Introduction
Worldwide, China has one of the highest incidences
of esophageal squamous cell carcinoma (ESCC), which is one of the
most common malignancies and seriously affects the daily lives of
individuals (1,2). Research in previous decades has
demonstrated that the development of human ESCC occurs due to
multiple genetic or epigenetic alterations (3). Epigenetic changes, including increased
DNA methylation, are considered to be a primary risk factor in
human ESCC development (3).
Epigenetic changes are an alternative way to inactivate tumor
suppressor genes, and have attracted increasing research (3). DNA methyltransferase 1 (DNMT1) is an
endogenous and highly expressed protein involved in alteration of
DNA methylation patterns following DNA replication, tumor
suppressor gene re-expression and DNA damage. DNMT1 may be as a
biomarker during the course of ESCC development (4). Aberrant DNA methylation frequently
occurs as a result of dysregulation of DNMTs, as has been
identified in various types of human cancer, including lung,
prostate, colorectal and breast cancer, which indicates that
aberrant methylation substantially contributes to cancer initiation
and progression (5–8). Therefore, preventing the methylation of
certain tumor suppressor genes may be a potential therapy for human
cancer.
Long non-coding RNAs, microRNAs and small RNAs are
increasingly being identified as potential inhibitors of DNA
methylation, which are able to regulate gene expression at the
transcriptional and post-transcriptional levels (9,10). Short
hairpin RNAs (shRNAs) are a type of small RNA, which are able to
affect histone modification, DNA methylation targeting and gene
silencing by binding directly to untranslated regions with high
affinity. The binding of shRNAs leads to inhibition of DNMT1 enzyme
activity, which may induce demethylation of genes, resulting in
hypomethylation of moderately methylated regions (11,12).
Several studies suggested that hypomethylation leads to activation
and altered gene expression of certain tumor suppressor genes,
including p16 (13,14). The deactivation of p16 by
DNMT1-mediated methylation may be the most frequent early event in
carcinogenesis (13,14).
In order to identify a novel potential therapy for
the treatment of human ESCC, the association between DNMT1
silencing and p16 expression, a tumor suppressor gene, was
investigated in vitro and in vivo. A series of
experiments were carried out in cell lines and a nude mouse model
to determine whether shRNA targeting DNMT1 (shRNA-DNMT1) was able
to decrease DNMT1 expression significantly and whether this results
in demethylation of the p16 gene.
Materials and methods
Culture of cell lines and transfection
with shRNA
The human ESCC lines KYSE150 and KYSE410 (American
Type Culture Collection, Manassas, VA, USA) were maintained in
RPMI-1640 medium with 10% heat-inactivated fetal bovine serum and
1% antibiotic-antimycotic solution (all Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified
atmosphere of 95% air and 5% CO2. shRNA sequences were
as follows: shRNA-DNMT1 forward, 5-GAT CCC CGC AGG CGG CTC AAA GAT
TTGTTC AAG AGA CAA ATC TTT GA GCC GCC TGC TTT TTC-3 and reverse,
5-TCG AGA AAA AGC AGG CGG CTC AAA GAT TTG TCT CTT GAA CAA ATC TTT
GAG CCG CCT GCG GG-3 were constructed by Shanghai GenePharma Co.,
Ltd. (Shanghai, China). The KYSE150 and KYSE410 cell lines were
transfected with shRNA-DNMT1 (50 µM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 72 h. Negative control shRNA (shRNA-NC; 50
µM) was used for comparison.
Animal experimentation
For in vivo tumor formation, KYSE150 and
KYSE410 cells (1.5×107) were isolated using trypsin-EDTA
(0.05% trypsin, 0.53 mM EDTA tetrasodium salt; Gibco; Thermo Fisher
Scientific, Inc.). Subsequently, 0.3 ml Protanal®LF
10/60 sodium alginate solution (1.5%; FMC Health and Nutrition,
Philadelphia, PA, USA) and 40 µl CaSO4 (21%) were added. Following
72 h posttransfection with shRNA-DNMT1 or shRNA-NC, cell clusters
were subcutaneously injected into the dorso-lumbar area of 10 male
nude mice (7-week-old; body weight, 20±2 g; Japan SLC, Inc.,
Hamamatsu, Japan). Subsequently, the 10 mice were divided into two
groups (n=5). Food and water were offered ad libitum under a
pathogen-free condition at 26–28°C with 12 h dark/light cycles. The
animals were sacrificed with an overdose of sodium pentobarbital
anesthetic (cat. no. P3761; dosage, 100 mg/kg; Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) 14 days following transplantation of
cells. Tumors (50–150 mm3) were excised using a scalpel
and surgical forceps. Excised tumor samples were froze in liquid
nitrogen and stored in a freezer at −80°C for subsequent western
blotting analysis and methylation-specific polymerase chain
reaction (MSP) analyses. Furthermore, harvested tumors were fixed
in paraformaldehyde for subsequently use in immunohistochemistry.
These experiments were approved by the Use Committee for Animal
Care of the Second Affiliated Hospital of Guilin Medical University
(Guilin, China), and conducted according to the Guide for the Care
and Use of Laboratory Animals (NIH publication no. 80–23, revised
1996).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the KYSE150 and KYSE410
cells using a UNIQ-10 column and TRIzol® Total RNA
Isolation kit (Sangon Biotech Co., Ltd., Shanghai, China) according
to the manufacturer's protocol. A 1 µg sample of total RNA was used
for reverse transcription in a reaction volume of 20 µl [RNA, 10.0
µl (0.2 µg/µl); 5X RT Buffer, 4.0 µl; Reverse Transcriptase Enzyme
mix, 1.0 µl; Primer mix, 1.0 µl; diethyl pyrocarbonate H2O 1.0 µl;
total volume, 20 µl] using cloned avian myoblastosis virus reverse
transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 10 min at 50°C; 10 min at
80°C; and then the reactions were cooled to 4°C. Oligo d (T) 20
(#18418012; Invitrogen; Thermo Fisher Scientific, Inc.) were used
as the reverse transcription primer. A total of 2 µl cDNA was used
for qPCR using an ExTaq RT-PCR version 2.1 kit (Takara Bio, Inc.,
Otsu, Japan). Gene-specific PCR primers for p16 and GAPDH are
listed in Table I, and PCR signals
were detected using a DNA Engine Opticon® 2 Continuous
Fluorescence Detection System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Thermocycling conditions for the qPCR analysis
were performed as follows: 94°C for 2 min; 94°C for 20 sec; 58°C
for 20 sec; followed by 40 cycles of 72°C for 20 sec. At the end of
the PCR cycles, melting curve analysis was performed using
fluorescent quantitative PCR (Stratagene, Mx3000P; Agilent
Technologies, Inc., Santa Clara, CA, USA). Agar gel electrophoresis
(2%) was performed to assess the purity of the PCR products.
Negative control reactions (lacking template) were routinely
included to monitor potential contamination of reagents. Relative
amounts of p16 mRNA were normalized to GAPDH mRNA as described by
Livak and Schmittgen (15).
Experiments were performed in triplicate.
 | Table I.Sequences of the primers used for
detection of p16 in vitro. |
Table I.
Sequences of the primers used for
detection of p16 in vitro.
| Gene | Primer sequence
(5′-3′) |
|---|
| p16 | Forward: ACA
AGCTTCCTTTCCGTCATGCCG |
|
| Reverse:
ACAAGCTTCCTTTCCGTCATGCCG |
| GAPDH | Forward:
CGGAGTCAACGGATTTGGTCGTAT |
|
| Reverse:
GCCTTCTCCATGGTGGTGAAGAC |
Protein isolation and western blot
analysis
To evaluate the change in target gene expression
in vivo and in vitro as a result of DNMT1 silencing,
proteins of cell lines and tumor samples from nude mice were
extracted using a Total Protein Extraction kit (#AR0103; Wuhan
Boster Biological Technology, Ltd., Wuhan, China). Subsequently,
protein concentration was determined using a BCA assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. A total of 20 µg protein lysate was separated using
SDS-PAGE on a 10% gel, followed by transfer on to nitrocellulose
membranes. Western blot analysis was performed as previously
described (16), and the signal was
detected using an RapidStep™ ECL detection reagent (EMD Millipore,
Billerica, MA, USA) according to the manufacturer's protocol. The
primary antibodies used were anti-human p16 (#sc-68393; dilution,
1:4,000), anti-mouse p16 (#sc-68393; dilution, 1:2,000) and
anti-GAPDH (#sc-32233; dilution, 1:2,000) (all Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The secondary antibody
bovine anti-rabbit immunoglobulin G-horseradish peroxidase was
provided by Santa Cruz Biotechnology (#sc-2374; dilution,
1:2,000).
Immunohistochemistry
Immunohistochemistry was used to detect the
distribution of p16. Esophagus' of nude mice were dissected and
fixed overnight in 4% paraformaldehyde using a 0.1 M phosphate
buffer solution (pH 7.4) at room temperature. Sections were
subsequently embedded in paraffin and sliced into 5-µm-thick serial
sections using a microtome (Leica Microsystems, Inc., Buffalo
Grove, IL, USA). The sections were deparaffinized by xylene at room
temperature followed by washing with deionized water. Subsequently,
samples were treated with sodium citrate buffer (pH 6.0; 100°C).
Then, slides were rinsed twice for 90 sec each with PBS (pH 7.4) at
room temperature followed by blocking with 3% bovine serum albumin
(#V900933; Sigma-Aldrich; Merck Millipore) for 30 min. The sections
were incubated with the primary anti-mouse p16 antibody (#ab151303;
dilution, 1:1,000 dilution; Abcam, Cambridge, UK) at 4°C overnight,
followed by washing three times with PBS (pH 7.4). Secondary
antibody anti-rabbit immunoglobulin G (#ab98489; dilution, 1:200;
Abcam) was added to the sections, which were incubated at 37°C for
50 min. Following washing three times with PBS (pH 7.4),
immunoreactivity was visualized with 3,3-diaminobenzidine (#D8001;
Merck KGaA, Darmstadt, Germany). Three images of the identified
adjacent areas were captured (magnification, ×400).
MSP
The methylation status of the p16 (NCBI reference
sequence of p16, NG_1029) promoter region was determined using MSP.
Primers were designed using the MethPrimer design tool (www.urogene.org/methprimer). Primers illustrated
in Table II, which distinguished
between unmethylated (U) and methylated (M) alleles, were designed
to amplify the specific sequence. Each PCR sample contained 20 ng
of sodium bisulfite-modified DNA, 250 pmol of each primer, 250 pmol
of deoxynucleoside triphosphate, 1X PCR buffer (#EM101-01; Tiangen
Biotech Co., Ltd., Beijing, China) and 1 U of ExTaq Hot Start
polymerase (Takara Bio, Inc.) in a final reaction volume of 20 µl.
Thermocycling conditions were as follows: Initial denaturation at
95°C for 3 min; 40 cycles of 94°C for 30 sec, 65°C (M) or 63°C (U)
for 30 sec and 72°C for 30 sec. For each set of MSPs, H2O was used
as a control. PCR products were separated on 4% agarose gels,
stained with ethidium bromide and visualized under UV illumination.
For cases with borderline results, PCR analyses were repeated using
three independent experiments.
 | Table II.Sequences of the primers used for
methylation-specific polymerase chain reaction. |
Table II.
Sequences of the primers used for
methylation-specific polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) |
|---|
| U | Forward:
TTATTAGAGGGTGGGGTGGATTGT |
|
| Reverse:
TTATTAGAGGGTGGGGTGGATTGT |
| M | Forward:
TTATTAGAGGGTGGGGCGGATCGC |
|
| Reverse:
TTATTAGAGGGTGGGGCGGATCGC |
Statistical analysis
Values are expressed as the mean ± standard
deviation. Statistical significance was assessed using Student's
t-test and one-way ANOVA followed by a Tukey's post hoc test using
SPSS software (version 19.0; IBM SPSS, Armonk, NY, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
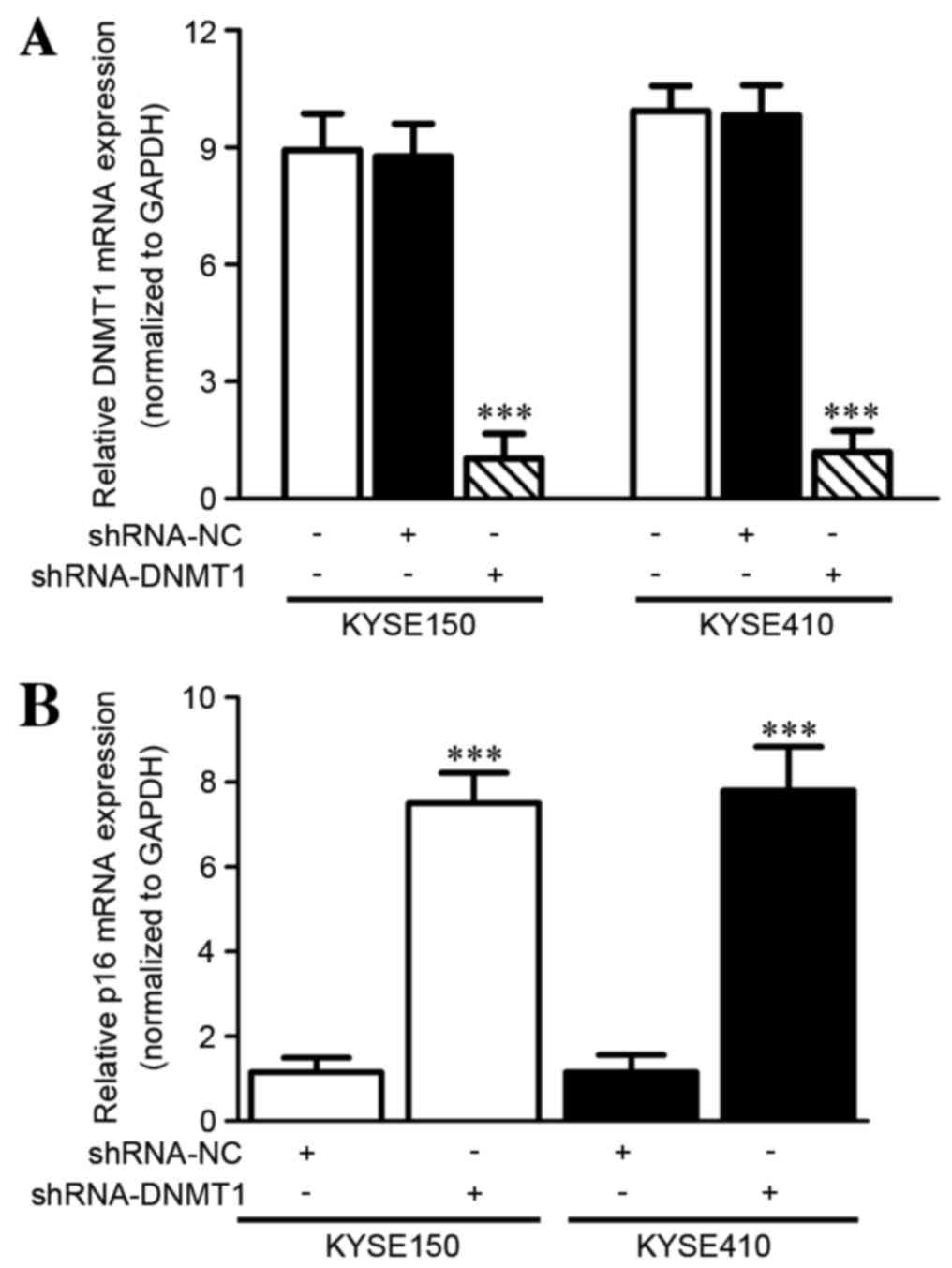
DNMT1 silencing increases the p16 mRNA
level in vitro as detected by RT-qPCR
In order to demonstrate the effect of silencing
DNMT1 on the expression of the tumor suppressor gene p16 in ESCC
cells, the p16 mRNA level was evaluated using RT-qPCR. RNA
interference (RNAi) of DNMT1 was conducted successfully with
significantly decreased DNMT1 mRNA expression in KYSE150 (P=0.0014)
and KYSE410 (P=0.001) cell lines following shRNA-DNMT1 transfection
compared with the negative control (paired Student's t-test;
Fig. 1A). Furthermore, the mRNA level
of p16 was detected in the KYSE150 and KYSE410 cell lines following
RNAi. p16 mRNA was significantly increased compared with the
control group (KYSE150, P<0.0001; KYSE410, P=0.001; paired
Student's t-test; Fig. 1B).
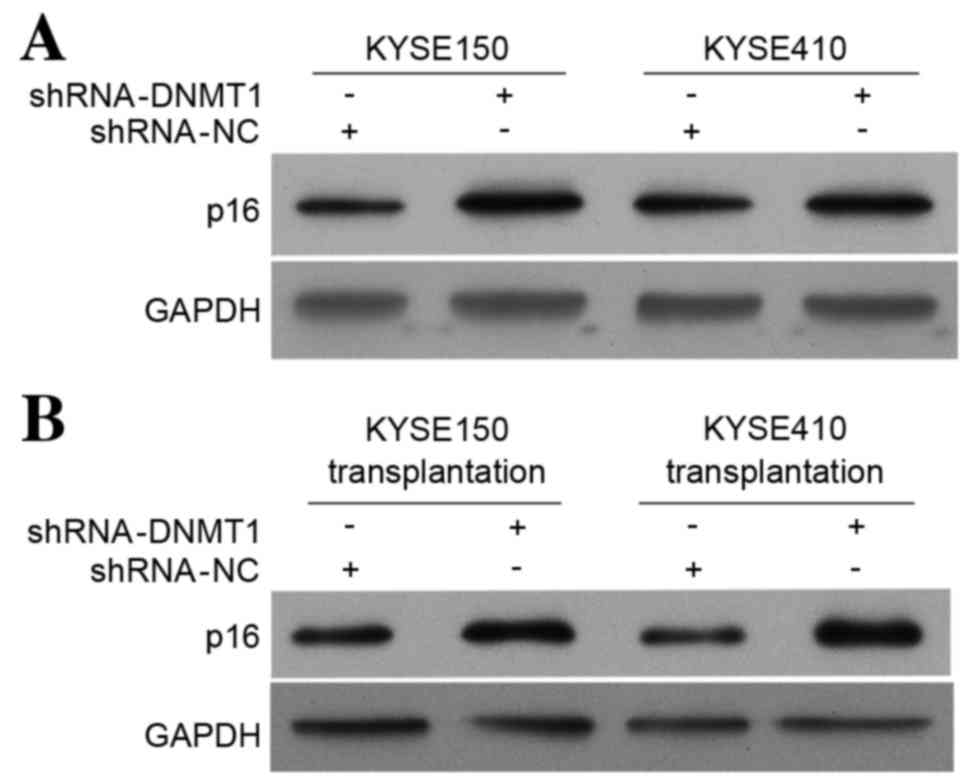
DNMT1 silencing increases the p16
protein level in vitro and in vivo as detected by western
blotting
Western blot analysis was used to investigate the
effect of DNMT1 silencing on p16 protein expression in vitro
and in vivo. The p16 protein level in the KYSE150 and
KYSE410 cell lines (Fig. 2A) and in
tumor tissues from nude mice transplanted with modified KYSE150 and
KYSE410 cell lines (Fig. 2B) were
increased compared with the control group. These results are
consistent with the results of the p16 RT-qPCR (Fig. 1B). These data suggest that silencing
DNMT1 upregulates expression of the tumor suppressor gene p16.
DNMT1 silencing affects p16
distribution in vivo as detected by immunohistochemistry
Using immunohistochemistry, it was identified that
p16 expression was increased in tumor tissues from nude mice
transplanted with the shRNA-DNMT1-treated KYSE150 and KYSE410 cells
respectively (Fig. 3) compared with
the group transfected with the shRNA-NC.
DNMT1 silencing inhibits methylation
of p16 in vitro as detected by MSP
In order to demonstrate the underlying molecular
mechanisms of DNMT knockdown-induced p16 upregulation in ESCC
cells, the methylation status of p16 was investigated using MSP
assays. It was demonstrated that unmethylated p16 levels were
increased in the KYSE150 and KYSE410 cell lines following
shRNA-DNMT1 transfection compared with their corresponding
controls, and no methylated p16 was revealed when RNAi of DNMT1 was
performed (Fig. 4A). These results
were consistent with results obtained from tumor tissues.
Methylation of p16 was inhibited in tumors isolated from nude mice
injected with the DNMT-knockdown KYSE150 and KYSE410 cells compared
with their corresponding controls (Fig.
4B). These results suggest that silencing DNMT1 downregulates
the methylation status of the tumor suppressor gene p16.
Discussion
In the present study the potential association
between DNMT1 silencing and expression of the tumor suppressor gene
p16 in vitro and in vivo was investigated.
shRNA-DNMT1 was transfected into two human ESCC lines, KYSE150 and
KYSE410, and the modified cancer cells were injected into separate
groups of nude mice. These results suggested that DNMT1 was
successfully and significantly decreased following transfection,
and that the transplantation technique is safe for animals.
Furthermore, the cancer cells and tumor tissues were harvested from
experimental animals, and the expression, distribution and
methylation of p16 were detected using RT-qPCR, western blot
analysis, immunohistochemistry and MSP. It is suggested that p16
was able to serve a protective role following demethylation and
subsequent activation induced by DNMT1 silencing.
DNMT1 is a maintenance enzyme in the DNMT family
that copies DNA methylation patterns during DNA replication
(17). DNMT1 is primarily expressed
in the S phase of the cell cycle and localizes to the DNA
replication fork (18). The role of
DNMT1 is to ensure the inheritance of the DNA methylation pattern
through cell division. Since aberrant expression of DNMT1 occurs in
the majority of cancer types, DNA methylation-based biomarkers
mediated by DNMT1 have been used as diagnostic tools, with
modification typically occurring in the CpG-rich island of the gene
promoter region (19–21). Therefore, it has been suggested that
DNA methylation patterns may be used to distinguish cancer cells
from normal cells (22).
It is well-known that p16, an important tumor
suppressor gene, is an essential G1 cell cycle
regulatory gene and its loss of function serves an important role
during ESCC development (23). p16
gene hypermethylation leads to decreased p16 protein expression,
and the inactivation of this gene may promote the development of
various types of cancer, including ESCC (23,24).
Previous research demonstrated that acid-induced increases in p16
gene promoter methylation led to decreases in p16 mRNA levels,
which may increase proliferation of the telomerase-immortalized
non-neoplastic human Barrett's cell line (25,26). This
may be dependent on DNMT1 activation (25,26). It
was therefore hypothesized that DNMT1-mediated DNA methylation of
the typically unmethylated p16 promoter region may be the critical
underlying molecular mechanism of p16 inactivation and is
frequently associated with the repression of gene transcription.
However, to date, few studies have evaluated the role of DNMT1 in
predicting the progression of human esophageal carcinoma through
p16 hypermethylation, using a longitudinal cohort of patients, and
in in vitro and in vivo experiments.
In conclusion, the results of the present study
demonstrated that silencing DNMT1 expression using shRNA in ESCC
cell lines may lead to increased active p16 protein levels by
decreasing p16 gene methylation. These results demonstrated that
RNAi of DNMT1 may be a potential therapy for the treatment of human
esophageal cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81160273 and
81160296) and the Scientific Research Innovation Fund for Graduate
Students (Guangxi, China; grant no. YCSZ2015214).
References
|
1
|
Zhao SL, Zhu ST, Hao X, Li P and Zhang ST:
Effects of DNA methyltransferase 1 inhibition on esophageal
squamous cell carcinoma. Dis Esophagus. 24:601–610. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Du Z, Ma K, Sun X, Li A, Wang H, Zhang L,
Lin F, Feng X and Song J: Methylation of RASSF1A gene promoter and
the correlation with DNMT1 expression that may contribute to
esophageal squamous cell carcinoma. World J Surg Oncol. 13:1412015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baba S, Yamada Y, Hatano Y, Miyazaki Y,
Mori H, Shibata T and Hara A: Global DNA hypomethylation suppresses
squamous carcinogenesis in the tongue and esophagus. Cancer Sci.
100:1186–1191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sato F and Meltzer SJ: CpG island
hypermethylation in progression of esophageal and gastric cancer.
Cancer. 106:483–493. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vendetti FP and Rudin CM: Epigenetic
therapy in non-small-cell lung cancer: Targeting DNA
methyltransferases and histone deacetylases. Expert Opin Biol Ther.
13:1273–1285. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin J, Haffner MC, Zhang Y, Lee BH,
Brennen WN, Britton J, Kachhap SK, Shim JS, Liu JO, Nelson WG, et
al: Disulfiram is a DNA demethylating agent and inhibits prostate
cancer cell growth. Prostate. 71:333–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Naselli F, Belshaw NJ, Gentile C, Tutone
M, Tesoriere L, Livrea MA and Caradonna F: Phytochemical
indicaxanthin inhibits colon cancer cell growth and affects the DNA
methylation status by influencing epigenetically modifying enzyme
expression and activity. J Nutrigenet Nutrigenomics. 8:114–127.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pathania R, Ramachandran S, Elangovan S,
Padia R, Yang P, Cinghu S, Veeranan-Karmegam R, Arjunan P,
Gnana-Prakasam JP, Sadanand F, et al: DNMT1 is essential for
mammary and cancer stem cell maintenance and tumorigenesis. Nat
Commun. 6:69102015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Veeck J and Esteller M: Breast cancer
epigenetics: From DNA methylation to microRNAs. J Mammary Gland
Biol Neoplasia. 15:5–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sarkar D, Leung EY, Baguley BC, Finlay GJ
and Askarian-Amiri ME: Epigenetic regulation in human melanoma:
Past and future. Epigenetics. 10:103–121. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katz TA, Vasilatos SN, Harrington E,
Oesterreich S, Davidson NE and Huang Y: Inhibition of histone
demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases
sensitivity to DNMT inhibitor-induced apoptosis in breast cancer
cells. Breast Cancer Res Treat. 146:99–108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mutze K, Langer R, Schumacher F, Becker K,
Ott K, Novotny A, Hapfelmeier A, Höfler H and Keller G: DNA
methyltransferase 1 as a predictive biomarker and potential
therapeutic target for chemotherapy in gastric cancer. Eur J
Cancer. 47:1817–1825. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan D, Ye S, Pan Y, Bao Y, Chen H and
Shao C: Long-term cadmium exposure leads to the enhancement of
lymphocyte proliferation via down-regulating p16 by DNA
hypermethylation. Mutat Res. 757:125–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ren J, Chu Y, Ma H, Zhang Y, Zhang X, Zhao
D, Li Z, Wang J, Gao YE, Xiao L, et al: Epigenetic interventions
increase the radiation sensitivity of cancer cells. Curr Pharm Des.
20:1857–1865. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang HQ, Fang N, Liu XM, Xiong SP, Liao
YQ, Jin WJ, Song RF and Wan YY: Antitumor antivity of chloroquine
in combination with cisplatin in human gastric cancer xenografts.
Asian Pac J Cancer P. 16:3907–3912. 2015. View Article : Google Scholar
|
|
17
|
Trievel RC: Structure and function of
histone methyltransferases. Crit Rev Eukaryot Gene Expr.
14:147–169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desjobert C, El Mai M, Gérard-Hirne T,
Guianvarc'h D, Carrier A, Pottier C, Arimondo PB and Riond J:
Combined analysis of DNA methylation and cell cycle in cancer
cells. Epigenetics. 10:82–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin RK, Wu CY, Chang JW, Juan LJ, Hsu HS,
Chen CY, Lu YY, Tang YA, Yang YC, Yang PC and Wang YC:
Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1
overexpression in lung cancer. Cancer Res. 70:5807–5817. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Venturelli S, Sinnberg TW, Berger A, Noor
S, Levesque MP, Böcker A, Niessner H, Lauer UM, Bitzer M, Garbe C
and Busch C: Epigenetic impacts of ascorbate on human metastatic
melanoma cells. Front Oncol. 4:2272014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qin W, Zhang K, Clarke K, Weiland T and
Sauter ER: Methylation and miRNA effects of resveratrol on mammary
tumors vs. normal tissue. Nutr Cancer. 66:270–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Delpu Y, Cordelier P, Cho WC and Torrisani
J: DNA methylation and cancer diagnosis. Int J Mol Sci.
14:15029–15058. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Taghavi N, Biramijamal F, Sotoudeh M,
Khademi H, Malekzadeh R, Moaven O, Memar B, A'rabi A and
Abbaszadegan MR: p16INK4a hypermethylation and p53, p16 and MDM2
protein expression in esophageal squamous cell carcinoma. BMC
Cancer. 10:1382010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Enders GH, Koh J, Missero C, Rustgi AK and
Harlow E: p16 inhibition of transformed and primary squamous
epithelial cells. Oncogene. 12:1239–1245. 1996.PubMed/NCBI
|
|
25
|
Hong J, Resnick M, Behar J, Wang LJ, Wands
J, DeLellis RA, Souza RF, Spechler SJ and Cao W: Acid-induced p16
hypermethylation contributes to development of esophageal
adenocarcinoma via activation of NADPH oxidase NOX5-S. Am J Physiol
Gastrointest Liver Physiol. 299:G697–G706. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hong J, Li D, Wands J, Souza R and Cao W:
Role of NADPH oxidase NOX5-S, NF-κB, and DNMT1 in acid-induced p16
hypermethylation in Barrett's cells. Am J Physiol Cell Physiol.
305:C1069–C1079. 2013. View Article : Google Scholar : PubMed/NCBI
|