl.
The primers used are as previously reported (10): Primers specific for methylated
sequence were STK11 forward 5′-ACGAAGTTGATTTTGATCGGGTC-3′
and reverse 5′-CGATACAAAATCTACGAACCGACG-3′, whereas those for the
unmethylated sequence were STK11 forward,
5′-GGATGAAGTTGATTTTGATTGGGTT-3′, and reverse,
5′-ACCCAATACAAAATCTACAAACCAACA-3′; GAPDH forward
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse
5′-GGCTGTTGTCATACTTCTCATGG-3′. All primers were synthesized and
purchased from Zymo Research (USA). PCR fragments were 122 bp in
length. The reaction consisted of initialization at 95°C for 10
min, followed by 35 cycles of denaturation at 95°C for 30 sec,
cooling at 57°C for 59 sec and extension at 72°C for 30 sec, and
final extension at 72°C for 10 min. The PCR products were analyzed
on a 1% agarose gel (Beijing Hengao Biotechnology, Beijing, China);
DNA bands were captured using a UV gel imaging system (EC3 Imaging
system, UVP LLC, Upland, CA, USA). The presence of methylated and
unmethylated bands in the PCR product indicated partial
methylation, the presence of only a methylated band indicated
methylation, and the presence of only an unmethylated band
indicated unmethylation.
Statistical analysis
Statistical analysis was performed using R 3.0.2.
All data are presented as the mean ± standard deviation.
Significance was assessed using analysis of variance followed by
Tukey Honestly Significant Difference test for all baseline
characteristics and hematological parameters, with the exception of
the TNM stage, AJCC stage, blood type and sex, which were analyzed
using Fisher's exact test. Survival curves of patients in the three
groups were plotted and the differences between the three curves
were estimated by log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Methylation status of STK11 promoter
in ccRCC
The methylation status of the STK11 promoter
in 42 ccRCC paraffin-embedded tissue samples was determined using
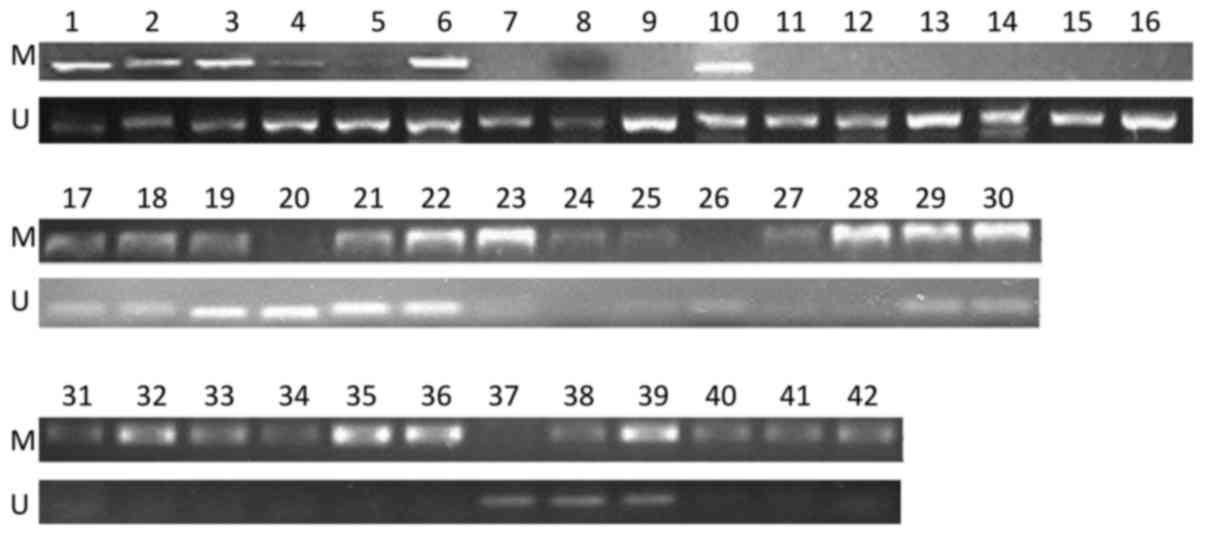
MSP. The data showed that, among the 42 samples, there were 12
(28.6%), 18 (42.9%) and 12 (28.6%) samples in the methylation group
(M group), partial methylation group (P group) and unmethylation
group (U group), respectively, based on the status of the
STK11 promoter (Fig. 1).
Patient demographic data
To investigate the effect of the methylation status
of the STK11 promoter on ccRCC, the 42 enrolled patients
were grouped into the M, P and U groups, according to the
methylation status of the STK11 promoter. The patient
demographic data of the 3 groups is shown in Table I. In general, with the exception of
the follow-up time, there were no significant differences in the
clinical characteristics among the 3 groups. Comparison of
hematological parameters among the three groups also showed no
significant difference (Table II).
These results revealed the equivalence of demographic and clinical
characteristics of the three groups.
 | Table I.Demographic data of clear cell renal
cell carcinoma patients in the M, P and U groups with regard to
serine-threonine kinase 11 promoter status (n=42). |
Table I.
Demographic data of clear cell renal
cell carcinoma patients in the M, P and U groups with regard to
serine-threonine kinase 11 promoter status (n=42).
|
| Group |
|
|---|
|
|
|
|
|---|
| Characteristic | M | P | U | P-value |
|---|
| Total, n | 12 | 18 | 12 |
|
| Sex, n (%) |
|
|
| 0.1783 |
|
Male | 10 (83.3) | 9 (50.0) | 9 (75.0) |
|
|
Female | 2 (16.7) | 9 (50.0) | 3 (25.0) |
|
| Age, years (SD,
range) | 50.8 (13.4,
33–82) | 44.4 (15.5,
14–68) | 49.9 (12.9,
27–67) | 0.413 |
| Height, cm (SD,
range) | 165.1 (8.7,
152–176) | 160.2 (9.5,
150–178) | 163.8 (7.8,
147–171) | 0.292 |
| Body weight, kg
(SD, range) | 62.4 (10.6,
47–86.5) | 59.4 (13.8,
41–85) | 65.2 (12.6,
47–82) | 0.474 |
| BMI,
kg/m2 (SD, range) | 22.7 (2.2,
20–28) | 23.0 (4.2,
17–31) | 24.2 (3.6,
17–29) | 0.565 |
| Tumor diameter, cm
(SD, range) | 6.9 (2.98,
3.5–13) | 6.7 (2.74,
2.5–11) | 6.71 (2.71,
2.9–11.5) | 0.981 |
| Follow-up time,
months (SD, range) | 47.17 (22.64,
20–94) | 95.25 (51.13,
14–153) | 92.56 (45.53,
19–150) | 0.010 |
 | Table II.Hematological parameters of clear
cell renal cell carcinoma patients in the M, P and U groups with
regard to serine-threonine kinase 11 promoter status (n=42). |
Table II.
Hematological parameters of clear
cell renal cell carcinoma patients in the M, P and U groups with
regard to serine-threonine kinase 11 promoter status (n=42).
|
| Group |
|
|---|
|
|
|
|
|---|
| Parameter | M | P | U | P-value |
|---|
| K+,
mmol/l (SD, range) | 4.43 (0.64,
3.7–5.9) | 4.15 (0.39,
3.5–4.9) | 4.43 (0.34,
3.9–4.9) | 0.382 |
| Na+,
mmol/l (SD, range) | 140.1 (2.9,
136–145) | 139.9 (2.3,
135–142) | 141.7 (5.5,
131–149) | 0.554 |
| Cl+,
mmol/l (SD, range) | 105.5 (5.1,
96–115) | 103.9 (4.1,
99–114) | 105.6 (7.8,
90–114) | 0.772 |
| Ca2+,
mmol/l (SD, range) | 2.31 (0.47,
1.03–2.91) | 2.38 (0.12,
2.09–2.55) | 2.12 (0.46,
1.02–2.5) | 0.219 |
| AST, U/l (SD,
range) | 23.6 (13.5,
13–57.2) | 22.1 (9.4,
6–41) | 22.8 (12.1,
8–51) | 0.934 |
| ALT, U/l (SD,
range) | 28.1 (25.9,
6–89.9) | 21.1 (14.2,
7–56) | 23.4 (12.4,
3–41) | 0.584 |
| TBA, mmol/l (SD,
range) | 4.0 (1.9,
1.1–7.1) | 5.7 (2.9,
2.2–11.5) | 6.6 (5.1,
1.7–20.8) | 0.226 |
| ALP, U/l (SD,
range) | 73.1 (21.1,
42–117.1) | 67.5 (35.2,
35–190) | 73.8 (27.8,
35–136) | 0.811 |
| GGT, U/l (SD,
range) | 36.2 (33.4,
10–132) | 30.6 (26.5,
2.4–93) | 37.8 (31.8,
1–99) | 0.787 |
| LDH, U/l (SD,
range) | 156 (31.3,
114–208) | 202 (48.8,
133–296.3) | 187 (91.8,
82–346) | 0.142 |
| AFU, nmol/ml·h (SD,
range) | 12.9 (5.97,
5–26) | 10.2 (4.92,
5–21) | 8.78 (5.53,
2–19.4) | 0.191 |
| ALB, g/l (SD,
range) | 40.3 (3.99,
34.6–47.4) | 43.1 (3.6,
37.5–48.6) | 40.5 (5.16,
30.3–48.6) | 0.116 |
| GLO, g/l (SD,
range) | 28.1 (5.9,
22.1–38) | 31.5 (4.72,
22.2–40.8) | 30.7 (5.75,
22.5–43.9) | 0.223 |
| DBIL, µmol/l (SD,
range) | 3.37 (2.39,
1.1–10.1) | 3.34 (1.88,
0.22–8.87) | 3.37 (2.23,
0.76–8.94) | 0.999 |
| IBIL, µmol/l (SD,
range) | 7.27 (3.02,
3.1–11.72) | 11.5 (8.6,
3.4–40.3) | 10.7 (5.4,
0.81–20.83) | 0.249 |
| BUN, mmol/l (SD,
range) | 5.03 (2.53,
2.13–10.11) | 5.1 (1.61,
2.82–8.01) | 5.11 (1.71,
2.49–8) | 0.994 |
| CRE, µmol/l (SD,
range) | 99.3 (26.1,
67.29–141) | 92.5 (21.1,
51.3–134) | 93.2 (28.7,
29–133) | 0.747 |
| UA, µmol/l (SD,
range) | 326 (89.8,
172.6–454) | 323 (94.1,
118–551) | 375 (144.5,
87–597) | 0.403 |
| CHO, mmol/l (SD,
range) | 4.34 (0.97,
2.97–5.4) | 4.91 (1.10,
3.61–7.52) | 4.39 (0.85,
3.14–6) | 0.233 |
| TG, mmol/l (SD,
range) | 1.15 (0.62,
0.54–2.51) | 1.52 (0.92,
0.53–3.96) | 2.02 (1.42,
0.65–5.19) | 0.126 |
| GLU, mmol/l (SD,
range) | 4.92 (0.77,
3.22–5.94) | 5.08 (0.73,
4.16–6.89) | 5.07 (0.58,
3.68–5.96) | 0.819 |
| HDL, mmol/l (SD,
range) | 1.17 (0.42,
0.59–2.26) | 1.25 (0.33,
0.82–1.87) | 1.09 (0.22,
0.71–1.47) | 0.413 |
| LDL, mmol/l (SD,
range) | 2.67 (0.82,
1.47–4.12) | 3.0 (0.83,
1.46–4.53) | 2.72 (0.93,
1.64–4.46) | 0.532 |
| WBC,
109/l (SD, range) | 8.04 (1.92,
4.6–11) | 8.11 (2.52,
5.1–13.5) | 8.73 (2.44,
5–14) | 0.725 |
| RBC,
1012/l (SD, range) | 4.64 (0.61,
3.35–5.71) | 4.48 (0.99,
2.79–6.76) | 4.77 (0.72,
3.79–6.32) | 0.647 |
| Hb, g/l (SD,
range) | 132 (17.4,
95.3–156) | 129 (25.3,
79–165) | 135 (21.8,
93–171) | 0.750 |
| PLT,
109/l (SD, range) | 285 (91.6,
179–475) | 272 (124.6,
128–553) | 235 (33.8,
198–286) | 0.420 |
|
|
|
|
|
Association between STK11 promoter
methylation and TNM/AJCC staging
To investigate whether the STK11 promoter
methylation status is associated with the disease stage of RCC, the
distributions of TNM and AJCC stages among the three groups were
investigated. As shown in Table
III, all stage distributions were significantly different
between the 3 groups. There was a statistically significant
difference in the distribution of the T (P=0.036), N (P=0.007) and
AJCC (P<0.001) stages among the M, P, and U groups. In addition,
significant or marginally significant trends were observed that the
M group had more patients with advanced stage disease than the P
and U groups (P<0.10 for T and N stages, P<0.05 for M and
AJCC stages; residual analysis). The data suggested that the
methylation status of the STK11 promoter was associated with
T, N and AJCC stages in RCC.
 | Table III.TNM and AJCC staging based on the
methylation status of the serine-threonine kinase 11 promoter
(n=42). |
Table III.
TNM and AJCC staging based on the
methylation status of the serine-threonine kinase 11 promoter
(n=42).
|
| Group, n (%) |
|
|---|
|
|
|
|
|---|
| Variable | M | P | U | P-value |
|---|
| Total, n | 12 | 18 | 12 |
|
| T stage |
|
|
| 0.036 |
| T1 | 4 (33.3) | 8 (44.4) | 8 (66.7) |
|
| T2 | 3 (25.0) | 10 (55.6) | 3 (25.0) |
|
| T3 | 3 (25.0) | 0 (0.0) | 1 (8.3) |
|
| T4 | 2 (16.7) | 0 (0.0) | 0 (0.0) |
|
| N stage |
|
|
| 0.007 |
| N0 | 5 (41.7) | 15 (83.3) | 11 (91.0) |
|
| N1 | 3 (25.0) | 3 (16.7) | 0 (0.0) |
|
| N2 | 4 (33.3) | 0 (0.0.) | 1 (9.0) |
|
| M stage |
|
|
| 0.154 |
| M0 | 9 (75.0) | 17 (94.4) | 12 (100.0) |
|
| M1 | 3 (25.0) | 1 (5.6) | 0 (0.0) |
|
| AJCC stage |
|
|
| <0.001 |
| I | 0 (0.0) | 7 (38.9) | 8 (66.7) |
|
| II | 3 (25.0) | 9 (50.0) | 3 (25.0) |
|
|
III | 3 (25.0) | 0 (0.0) | 0 (0.0) |
|
| IV | 6 (50.0) | 2 (11.1) | 1 (8.3) |
|
STK11 promoter methylation and
survival
Since the association between methylation status and
tumor stage was observed, whether the methylation status has an
effect on the survival of RCC patients was then investigated. The
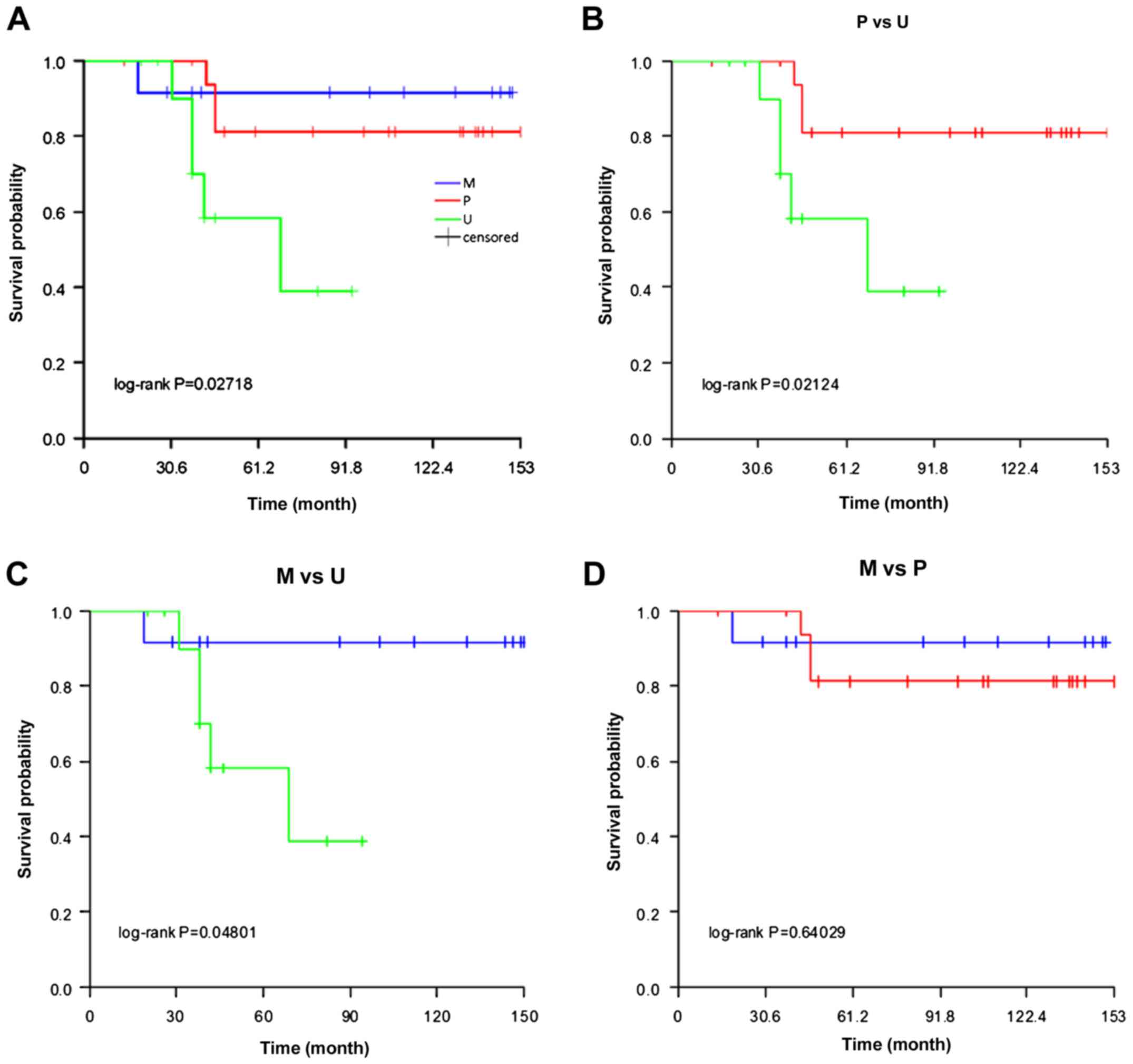
results of Kaplan-Meier survival analysis showed that there was a
significant survival difference among the three groups (log-rank
test, P<0.05; Fig. 2A). Additional
analysis revealed that the survival times of patients in the P
(P=0.021) and U (P=0.048) groups were significantly increased
compared with the M group (Fig. 2B and
C). However, there was no significant difference in survival
time between the U and P groups (P=0.640; Fig. 2D). The data suggest that the
methylation status of the STK11 promoter has an impact on
the survival of RCC patients.
Discussion
In the present study, STK11 promoter
methylation was analyzed using specimens from 42 ccRCC patients and
found an association between methylation status and cancer stage.
The results showed that 28.6 and 42.9% of ccRCC samples had
methylation and partial methylation at the STK11 promoter,
respectively. Additional analyses found the methylation status of
the STK11 promoter was associated with the T, N and AJCC
stages in RCC. In addition, the M group had an increased number of
patients at an advanced stage compared with the P and U groups.
Furthermore, survival analyses among three groups showed that the
survival time was significantly longer in both P and U groups
compared with the in M group, indicating that the methylation
status of the STK11 promoter has an impact on the survival
of RCC patients. To the best of our knowledge, the present study is
the first to report the methylation frequency of the STK11
promoter in ccRCC and its impact on the tumor stage and survival of
ccRCC patients.
STK11 has multiple biological and
physiological functions in cells, since knockout mice studies have
shown that inactivation of STK11 has severe consequences,
including tumorigenesis. Although STK11 was identified
almost two decades ago (5), the
studies focusing on its roles in the pathogenesis of RCC remain
rare. In 1999, Avizienyte et al (15) detected no mutation in 19 RCC
specimens. In 2014, Yalniz et al (16) reported an overall mutation frequency
of 51.6% (32/62) in RCC patients. In 2013, Duivenvoorden et
al (18) conducted a study to
investigate the tumor suppressor function of STK11 in ccRCC
in vitro and in vivo. Knockdown of STK11 in
the ccRCC 786-O cell line increased the cell proliferation,
invasion and vascular endothelial growth factor secretion. In
addition, the growth of STK11 knockdown cell xenografts was
significantly increased compared with the control. These results
suggested a tumor suppressor function of STK11 in ccRCC. In
addition, this study also investigated the expression of
STK11 at the mRNA and protein levels, as well as performing
immunohistochemistry staining. It was found that under-expression
of STK11 in ccRCC is a comment event. However, this study did not
further investigate the mechanism for the under-expression of
STK11 in ccRCC (16).
A previous study has already shown that mutation of
the STK11 gene may contribute to the inactivation of
STK11 (24). In the present
study, to determine if epigenetic alteration may also contribute to
inactivation of STK11, the methylation status of the
STK11 promoter region in 42 ccRCC specimens was
investigated. Hypermethylation of the STK11 promoter has
been demonstrated in previous studies. In a cell line study,
Esteller et al (23) showed
that three colorectal and one cervical carcinoma cell lines were
methylated at STK11. As for sporadic primary tumors, studies
showed the methylation frequency of the STK11 promoter in
various tumors is rare. In the study by Esteller et al
(23), a series of primary tumors
were also investigated. Among colorectal, breast, gastric,
pancreatic, thyroid, bladder and testicular carcinomas, only
colorectal carcinoma (7.7%; 1/13) and testicular tumor (10.7%;
3/28) exhibited methylated at STK11 (23). In another study by Trojan et al
(21), an overall methylation
frequency of 8% (4/48) was observed in colorectal cancer. Lee et
al (22) reported that promoter
methylation was detected in 13.2% (21/159) of Korean patients with
non-small cell lung cancer. Notably, in contrast to these studies,
the present study showed a significantly increased methylation rate
(28.6%; 12/42) in patients with ccRCC. This finding may indicate
that epigenetic alteration plays a more important role in the
pathogenesis of RCC compared with other cancer types. However,
whether this relatively high methylation frequency of STK11
in ccRCC is a general phenomenon or may be attributed to the
enrollment bias of the present study should be further verified in
a subsequent study.
In the present study, the correlation between the
methylation status of the STK11 promoter and the tumor stage
and survival of ccRCC patients was further analyzed. The results
showed that the methylation status of the STK11 promoter was
associated with the tumor progress in RCC patients. Patients in the
M group (with methylated at STK11) had a increased
percentage of patients with advanced stages, using either the TNM
or AJCC staging systems, compared with patients in the P and U
groups. It is notable that the results of the survival analysis
further support this observation. The survival time of patients
with methylated STK11 (M group) was significantly lower than
those in the U and P groups. The follow-up time of the M group was
also significantly shorter than those of the U and P groups, which
may be due to the fact that M group had a shorter survival time.
These findings indicated that the methylation of STK11 may
be important in the pathogenesis of RCC and may be a risk factor
for the prognosis of RCC. However, this conclusion should be
further verified in a subsequent study with a large sample size to
exclude the possibility of enrollment bias.
There are certain limitations in the present study.
The expression level of mRNA and protein was not further
investigated in these tumor samples to confirm the epigenetic
inactivation of STK11. Secondly, the sample size of the
present study was small. Thirdly, the surrounding normal tissues of
the ccRCC tumor specimens were not simultaneously analyzed to
identify the methylation difference in STK11 between normal
and tumor tissues. These limitations should be addressed in
subsequent studies.
In summary, the present study investigated the
methylation status of STK11 and its association with tumor
stage and survival of ccRCC patients. The methylation frequency of
STK11 was 28.4% in 42 ccRCC specimens. Patients in the M
group had an increased percentage of patients with advanced stage
RCC and a decreased survival time compared with the P and U groups.
The present findings suggested the methylation status of
STK11 may be important in the tumorigenesis of ccRCC.
Acknowledgements
The present study was supported by the Science and
Technology Foundation of Guangzhou (grant no. 2014A020212580).
Glossary
Abbreviations
Abbreviations:
|
RCC
|
renal cell carcinoma
|
|
ccRCC
|
clear cell RCC
|
|
STK11
|
serine-threonine kinase 11
|
|
LKB1
|
liver kinase B1
|
|
TNM
|
tumor-node-metastasis
|
|
AJCC
|
American Joint Committee on Cancer
|
|
MSP
|
methylation-specific polymerase chain
reaction
|
References
|
1
|
McLaughlin JK, Lipworth L and Tarone RE:
Epidemiologic aspects of renal cell carcinoma. Semin Oncol.
33:527–533. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qin C, Sun LJ, Cui L, Cao Q, Zhu J, Li P,
Zhang GM, Mao X, Shao PF, Wang ML, et al: Application of the
revised tumour node metastasis (TNM) staging system of clear cell
renal cell carcinoma in eastern China: Advantages and limitations.
Asian J Androl. 15:550–557. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cairns P: Renal cell carcinoma. Cancer
Biomark. 9:461–473. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jenne DE, Reimann H, Nezu J, Friedel W,
Loff S, Jeschke R, Müller O, Back W and Zimmer M: Peutz-Jeghers
syndrome is caused by mutations in a novel serine threonine kinase.
Nat Genet. 18:38–43. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao RX and Xu ZX: Targeting the LKB1
tumor suppressor. Curr Drug Targets. 15:32–52. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sanchez-Cespedes M, Parrella P, Esteller
M, Nomoto S, Trink B, Engles JM, Westra WH, Herman JG and Sidransky
D: Inactivation of LKB1/STK11 is a common event in adenocarcinomas
of the lung. Cancer Res. 62:3659–3662. 2002.PubMed/NCBI
|
|
8
|
Kim CJ, Cho YG, Park JY, Kim TY, Lee JH,
Kim HS, Lee JW, Song YH, Nam SW, Lee SH, et al: Genetic analysis of
the LKB1/STK11 gene in hepatocellular carcinomas. Eur J Cancer.
40:136–141. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bignell GR, Barfoot R, Seal S, Collins N,
Warren W and Stratton MR: Low frequency of somatic mutations in the
LKB1/Peutz-Jeghers syndrome gene in sporadic breast cancer. Cancer
Res. 58:1384–1386. 1998.PubMed/NCBI
|
|
10
|
Nakau M, Miyoshi H, Seldin MF, Imamura M,
Oshima M and Taketo MM: Hepatocellular carcinoma caused by loss of
heterozygosity in Lkb1 gene knockout mice. Cancer Res.
62:4549–4553. 2002.PubMed/NCBI
|
|
11
|
Robinson J, Nye E, Stamp G and Silver A:
Osteogenic tumours in Lkb1-deficient mice. Exp Mol Pathol.
85:223–226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su GH, Hruban RH, Bansal RK, Bova GS, Tang
DJ, Shekher MC, Westerman AM, Entius MM, Goggins M, Yeo CJ and Kern
SE: Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers
gene in pancreatic and biliary cancers. Am J Pathol. 154:1835–1840.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Avizienyte E, Roth S, Loukola A, Hemminki
A, Lothe RA, Stenwig AE, Fosså SD, Salovaara R and Aaltonen LA:
Somatic mutations in LKB1 are rare in sporadic colorectal and
testicular tumors. Cancer Res. 58:2087–2090. 1998.PubMed/NCBI
|
|
14
|
Koivunen JP, Kim J, Lee J, Rogers AM, Park
JO, Zhao X, Naoki K, Okamoto I, Nakagawa K, Yeap BY, et al:
Mutations in the LKB1 tumour suppressor are frequently detected in
tumours from Caucasian but not Asian lung cancer patients. Br J
Cancer. 99:245–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Avizienyte E, Loukola A, Roth S, Hemminki
A, Tarkkanen M, Salovaara R, Arola J, Bützow R,
Husgafvel-Pursiainen K, Kokkola A, et al: LKB1 somatic mutations in
sporadic tumors. Am J Pathol. 154:677–681. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yalniz Z, Tigli H, Tigli H, Sanli O, Dalay
N and Buyru N: Novel mutations and role of the LKB1 gene as a tumor
suppressor in renal cell carcinoma. Tumour Biol. 35:12361–12368.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fenton H, Carlile B, Montgomery EA,
Carraway H, Herman J, Sahin F, Su GH and Argani P: LKB1 protein
expression in human breast cancer. Appl Immunohistochem Mol
Morphol. 14:146–153. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duivenvoorden WC, Beatty LK, Lhotak S,
Hill B, Mak I, Paulin G, Gallino D, Popovic S, Austin RC and
Pinthus JH: Underexpression of tumour suppressor LKB1 in clear cell
renal cell carcinoma is common and confers growth advantage in
vitro and in vivo. Br J Cancer. 108:327–333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gan RY and Li HB: Recent progress on liver
kinase B1 (LKB1): Expression, regulation, downstream signaling and
cancer suppressive function. Int J Mol Sci. 15:16698–16718. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Trojan J, Brieger A, Raedle J, Esteller M
and Zeuzem S: 5′-CpG island methylation of the LKB1/STK11 promoter
and allelic loss at chromosome 19p13.3 in sporadic colorectal
cancer. Gut. 47:272–276. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee SM, Choi JE, Na YK, Lee EJ, Lee WK,
Choi YY, Yoon GS, Jeon HS, Kim DS and Park JY: Genetic and
epigenetic alterations of the LKB1 gene and their associations with
mutations in TP53 and EGFR pathway genes in Korean non-small cell
lung cancers. Lung Cancer. 81:194–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esteller M, Avizienyte E, Corn PG, Lothe
RA, Baylin SB, Aaltonen LA and Herman JG: Epigenetic inactivation
of LKB1 in primary tumors associated with the Peutz-Jeghers
syndrome. Oncogene. 19:164–168. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sato N, Rosty C, Jansen M, Fukushima N,
Ueki T, Yeo CJ, Cameron JL, Iacobuzio-Donahue CA, Hruban RH and
Goggins M: STK11/LKB1 Peutz-Jeghers gene inactivation in
intraductal papillary-mucinous neoplasms of the pancreas. Am J
Pathol. 159:2017–2022. 2001. View Article : Google Scholar : PubMed/NCBI
|