Introduction
Renal cell carcinoma (RCC), which originates from
the epithelium of renal parenchyma uriniferous tubule, is one of
the most commonly occurring types of cancer in the world (1,2). Each
year, >240,000 new cases are diagnosed and >100,000
RCC-associated mortalities occur (2).
The majority of cases of RCC (~75%) are clear cell RCC (CCRCC)
(3). Currently, nephrectomy remains
the mainstay of CCRCC therapy, but ~20% of patients treated with
nephrectomy are subject to recurrence and metastasis (4). Patients with metastatic CCRCC usually
have a poor prognosis and obtain little benefit from conventional
chemoradiotherapy.
In previous years, targeted therapy has been
developed as a promising approach against metastatic advanced-stage
CCRCC (5–7). The key to targeted therapy is to
identify the crucial molecular targets that usually mediate
aberrant signaling pathways in carcinogenesis and tumor
progression. The Notch signaling pathway performs a pivotal role in
the regulation of cell proliferation, differentiation and apoptosis
(3). The four mammalian Notch
receptors, termed Notch 1–4, are the key components of the Notch
pathway (3). Notch receptors receive
signals from cell surface-tethered ligands (including Delta-like 1,
3 and 4, and Jagged 1 and 2) on neighboring cells through specific
binding (3), and then induce a
two-step proteolytic cleavage by which the Notch intracellular
domain is released to activate the transcription of downstream
target genes in the nucleus (8).
Dysregulation of the Notch signaling pathway has been demonstrated
to be associated with multiple types of human cancer, including
CCRCC (9). One of the most frequently
detected members of the Notch receptor family in tumor tissues is
Notch 1, and its overexpression can promote cancer genesis through
multiple processes such as cell growth and apoptosis (10–12).
Therefore, Notch 1 may be a potential target for CCRCC therapy.
On this basis, in the present study, an association
analysis based on 52 CCRCC cases and 30 normal controls was
performed to investigate the association between Notch 1 protein
expression and CCRCC development. The important role Notch 1
performs in CCRCC cell proliferation and apoptosis was then
confirmed through silencing Notch 1 expression in human CCRCC 786-O
cells with Notch 1-specific small interfering RNA (siRNA), and the
underlying mechanisms were elucidated in vitro.
Materials and methods
Subject recruitment and sample
collection
Patients who received curative surgical resection
for primary renal tumors at the Department of Surgery of Zhangzhou
Affiliated Hospital of Fujian Medical University (Fujian, China)
between October 2005 and April 2012 were recruited. Patients were
eligible for the present study if they were diagnosed as CCRCC by
the postoperative pathological analysis and had not received
chemotherapy, radiotherapy and immunotherapy prior to surgery.
Finally, a total of 52 CCRCC cases (age, 24–81; 34 male and 18
female) were included in the present study. Pathological stage of
each case was evaluated according to the tumor-node-metastasis
(TNM) classification (13) and
Fuhrman grade (14). Detailed
clinical data of the cases are presented in Table I. Renal tumor and pericarcinoma tissue
samples were obtained from surgically removed tissues of the 52
patients. A total of 30 normal renal tissue samples were collected
as control samples from other patients who underwent surgical
resection for trauma. Informed consent was obtained from all
individual patients included in the present study. The present
study was reviewed and approved by the Ethics Committee of
Zhangzhou Affiliated Hospital of Fujian Medical University (Fujian,
China).
 | Table I.Characteristics of patients with clear
cell renal cell carcinoma included in the present study. |
Table I.
Characteristics of patients with clear
cell renal cell carcinoma included in the present study.
| Characteristics | Patients, n (%) |
|---|
| Sex |
|
| Male | 34 (65.4) |
|
Female | 18 (34.6) |
| Age, range (mean ±
standard deviation) | 24-81(55.5±13.9) |
| <60
years | 32 (61.5) |
| ≥60
years | 20 (38.5) |
| Tumor-node-metastasis
stage |
|
|
I+II | 33 (63.5) |
|
III+IV | 19 (36.5) |
| Fuhrman grade |
|
|
1+2 | 35 (67.3) |
|
3+4 | 17 (32.7) |
| Tumor size |
|
| <5.0
cm | 16 (30.8) |
| ≥5.0
cm | 36 (69.2) |
Tissue microarray and
immunohistochemistry (IHC) assays
The tissue microarray was constructed according to
our previous description (15).
Briefly, target tissue cores were obtained from donor paraffin
blocks and then arranged in a new recipient paraffin block (tissue
array block), which was subsequently sectioned into numerous thin
slices with a thickness of 4 µm for each section. Tissue sections
were randomly selected and attached to the L-lysine-treated glass
slides for IHC assays, which were processed by utilizing the
UltraSensitive™ S-P kit (Maixin-Bio Co., Ltd., Fuzhou, China)
according to the manufacturer's protocol. These sections were
blocked with normal goat serum (Maixin-Bio Co., Ltd.) for 20 min at
room temperature. IHC staining of sections was performed with the
primary antibody rabbit anti-human polyclonal antibody to Notch 1
(cat. no. N4788; dilution, 1:200; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Sections treated with PBS instead of the
primary antibody were used as the negative controls, and purchased
positive tissue sections (Maixin-Bio Co., Ltd.) were used as the
positive controls. In this process, 3,3-diaminobenzidene (DAB) was
employed as the stain to treat sections for 3–15 min at room
temperature. Brown yellow staining indicated the positive Notch 1
expression. Cells with light or brown yellow staining particles in
the cytomembrane or cytoplasm were the positive cells. A
semi-quantitative scoring system (Olympus BX41, magnification,
×200; Olympus Co., Tokyo, Japan) was employed to evaluate the IHC
results. Scoring for color density and percentage of stained cells
constituted the basis of the evaluation system. Color density was
scored as follows: No staining, 0; slight staining, 1; moderate
staining, 2; and strong staining, 3. With regard to scoring for
percentage of stained cells, 0, <50, 50–75 and >75% cells
staining were scored as 0, 1, 2 and 3, respectively. The sum value
of the two scores 0, 1–2, 3–4 or 5–6 were further scored as -, +,
++ and +++, respectively. The symbol - was considered as negative;
+ and ++ indicated low expression of Notch 1 protein, while +++
indicated high expression.
Cells and cell culture
The CCRCC 786-O cell line was purchased from the
Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China)
and maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 15% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
and 100 µg/ml streptomycin in an atmosphere with saturation
humidity and 5% CO2 at 37°C. Cells grew as a monolayer
and adhered to the wall, being regularly passaged every 2–3 days.
Cells in the logarithmic phase were used in subsequent
experiments.
RNA interference
RNA interference was employed to silence the
expression of Notch 1 in 786-O cells. The siRNA targeted to human
Notch 1 gene was designed referring to the principles described in
previous studies (16–19). The Notch 1-specific siRNA sequences
were 5′-GCAGCUGCACUUCAUGUACGU-3′ (sense) and
5′-CGUCGACGUGAAGUACAUGCA-3′ (antisense). A nonspecific control
siRNA whose sequences were 5′-UUCUCCGAACGUGUCACGUTT-3′ (sense) and
5′-ACGUGACACGUUCGGAGAATT-3′ (antisense) was also designed. The
siRNAs were synthesized by Shanghai Gene Pharma Co., Ltd.
(Shanghai, China). Transfection assays were performed utilizing
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol. The
prepared 786-O cells (1×105/well) were seeded on 24-well
plates and transfected with the non-specific control siRNA and the
Notch 1-specific siRNA (40, 80 and 120 nmol/l). Transfection
efficiency was measured using the parallel transfection of a random
sequence marked with green fluorescence; the transfection
efficiency was calculated after 6 h of transfection by the formula:
Transfection efficiency = the count of cells with fluorescence
(positive)/the count of total cells.
Reverse transcription-polymerase chain
reaction (RT-PCR) assays
At 24 h post-transfection, 786-O cells in each group
were harvested and washed with 0.02 M PBS (pH 7.4). Total RNA of
these cells was extracted using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The acquired RNA precipitate was redissolved by 20–24 µl sterile
deionized water and its concentration was determined using a UV2100
spectrophotometer (Shanghai Puxi Instrument Co., Ltd., Shanghai,
China). The complementary DNA (cDNA) was synthesized in a 25 µl
mixture containing 0.4 µg total RNA, 1.0 µl oligo (dT) primer, 5.0
µl 5X reaction buffer, 2.0 µl dNTP mix (10 mM), 1.0 µl RNasin, 1.0
µl avian myeloblastosis virus (AMV) reverse transcriptase (20 U/µl)
and diethyl pyrocarbonate-treated water, at 42°C for 30 min. At the
end of synthesis, the mixture was incubated at 99°C for 5 min to
inactivate the AMV reverse transcriptase. Subsequently, the
synthesized cDNA was used as the template to amplify Notch 1, and
β-actin was used as the internal control. Primers of Notch1 and
β-actin were designed utilizing Primer 5.0 (Premier Biosoft
International, Palo Alto, CA, USA). The primer sequences were as
follows: Notch 1 forward, 5′-CCGTCATCTCCGACTTCATC-3′ and reverse,
5′-GGACTTGCCCAGGTCATCTAC-3′; β-actin forward,
5′-CTCGTCATACTCCTGCTTGCT-3′ and reverse,
5′-CGGGACCTGACTGACTACCTC-3′. The PCR amplification system contained
5.0 µl 10X PCR buffer, 4.0 µl dNTP Mix (10 mM), 2.0 µl of each
primer, 1.0 µl cDNA, 1.0 µl Taq DNA polymerase (Invitrogen; Thermo
Fisher Scientific, Inc.) and 31.0 µl ddH2O. PCR
thermocycling conditions were as follows: 5 min pre-degeneration at
95°C, 45 min thermal-cycle program, including 30 cycles of 95°C for
30 sec, 58°C for 30 sec and 72°C for 30 sec, and 10 min extension
at 72°C. PCR products were separated by 1.5% agarose gel
electrophoresis supplemented with ethidium bromide (cat. no.
sc-286960; Santa Cruz Biotechnology, Inc.) for visualization, and
semi-quantified using the GelDocEZ system (Bio-Rad Laboratories,
Inc., Hercules, CA, USA).
Western blot analysis
At 24 h post-transfection, 786-O cells were
harvested. Subsequent to washing twice with cooled TBS solution,
1×106 cells were mixed with 100 µl lysis buffer
(included in MicroRoto for cell lysis and protein extraction kit;
Bio-Rad Laboratories, Inc.) and 1 µl enzyme inhibitor, and
incubated at 4°C for 30 min to obtain lysate. Following
centrifugation at 12,000 × g for 10 min at 10°C, the supernatant of
the lysate in the middle layer was collected and total protein
content of each collected lysate sample was determined by Bradford
method. Subsequently, proteins were separated on a 12% SDS-PAGE gel
and blotted to nitrocellulose (NC) membranes. The NC membranes were
blocked with the 0.02 M TBS buffer (pH 7.4) containing 5% skimmed
milk for 60 min at 37°C, and then probed with specific primary
antibodies, including mouse anti-human monoclonal antibodies of
Notch 1 (cat. no. sc-71719; dilution, 1:400; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), B-cell lymphoma-2 (Bcl-2;
cat. no. sc-23960; dilution, 1:500; Santa Cruz Biotechnology, Inc.)
and procaspase-3 (cat. no. sc-271028; dilution, 1:500; Santa Cruz
Biotechnology, Inc.), and rabbit anti-human polyclonal antibodies
of RAC-alpha serine/threonine-protein kinase (Akt; cat. no. 14702;
dilution, 1:500; Cell Signaling Technology, Inc., Danvers, MA,
USA), phosphorylated (p)-Akt (cat. no. 13461; dilution, 1:250; Cell
Signaling Technology, Inc.), p-mammalian target of rapamycin (mTOR)
(cat. no. 5536; dilution, 1:400; Cell Signaling Technology, Inc.),
p-P70S6K (cat. no. 9862; dilution, 1:500; Cell Signaling
Technology, Inc.) and β-actin (cat. no. sc-81760; dilution,
1:2,500; Santa Cruz Biotechnology, Inc.) following the
manufacturer's protocol. All the incubations with primary
antibodies were maintained at 37°C for 60 min. Subsequent to
washing with TBST, the membranes were incubated with corresponding
secondary antibodies, including goat anti-mouse (cat. no.
sc-362267; dilution, 1:5,000-7,500; Santa Cruz Biotechnology, Inc.)
and goat anti-rabbit antibodies (cat. no. sc-2040; dilution,
1:5,000–7,500; Santa Cruz Biotechnology, Inc.) for 60 min at 37 °C,
followed by incubation with enhanced chemiluminescence reagent
(Santa Cruz Biotechnology, Inc.). Finally, target protein content
was determined by assessing chemiluminescence with AlphaDigiDoc
imaging analysis system (version 7.1; Alpha Innotec, Kasendorf,
Germany). Each experiment was repeated three times.
Cell proliferation analysis
786-O cells were seeded on 96-well plates
(1×105 cells/well) and then transfected with negative
control siRNA and Notch 1 siRNA (40, 60, 80, 100 and 120 nmol/l).
Cell proliferation was detected by MTT assays. Briefly, at 24 h
post-transfection, 786-O cells in each well were treated with 10 µl
MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) and incubated for an
additional 4 h at 37°C. At the end of incubation, the plates were
centrifuged at 800 × g for 5 min at room temperature and the
supernatant was removed. Following the addition of 100 µl dimethyl
sulfoxide (Sigma-Aldrich; Merck KGaA) in each well, cells were
detected on a microplate reader Stat Fax-2100 (Awareness
Technologies, Westport, CT, USA) at a dual-wavelength of 492 and
630 nm, and cell proliferation rate was calculated according to the
formula: Cell proliferation rate (%) = (Aexperiment -
Ablank)/(Acontrol - Ablank). Each
sample in these assays was performed in triplicate.
Apoptosis assays
Cell apoptosis was investigated by terminal
deoxynucleotidyl-transferase-mediated dUTP nick end labeling
(TUNEL) assays using an in situ cell death detection kit
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol. In brief, following treatment with control
siRNA and Notch 1 siRNA (40, 80 and 120 nmol/l), 786-O cells were
washed and dried in air. Cells (5×104) were then fixed
with 4% paraformaldehyde supplemented in PBS at room temperature
for 25 min. Subsequent to washing twice with PBS, cells were
incubated with 0.2% Triton X-100 PBS for 5 min at room temperature,
followed by 5–10 min incubation with equilibration buffer
(Invitrogen; Thermo Fisher Scientific, Inc.) at room temperature.
Subsequently, TUNEL reaction solution was added to each sample and
incubated at 37°C for 60 min. When reaction was terminated, samples
were immersed in 0.3% H2O2 for 3–5 min and
then rinsed with PBS. Following the addition of horseradish
peroxidase diluted with PBS, the samples were incubated at room
temperature for 30 min. Finally, samples were incubated with
3,3′-diaminobenzidine mixture till light brown appeared in the
background. Subsequent to rinsing with deionized water, samples
were observed under a microscope (Olympus BX41; magnification,
×200). Three fields of view were selected for each sample.
Apoptotic rate was calculated as follows: Apoptotic rate (%) =
number of apoptotic cells/number of total cells. Experiments were
repeated in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS
version 11.5 software (SPSS, Inc., Chicago, IL, USA). Association
analysis between Notch 1 protein expression and CCRCC was performed
with χ2 test. For other analysis, comparison among three
or more groups was performed by one-way analysis of variance,
followed by Student-Newman-Keuls post-hoc test for comparison
between two groups. Measurement data are presented as the mean ±
standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
Notch 1 protein expression is
positively associated with CCRCC carcinogenesis and
progression
Tissue samples obtained from 52 cases were eligible
for the present study and tissue microarrays were successfully
constructed with the exception of 5 pericarcinoma tissue samples.
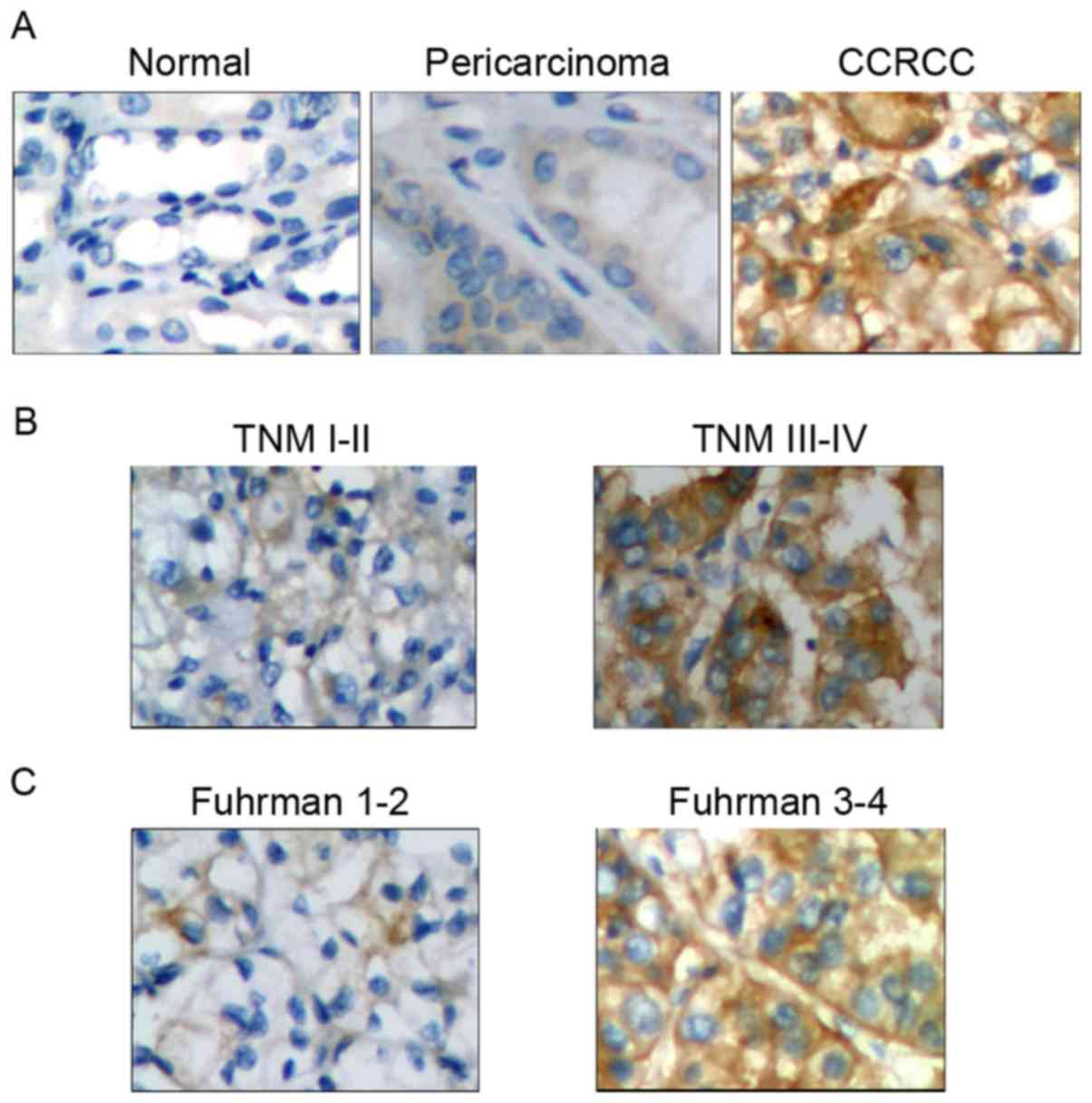
IHC and association analyses suggested that Notch 1 protein
expression was positive in 82.7% of CCRCC tumor tissues, and the
positive incidence was significantly higher compared with that in
pericarcinoma (42.6%; P<0.01) and normal (16.7%; P<0.01)
renal tissues (Table II). Additional
analysis indicated that high expression of Notch 1 protein was
associated with TNM stage (χ2=6.267; P<0.05), Fuhrman
grade (χ2=7.90; P<0.01) and tumor size
(χ2=4.160; P<0.05), but not associated with sex
(χ2=0.036; P>0.05) and age (χ2=0.054;
P>0.05) (Table III).
Representative IHC results are presented in Fig. 1. Therefore, Notch 1 protein expression
was closely associated with CCRCC carcinogenesis and
progression.
 | Table II.Notch 1 protein expression was
associated with the occurrence of CCRCC. |
Table II.
Notch 1 protein expression was
associated with the occurrence of CCRCC.
|
|
| Notch 1 protein
expression, n (%) |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | Patients, n | Positive | Negative | χ2 |
P-valuea |
|---|
| CCRCC | 52 | 43 (82.7) | 9
(17.3) |
|
|
| Pericarcinoma | 47 | 20 (42.6) | 27 (57.4) | 17.188 | <0.01 |
| Normal | 30 | 5
(16.7) | 25 (83.3) | 34.170 | <0.01 |
 | Table III.Clinical significance of Notch 1
protein expression in clear cell renal cell carcinoma cases
(n=43). |
Table III.
Clinical significance of Notch 1
protein expression in clear cell renal cell carcinoma cases
(n=43).
|
| Notch 1 protein
expression, n (%) |
|
|
|---|
|
|
|
|
|
|---|
| Group | Low | High | χ2 | P-value |
|---|
| Sex |
|
| 0.036 | >0.05 |
|
Male | 8 (18.6) | 21 (48.9) |
|
|
|
Female | 5 (11.6) | 9 (20.9) |
|
|
| Age |
|
| 0.054 | >0.05 |
| <60
years | 9 (20.9) | 18 (41.9) |
|
|
| ≥60
years | 4 (9.3) | 12 (27.9) |
|
|
|
Tumor-node-metastasis stage |
|
| 6.267 | <0.05 |
|
I+II | 11 (25.6) | 13 (30.2) |
|
|
|
III+IV | 2 (4.7) | 17 (39.5) |
|
|
| Fuhrman grade |
|
| 7.900 | <0.01 |
|
1+2 | 12 (27.9) | 14 (32.6) |
|
|
|
3+4 | 1 (2.3) | 16 (37.2) |
|
|
| Tumor size |
|
| 4.160 | <0.05 |
| <5.0
cm | 7 (17.1) | 7 (17.1) |
|
|
siRNA against Notch 1 interferes with
Notch 1 expression in CCRCC 786-O cells
The effectiveness of the transfections was evaluated
by transfection efficiency, which was determined by the rate of
positive cells to total cells in the parallel transfection group.
Positive cells with green fluorescence and total cells at the same
visual field (Fig. 2A) were counted
under an inverted fluorescence microscope after 6 h of
transfection. The transfection efficiency was 32.6±4.8%, indicating
the success of the interference assays. At 24 h post-transfection,
mRNA and protein expression of Notch 1 in 786-O cells was detected
by RT-PCR and western blot analysis, respectively, to confirm the
interfering effect of siRNAs against Notch 1. As a result, mRNA and
protein expression of Notch 1 was significantly inhibited by the
treatment of Notch 1-specific siRNA compared with the control, and
the inhibition was dose-dependent. Notch 1 mRNA expression
exhibited a 38.0% (P<0.05), 53.8% (P<0.01) and 70.4%
(P<0.01) decrease in 786-O cells treated with 40, 80 and 120
nmol/l Notch 1-specific siRNA compared with in those treated with
negative control siRNA, respectively (Fig. 2B). Notch 1 protein expression
exhibited a reduction of 37.4% (P<0.05), 65.7% (P<0.01) and
87.9% (P<0.01), respectively, in the three Notch 1 siRNA-treated
786-O cells compared with the control (Fig. 2C). Therefore, the Notch 1-specific
siRNA was successfully transfected into 786-O cells and
dose-dependently inhibited the expression of Notch 1.
Inhibition of Notch 1 expression
inhibits cell proliferation and induces cell apoptosis accompanied
with decreased Bcl-2 and procaspase-3 expression in CCRCC 786-O
cells
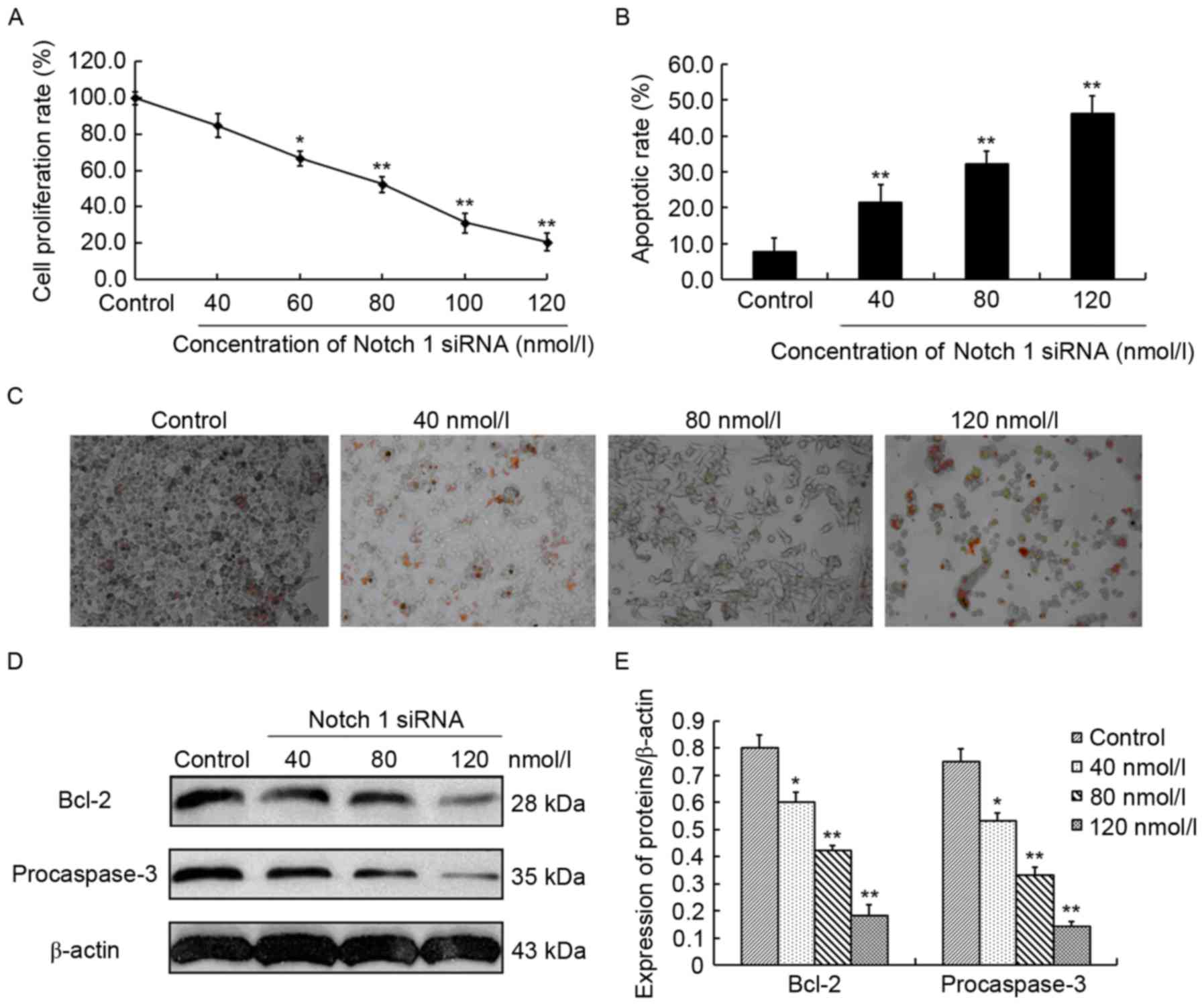
The effect of Notch 1 siRNA on the proliferation of
CCRCC 786-O cells was evaluated by MTT assays. For 786-O cells
treated with 40, 60, 80, 100 and 120 nmol/l Notch 1 siRNA, the cell
proliferation rate was 84.7±5.5, 66.6±3.2, 52.4±4.5, 31.2±2.4 and
20.5±2.9%, respectively, suggesting a 15.2% (P>0.05), 33.3%
(P<0.05), 47.6% (P<0.01), 68.8% (P<0.01) and 79.4%
(P<0.01) decrease compared with that of control siRNA-treated
786-O cells (99.8±3.47%) (Fig. 3A).
This indicated that Notch 1 siRNA dose-dependently suppressed the
proliferation of 786-O cells. However, in the TUNEL assays, 786-O
cells treated with 40, 80 and 120 nmol/l Notch 1 siRNA exhibited a
marked dose-dependent increase in cell apoptotic rate, which was
21.5±4.8, 32.3±3.5 and 46.3±4.7%, respectively, and was
2.8-(P<0.01), 4.2-(P<0.01) and 6.1-(P<0.01) fold higher
compared with the control siRNA-treated 786-O cells (Fig. 3B). Representative results are
presented in Fig. 3C. This
demonstrated that Notch 1 siRNA dose-dependently induced apoptosis
in 786-O cells. Together, these findings indicated that inhibition
of Notch 1 expression could inhibit cell proliferation and induce
cell apoptosis in 786-O cells. To identify the underlying molecular
mechanisms, the alterations of two important apoptosis-associated
molecules, Bcl-2 and procaspase-3, were investigated by western
blot analysis. Bcl-2 is an anti-apoptotic protein and procaspase-3
is an anti-apoptotic effector (8). In
the present study, Bcl-2 expression exhibited 25.0% (P<0.05),
47.5% (P<0.01) and 77.5% (P<0.01) reduction in the Notch1
siRNA-treated (40, 80 and 120 nmol/l) 786-O cells compared with the
control siRNA-treated 786-O cells, respectively (Fig. 3D and E). In addition, procaspase-3
expression significantly decreased by 29.3% (P<0.05), 56.0%
(P<0.01) and 81.3% (P<0.01) in the three Notch1 siRNA-treated
786-O cells compared with the control, respectively (Fig. 3D and E). The aforementioned findings
indicated that Notch 1 was involved in the regulation of cell
proliferation and apoptosis in 786-O cells.
Inhibition of Notch 1 expression
inactivates Akt/mTOR signaling in CCRCC 786-O cells
To elucidate the molecular mechanism underlying
Notch 1-mediated cell survival and proliferation in 786-O cells,
alterations of the Akt/mTOR pathway were monitored. Akt, p-Akt,
p-mTOR and p-P70 ribosomal S6-kinase (P70S6K) are the important
components of the Akt/mTOR pathway (20) and their protein expression was
investigated in 786-O cells following treatment with Notch 1 siRNA
(at the concentrations of 40, 80 and 120 nmol/l) and control siRNA.
No significant differences were identified between the Akt levels
in Notch 1 siRNA-treated 786-O cells compared with control
siRNA-treated 786-O cells, whereas p-Akt, p-mTOR and p-P70S6K
expression exhibited a dose-dependent decrease in Notch 1
siRNA-treated 786-O cells compared with the control. The decrease
was 65.8% (P<0.01), 76.3% (P<0.01) and 89.5% (P<0.01) for
p-Akt, 18.4% (P>0.05), 46.9% (P<0.01) and 75.5% (P<0.01)
for p-mTOR, and 40.0% (P<0.05), 71.4% (P<0.01) and 88.6%
(P<0.01) for p-P70S6K at 40, 80 and 120 nmol/l Notch 1 siRNA,
respectively (Fig. 4). These findings
demonstrated that inhibition of Notch 1 expression could suppress
the activity of the Akt/mTOR signaling pathway in 786-O cells.
Discussion
At present, the global incidence of RCC is
increasing year by year, and no therapeutic strategies can
completely cure the disease, particularly advanced RCC (2,5). Targeted
therapy has been proposed to overcome the difficulty due to the
gradually revealed molecular mechanisms underlying RCC development
(5). Several molecules have been
identified as the potential therapeutic targets of RCC, including
VEGF and mTOR (21). The present
study investigated the potential of Notch 1 as a target of CCRCC
treatment. The results demonstrated that Notch 1 protein expression
was associated with CCRCC carcinogenesis and progression. Therefore
Notch 1 may be a valuable target for CCRCC treatment.
The Notch signaling pathway is a highly-conserved
regulator in mediating cell to cell interactions (3). It, together with numerous other
signaling pathways, composes a complicated signaling network to
precisely regulate multiple biological processes, including cell
growth, differentiation and apoptosis (22). Notch 1 is an essential component of
this pathway and may perform a vital role in these biological
processes (3). In the present study,
Notch 1 protein expression in CCRCC tumor, pericarcinoma and normal
renal tissues was compared and the results demonstrated that Notch
1 protein expression was closely associated with CCRCC
carcinogenesis. In addition, patients with higher TNM stage or
Fuhrman grade or larger tumor size exhibited higher Notch 1 protein
expression, indicating that Notch 1 protein was positively
correlated with CCRCC progression. On this basis, it was
hypothesized Notch 1 may be a potential target for CCRCC
therapy.
To confirm the aforementioned hypothesis, the effect
of Notch 1 on cell proliferation and apoptosis was examined by
silencing Notch 1 expression in CCRCC 786-O cells in vitro.
Following the interference of Notch 1 expression with increasing
concentrations of Notch 1-specific siRNA, 786-O cells exhibited a
dose-dependent decrease in proliferation and a dose-dependent
increase in apoptosis accompanied with a dose-dependent decrease in
expression of Bcl-2 and procaspase-3. These findings demonstrated
that Notch 1 was involved in the regulation of cell survival and
proliferation in CCRCC 786-O cells, which was consistent with
previous studies on other cancers, including glioma, leukemia and
pancreatic cancers (23–25). Bcl-2 is a key anti-apoptotic protein
and usually acts as the switch of cell apoptosis (26). If Bcl-2 expression is downregulated,
cells switch to the apoptotic process, or else to the
anti-apoptotic process. Procaspase-3 is an anti-apoptotic effector.
When cells switch to the apoptotic process, procaspase-3 will be
cleaved into active capspase-3, which subsequently induces the
caspase cascades to facilitate cell apoptosis. Conversely, when
cells switch to the anti-apoptotic process, procaspase-3 will be
accumulated in cells (8). In the
present study, Bcl-2 and procaspase-3 expression exhibited a marked
dose-dependent decrease in 786-O cells following treatment with
increasing concentrations of Notch 1-specific siRNA, which
indicated that inhibition of Notch 1 by Notch 1 siRNA downregulated
the Bcl-2 expression, resulting in the initiation of apoptotic
process and the cleavage of procaspase-3, and ultimately leading to
apoptosis in 786-O cells. In this regard, Notch 1 was a potential
target against cell survival and proliferation for CCRCC 786-O
cells.
To confirm the potential of Notch 1 as the
therapeutic target in CCRCC, the underlying molecular mechanism
that Notch 1 may be involved in during CCRCC development was
investigated. It has been demonstrated that the Akt/mTOR signaling
pathway performs a central role in mediating cell cycle and
apoptosis signaling and is implicated in Notch 1-mediated cell
survival and proliferation in glioma (27,28). Akt,
also termed protein kinase B, is a serine/threonine protein kinase
and could be phosphorylated by phosphatidylinositol 3-kinase
(8). A previous study demonstrated
that p-Akt could upregulate Bcl-2 through cyclic AMP response
element binding protein to inhibit cell apoptosis (29). In the present study, p-Akt expression
was inhibited in a dose-dependent manner in 786-O cells by the
treatment of increasing concentrations of Notch 1 siRNA, while the
total Akt expression was not affected, suggesting that Notch 1
siRNA could inhibit Akt phosphorylation, which thereby reduced the
activity of the Akt signaling pathway and contributed to cell
apoptosis. This was consistent with previous studies (27,30). mTOR
is an important downstream effector of the Akt signaling pathway
and also is a crucial target for RCC therapy (31). It has been indicated that
phosphorylated mTOR could induce cell cycle transition between the
G1 and S phase to facilitate cell proliferation and
control protein synthesis through the regulation of P70S6K
(20,32). In the present study, it was revealed
that protein expression of p-mTOR and p-P70S6K exhibited a marked
dose-dependent decrease along with dose-dependently decreased p-Akt
in the Notch 1-specific siRNA-treated 786-O cells. Together, the
findings from the present study demonstrated that the inhibition of
Notch 1 expression by Notch 1-specific siRNA inactivated Akt/mTOR
signaling, inhibited cell proliferation and induced cell apoptosis
in 786-O cells. This is consistent with the result in a previous
study by Mungamuri et al (33)
on T-cell acute lymphocyte leukemia. Collectively, the present
study confirmed that Notch 1 was involved in the regulation of cell
survival and proliferation in 786-O cells through the Akt/mTOR
signaling-dependent pathway, and Notch 1 was a valuable target for
CCRCC treatment.
In summary, association analysis results from the
present study demonstrated that Notch 1 expression was closely
associated with CCRCC carcinogenesis and progression. In addition,
inhibition of Notch 1 expression inhibited cell proliferation and
induced cell apoptosis in CCRCC 786-O cells, suggested that Notch 1
was involved in CCRCC development. Furthermore, treatment with
Notch 1-specific siRNA decreased expression of Bcl-2 and
procaspase-3 in a dose-dependent manner in CCRCC 786-O cells, which
was accompanied with a dose-dependent decrease in the expression of
p-Akt, p-mTOR and p-P70S6K. This indicated that Notch 1 was
involved in the regulation of cell survival and proliferation in
786-O cells through the Akt/mTOR signaling-dependent pathway. Due
to the complexity of cancer-associated cell proliferation and
apoptosis, the underlying molecular mechanisms are not limited to
the aforementioned findings. However, these findings confirmed that
Notch 1 is a valuable target against cell survival and
proliferation in CCRCC, and Notch 1-specific siRNAs may be
developed as target agents to be used as single agents or in
combination with other chemotherapeutics in CCRCC treatment.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Fujian Province (grant no. 2013J01393).
References
|
1
|
Cohen HT and McGovern FJ: Renal-cell
carcinoma. N Engl J Med. 353:2477–2490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ljungberg B, Campbell SC, Choi HY, Jacqmin
D, Lee JE, Weikert S and Kiemeney LA: The epidemiology of renal
cell carcinoma. Eur Urol. 60:615–621. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sjölund J, Johansson M, Manna S, Norin C,
Pietras A, Beckman S, Nilsson E, Ljungberg B and Axelson H:
Suppression of renal cell carcinoma growth by inhibition of Notch
signaling in vitro and in vivo. J Clin Invest. 118:217–228. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Athar U and Gentile TC: Treatment options
for metastatic renal cell carcinoma: A review. Can J Urol.
15:3954–3966. 2008.PubMed/NCBI
|
|
5
|
Randall JM, Millard F and Kurzrock R:
Molecular aberrations, targeted therapy, and renal cell carcinoma:
Current state-of-the-art. Cancer Metastasis Rev. 33:1109–1124.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Holland WS, Tepper CG, Pietri JE, Chinn
DC, Gandara DR, Mack PC and Lara PN Jr: Evaluating rational
non-cross-resistant combination therapy in advanced clear cell
renal cell carcinoma: Combined mTOR and AKT inhibitor therapy.
Cancer Chemother Pharmacol. 69:185–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramp U, Caliskan E, Mahotka C, Krieg A,
Heikaus S, Gabbert HE and Gerharz CD: Apoptosis induction in renal
cell carcinoma by TRAIL and gamma-radiation is impaired by
deficient caspase-9 cleavage. Br J Cancer. 88:1800–1807. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu WK, Wang XJ, Cheng AS, Luo MX, Ng SS,
To KF, Chan FK, Cho CH, Sung JJ and Yu J: Dysregulation and
crosstalk of cellular signaling pathways in colon carcinogenesis.
Crit Rev Oncol Hematol. 86:251–277. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bolós V, Grego-Bessa J and de la Pompa JL:
Notch signaling in development and cancer. Endocr Rev. 28:339–363.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Li Y, Ahmad A, Banerjee S, Azmi
AS, Kong D, Wojewoda C, Miele L and Sarkar FH: Down-regulation of
Notch-1 is associated with Akt and FoxM1 in inducing cell growth
inhibition and apoptosis in prostate cancer cells. J Cell Biochem.
112:78–88. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ai Q, Ma X, Huang Q, Liu S, Shi T, Zhang
C, Zhu M, Zhang Y, Wang B, Ni D, et al: High-level expression of
Notch1 increased the risk of metastasis in T1 stage clear cell
renal cell carcinoma. PLoS One. 7:e350222012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu S, Ma X, Ai Q, Huang Q, Shi T, Zhu M,
Wang B and Zhang X: NOTCH1 functions as an oncogene by regulating
the PTEN/PI3K/AKT pathway in clear cell renal cell carcinoma. Urol
Oncol. 31:938–948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Terrone C, Cracco C, Porpiglia F, Bollito
E, Scoffone C, Poggio M, Berruti A, Ragni F, Cossu M, Scarpa RM and
Rossetti SR: Reassessing the current TNM lymph node staging for
renal cell carcinoma. Eur Urol. 49:324–331. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wan F, Zhu Y, Han C, Xu Q, Wu J, Dai B,
Zhang H, Shi G, Gu W and Ye D: Identification and validation of an
eight-gene expression signature for predicting high Fuhrman grade
renal cell carcinoma. Int J Cancer. 140:1199–1208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai L, Ma X, Huang Y, Zou Y and Chen X:
Aberrant histone methylation and the effect of Suv39H1 siRNA on
gastric carcinoma. Oncol Rep. 31:2593–2600. 2014.PubMed/NCBI
|
|
16
|
Masuda S, Kumano K, Shimizu K, Imai Y,
Kurokawa M, Ogawa S, Miyagishi M, Taira K, Hirai H and Chiba S:
Notch1 oncoprotein antagonizes TGF-beta/Smad-mediated cell growth
suppression via sequestration of coactivator p300. Cancer Sci.
96:274–282. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amarzguioui M and Prydz H: An algorithm
for selection of functional siRNA sequences. Biochem Biophys Res
Commun. 316:1050–1058. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ui-Tei K, Naito Y, Takahashi F, Haraguchi
T, Ohki-Hamazaki H, Juni A, Ueda R and Saigo K: Guidelines for the
selection of highly effective siRNA sequences for mammalian and
chick RNA interference. Nucleic Acids Res. 32:936–948. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reynolds A, Leake D, Boese Q, Scaringe S,
Marshall WS and Khvorova A: Rational siRNA design for RNA
interference. Nat Biotechnol. 22:326–330. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inoki K, Corradetti MN and Guan KL:
Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet.
37:19–24. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vano YA, Tartour E, Fournier LS,
Beuselinck B, Mejean A and Oudard S: Prognostic factors in patients
with advanced renal cell carcinoma treated with VEGF-targeted
agents. Expert Rev Anticancer Ther. 14:523–542. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Maillard I and Pear WS: Notch and cancer:
Best to avoid the ups and downs. Cancer Cell. 3:203–205. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yao J, Zheng K, Li C, Liu H and Shan X:
Interference of Notch1 inhibits the growth of glioma cancer cells
by inducing cell autophagy and down-regulation of Notch1-Hes-1
signaling pathway. Med Oncol. 32:6102015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grabher C, von Boehmer H and Look AT:
Notch 1 activation in the molecular pathogenesis of T-cell acute
lymphoblastic leukaemia. Nat Rev Cancer. 6:347–359. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kunnimalaiyaan S, Trevino J, Tsai S,
Gamblin TC and Kunnimalaiyaan M: Xanthohumol-mediated suppression
of Notch1 signaling is associated with antitumor activity in human
pancreatic cancer cells. Mol Cancer Ther. 14:1395–1403. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen C, Cui J, Zhang W and Shen P:
Robustness analysis identifies the plausible model of the Bcl-2
apoptotic switch. FEBS Lett. 581:5143–5150. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao N, Guo Y, Zhang M, Lin L and Zheng Z:
Akt-mTOR signaling is involved in Notch-1-mediated glioma cell
survival and proliferation. Oncol Rep. 23:1443–1447.
2010.PubMed/NCBI
|
|
28
|
Sangphech N, Osborne BA and Palaga T:
Notch signaling regulates the phosphorylation of Akt and survival
of lipopolysaccharide-activated macrophages via regulator of G
protein signaling 19 (RGS19). Immunobiology. 219:653–660. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pugazhenthi S, Nesterova A, Sable C,
Heidenreich KA, Boxer LM, Heasley LE and Reusch JE: Akt/protein
kinase B up-regulates Bcl-2 expression through cAMP-response
element-binding protein. J Biol Chem. 275:10761–10766. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Palomero T, Sulis ML, Cortina M, Real PJ,
Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, et
al: Mutational loss of PTEN induces resistance to NOTCH1 inhibition
in T-cell leukemia. Nat Med. 13:1203–1210. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho D: Novel targeting of
phosphatidylinositol 3-kinase and mammalian target of rapamycin in
renal cell carcinoma. Cancer J. 19:311–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bjornsti MA and Houghton PJ: The TOR
pathway: A target for cancer therapy. Nat Rev Cancer. 4:335–348.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mungamuri SK, Yang X, Thor AD and
Somasundaram K: Survival signaling by Notch1: Mammalian target of
rapamycin (mTOR)-dependent inhibition of p53. Cancer Res.
66:4715–4724. 2006. View Article : Google Scholar : PubMed/NCBI
|