Introduction
Melanoma is an aggressive cancer with an extremely
poor prognosis. In 2015, the number of newly diagnosed cases of
melanoma in the United States was 73,870, and the number of deaths
was approximately 9,940 (1). In
China, the number of newly diagnosed cases of melanoma in 2015 was
8,000, and the number of deaths was approximately 3200 (2).
BRAF is an important serine/threonine kinase in the
mitogen-activated protein kinase (MAPK) signaling pathway, which
plays a critical role in cell proliferation and apoptosis (3,4). Abnormal
expression or activation of BRAF has been found in a variety of
tumors (5). In melanoma, BRAF
mutations have been detected in 66% of Caucasian patients (6) and 25.5% of non-Caucasian patients
(7). Moreover, BRAF p.V600E
(1799T>A) is a hotspot mutation that is detected in melanoma,
and this mutation can increase BRAF kinase activity by 10–20-fold
(8) and accounts for 80–90% of all
detected BRAF mutations (7,9). Patients can be treated effectively
because the selective inhibitors vemurafenib plus cobimetinib
(10) or dabrafenib plus trametinib
(11) have been classified as the
primary first-line therapy by the National Comprehensive Cancer
Network (NCCN) for the treatment of advanced BRAF V600-mutant
melanoma. However, the clinical significance of other BRAF
mutations, which account for approximately 10 to 20% of cases, is
largely unknown.
Recent studies suggest that the effects of BRAF
mutations can be divided into kinase-activating (e.g., p.V600E) and
kinase-impairing (e.g., p.D594G or G596N) mutations (12). In contrast to BRAF V600E, which causes
hyperactivation of downstream kinase pathways, kinase-impairing
mutations lead to a reduction in BRAF kinase activity or
alternatively activation of silent and wild-type CRAF to elevate
MEK activity (13,14).
Because BRAF kinase-impairing mutations are
different from kinase-activating (e.g., p.V600E) mutations, we
retrospectively collected samples and clinical data from a
non-Caucasian patient population and compared the clinical and
pathological characteristics as well as clinical outcomes of
patients bearing BRAF kinase-activating mutated melanoma with those
of patients with BRAF wild-type melanoma. Our goal was to shed
light on different therapeutic strategies for treating BRAF-mutated
tumors.
Materials and methods
Patients and tissue samples
This study used samples from 1,554 melanoma patients
who were hospitalized between July 2012 and July 2015 at Beijing
Cancer Hospital and Institute. All samples were analyzed by
hematoxylin and eosin (H&E) staining and by
immunohistochemistry to confirm the diagnosis of melanoma. Clinical
data, including age, sex, stage, thickness (Breslow), ulceration,
and survival status (follow-up was continued until December 2016),
were collected. A total of 912 valid follow-up datas were obtained.
This study was approved by the Medical Ethics Committee of the
Beijing Cancer Hospital and Institute and was conducted according
to the Declaration of Helsinki Principles.
Mutation screening
Genomic DNA was extracted from melanoma tissue
sample sections using DNA FFPE Tissue kit (Qiagen, Hilden,
Germany). To detect hotspot mutations, we amplified exon 15 of the
BRAF gene by Nested PCR. The primer sequences are listed in
Table I. We purified PCR products
with QIAquick (Qiagen) and sequenced them on an ABI3130 automated
sequencer (Applied Biosystems, Foster City, CA, USA). All mutations
were confirmed by repeat bidirectional sequencing.
 | Table I.Primers used in Nested PCR. |
Table I.
Primers used in Nested PCR.
| Gene | Exon | Primer set 1 | Primer set 2 |
|---|
| BRAF | 15 | F:
5′-TTATTGACTCTAAGAGGAAAGATGAAG-3′ | F:
5′-TTATTGACTCTAAGAGGAAAGATGAAG-3′ |
|
|
| R:
5′-TGATTTTTGTGAATACTGGGAAC-3′ | R:
5′-GGCCAAAAATTTAATCAGTGGA-3′ |
Statistical analysis
All statistical analyses were performed with a
significance level of 0.05 (two-sided) using SPSS 20.0 software.
Fisher's exact test or the χ2 test was used when
comparing clinical and pathological characteristics according to
BRAF mutational status. Overall survival (OS) analysis was
performed according to the Kaplan-Meier method, and survival curves
were compared using the log-rank test. A Cox proportional hazard
model was adopted in the multivariate analysis.
Results
Prevalence of BRAF mutations in
Chinese melanoma patients
Of the 1,554 melanoma samples analyzed, 380 BRAF
mutations were detected in 377 patients (3 patients carried two
mutations simultaneously). The prevalence of BRAF mutations in this
group of Chinese melanoma patients was 24.3% (377/1554). BRAF
p.V600E (1799T>A) accounted for 87.5% (330/377) of the detected
mutations, which was consistent with previous studies. The
remaining 12.5% (47/377) of patients harbored non-p.V600E
mutations, of which p.V600K was the most frequent [5.3% (20/377)],
and 7 patients harbored p.D594G [1.9% (7/377)]. Twenty other
mutation types were also found, such as p.K601E, p.D594N, and
p.G596R.
BRAF mutations categorized by
serine-threonine kinase activity
To better understand the BRAF mutation spectrum in
melanomas, we categorized the BRAF mutations according to the
reported effects of the mutation on serine-threonine kinase
activity (Table II) (12). As showed in Table II, the overall mutation frequency was
93.4% (355/380) for kinase-activating mutations, 3.4% (13/380) for
kinase-impairing mutations, and 3.2% (12/380) for mutations with
unknown effects. Interestingly, the kinase-impairing mutations
detected in our cohort were all focused on BRAF codon 594 or 596,
which is of potential therapeutic importance.
 | Table II.BRAF mutations categorized by
serine-threonine kinase activity. |
Table II.
BRAF mutations categorized by
serine-threonine kinase activity.
| Kinase activity | BRAF mutations | No. |
|---|
| Kinase-activating
mutations | L597R | 1 |
|
| A598V | 1 |
|
| V600E | 330 |
|
| V600K | 20 |
|
| V600R | 1 |
|
| K601E | 2 |
|
| Total | 355 (93.4%) |
| Kinase-impairing
mutations | D594E | 1 |
|
| D594G | 7 |
|
| D594N | 2 |
|
| G596R | 2 |
|
| G596D | 1 |
|
| Total | 13 (3.4%) |
| Mutations with
unknown effects | V590A | 1 |
|
| I592T | 1 |
|
| V599 ins | 1 |
|
| S602P | 2 |
|
| S602F | 1 |
|
| S605N | 1 |
|
| S607F | 1 |
|
| Q612stop | 1 |
|
| S616F | 1 |
|
| 1795 ins ACT | 1 |
|
| 1793 ins CTA | 1 |
|
| Total | 12 (3.2%) |
| Total |
| 380a |
Correlation of BRAF mutational status
with the clinicopathologic features of melanoma
To better compare the clinical and pathological
characteristics of patients bearing different BRAF kinase
activation status, we conducted a long-term follow-up to these
patients. A total of 912 valid follow-up datas were obtained. These
patients were divided into three groups: the BRAF wild-type group
(n=752), BRAF V600E group (n=147), and BRAF D594/G596 group (n=13)
(Table III). In our cohort, the
median age was significantly different among the three groups
(P<0.001). The median age of patients with BRAF V600E was 48
years old, whereas that of patients with BRAF D594/G596 was 57
years old; the difference between these two groups was significant
(P<0.0001; Table III).
 | Table III.Association of BRAF gene mutation with
clinicopathological features. |
Table III.
Association of BRAF gene mutation with
clinicopathological features.
| Characteristics | BRAF wide-type
(n=752) n (%) | BRAF V600E mut
(n=147) n (%) | BRAF 594 or 596 mut
(n=13) n (%) | P-valuea | P-valueb | P-valuec |
|---|
| Sex |
|
|
| 0.091 | 0.109 | 0.303 |
| Male | 397 (52.8) | 67 (45.6) | 4 (30.8) |
|
|
|
|
Female | 355 (47.2) | 80 (54.4) | 9 (69.2) |
|
|
|
| Age (years) |
|
|
| <0.001 | <0.001 | 0.017 |
|
Median | 55 | 49 | 58 |
|
|
|
|
Range | 7–92 | 7–84 | 25–75 |
|
|
|
| Subtype |
|
|
| <0.001 | <0.001 | <0.001 |
|
Acral | 395 (52.5) | 58 (39.5) | 3 (23.1) |
|
|
|
|
Mucosal | 208 (27.7) | 14 (9.5) | 7 (53.8) |
|
|
|
|
Non-acral cutaneous | 149 (19.8) | 75 (51) | 3 (23.1) |
|
|
|
| Clinical stage |
|
|
| <0.001 | <0.001 | 0.801 |
| I | 8 (1.1) | 2 (1.4) | 0 (0) |
|
|
|
| II | 439 (58.4) | 39 (26.5) | 5 (38.5) |
|
|
|
|
III | 165 (21.9) | 29 (19.7) | 2 (15.4) |
|
|
|
| IV | 140 (18.6) | 77 (52.4) | 6 (46.2) |
|
|
|
| Ulceration |
|
|
| 0.034 | 0.030 | 0.043 |
|
Yes | 426 (56.6) | 90 (66.7) | 5 (38.5) |
|
|
|
| No | 326 (43.4) | 45 (33.3) | 8 (61.5) |
|
|
|
| Thickness (mm) |
|
|
| 0.017 | 0.008 | 0.124 |
|
≤1.0 | 71 (9.4) | 12 (8.2) | 0 (0) |
|
|
|
|
1.1–2.0 | 86 (11.4) | 9 (6.1) | 3 (23.1) |
|
|
|
|
2.1–4.0 | 196 (26.1) | 26 (17.7) | 2 (15.4) |
|
|
|
|
>4.0 | 399 (53.1) | 100 (6.8) | 8 (61.5) |
|
|
|
We found that BRAF V600E mutations were more
frequent in non-acral cutaneous melanoma (51.0%), whereas BRAF
D594/G596-mutated tumors occurred more frequently in mucosal
melanomas (53.8%); the difference between these two groups was
significant (P<0.001; Table
III). Clinical stage is an important clinical feature of
melanoma. Among the 147 patients with BRAF V600E mutations, the
percentages of patients at stages I, II, III, and IV were 1.4 (2
cases), 26.5 (39 cases), 19.7 (29 cases), and 52.4% (77 cases),
respectively, which was not significantly different from the
respective percentages for BRAF D594/G596 mutations (P=0.801;
Table III). In our cohort, the
overall ulceration rate was 57.1% (521/912). The frequencies of
ulceration in the BRAF wild-type group, BRAF V600E group, and BRAF
D594/G596 group were approximately 56.6, 66.7, and 38.5%,
respectively. The ulceration rate in patients with BRAF V600E
mutations was significantly higher than that in patients with BRAF
D594/G596 mutations (P=0.043).
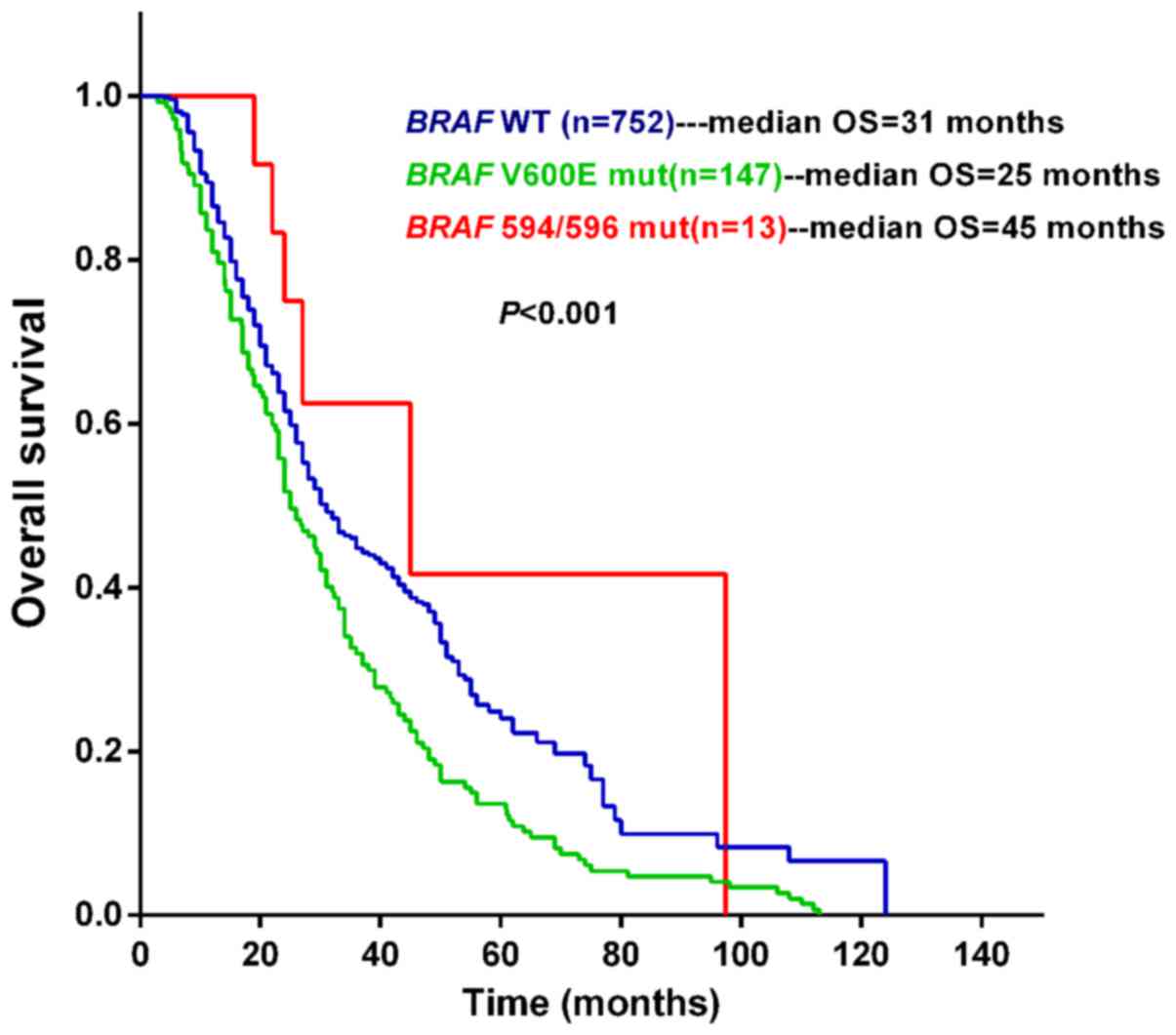
Prognostic significance of BRAF
mutational status for OS in melanoma
We further analyzed the correlation between BRAF
mutational status and OS time with a median follow-up period of
32.2 (range: 3.0–124.0) months. BRAF V600E-mutated patients had a
shorter median survival time (median OS =25.0 months) than patients
with wild-type BRAF (median OS =31.0 months) or with BRAF D594/G596
mutations (median OS =45 months). The median OS was significantly
different among these three groups (P<0.0001; Fig. 1). BRAF V600E was again associated with
a poor prognosis. Patients with BRAF D594/G596 mutations had longer
OS times than patients with BRAF V600E mutations (median OS: 45 vs.
25 months; HR: 0.45 (95% CI: 0.31–0.97), P=0.043; Table IV). In the Cox regression analysis,
BRAF mutational status, stage, thickness and pathological subtype
had a combined effect on the patients' prognosis.
 | Table IV.HR for death in relation to BRAF
mutation status. |
Table IV.
HR for death in relation to BRAF
mutation status.
| Groups | HR (95% CI of
ratio) | P-value |
|---|
| BRAF V600E vs. BRAF
WT | 1.45
(1.27–1.92) | <0.0001 |
| BRAF 594/596 mut
vs. BRAF WT | 0.61
(0.35–1.25) | 0.214 |
| BRAF 594/596 mut
vs. BRAF V600E | 0.45
(0.31–0.97) | 0.043 |
These data suggest that BRAF D594/G596 mutations may
have positive prognostic significance for melanoma patients,
whereas BRAF V600E mutations have a negative association with the
OS.
Discussion
Targeted therapy and immunotherapy have shown
promising clinical efficacy in treating melanoma (9–11,15,16).
Selective inhibitor combination therapy has been approved to treat
melanoma patients harboring the BRAF activation mutation p.V600E
(9–11). However, mutations in the BRAF gene can
result in activation or impairment of downstream kinases, leading
to entirely different active states of the MAPK pathway through
different molecular mechanisms. BRAF V600E is a kinase-activating
mutation that stimulates downstream kinase pathways. Whereas BRAF
kinase-impairing mutations increase MEK activity by activating CRAF
(17).
However, the clinical significance of BRAF
kinase-impairing mutations is largely unknown. Selecting treatment
options for patients with non-V600E/K mutations including these
kinase-impairing mutations is difficult. The BRAF mutations that
these patients carry are rare, and there is a lack of retrospective
studies or clinical guidelines to determine the impact of these
mutations on disease progression and survival. These reasons
prompted us to hypothesize that kinase-impairing mutations were of
potential importance in targeted therapies. In this study, we
examined the largest number of samples from non-Caucasian patients
to date to describe the characteristics of BRAF mutations.
Moreover, we categorized patients according to BRAF kinase activity
and compared the clinical characteristics among groups. This
approach differs from previous studies, making our results
important for identifying therapeutic strategies for treating
melanoma.
Consistent with other studies (7), BRAF mutations were detected in 24.3%
(377/1,554) of malignant melanomas in non-Caucasian patients, which
was less than the rate of 60% reported in a Caucasian population
(18), again demonstrating that there
is a large difference in the genetic features of melanoma between
non-Caucasians and Caucasians. In this study, most of the BRAF gene
mutations were concentrated among three mutation types, i.e., BRAF
V600E, BRAF V600K, and BRAF D594G, which accounted for 86.8, 5.3,
and 1.8% of all detected mutations, respectively. Mutations in
codon 594 also appeared in other forms, such as D594E (n=1) and
D594 N (n=2). In our study, the mutation probability for codon 594
was 0.64% (10/1554), which was lower than that reported in a
previous study (12). The reason for
this difference may be differences in the sample size (our sample
size was 1,554, and that of the previous study was 152).
Several previous studies have identified a variety
of kinase-inactivating mutations, such as T598I, D593V, and others
(19). In our cohort,
kinase-impairing mutations were found in 13 non-Caucasian patients,
and these mutations were all in BRAF codon 594 or 596.
Kinase-inactivating mutations are concentrated in these loci, and
these may be characteristics of non-Caucasian melanoma
patients.
Patients with BRAF V600E mutations were younger, and
the proportion of patients with non-acral skin melanoma was higher
than that of other subtypes. These results are consistent with
those of Bauer's group (20). We also
found that patients with BRAF D594/596 mutations were older and
that these mutations were more common in mucosal melanoma patients.
In addition, patients with BRAF V600E mutations were more likely to
ulcerate, whereas the incidence of ulceration in the BRAF D594/G596
group was significantly lower than that of the BRAF V600E mutation
group. Thus, there were significant differences in pathological
characteristics between BRAF V600E-mutated melanomas and BRAF
D594/G596-mutated melanomas. A recent study reported a similar
conclusion for mCRC patients (21),
but such a finding has not been reported in melanoma research.
The evidence from the clinical outcomes analysis
suggests that BRAF D594/G596 mutations may predict a longer
survival time. However, standardized treatment options are not
available for these types of mutations. BRAF D594/G596-mutated
melanomas may be not sensitive to specificity BRAF inhibitors such
as Vemurafenib due to the reduction in BRAF kinase activity
(13,14). In vitro studies, D594 G mutated
melanoma lines were highly resistant to the MEK inhibitor U0126
(17). This indicated that BRAF
D594/G596-mutated melanomas may be insensitive to either BRAF or
MEK inhibitors. Smalley et al reported that sorafenib (BAY
43–9006, Nexxavar), a multi-kinase inhibitor including inhibit
CRAF, was better at reducing the growth of melanoma xenografts with
D594G mutation than those with V600E mutation (17). These indicated that CRAF inhibitors
including but not limited to sorafenib may be a possible treatment
strategy for BRAF D594/G596-mutated melanomas. In future studies,
we will further study possible mechanisms and therapeutic
strategies in melanoma cell models and animal models.
In conclusion, we identified a rare and unexplored
subtype of melanoma with different molecular features, pathological
characteristics, and clinical outcomes compared with BRAF
V600E-mutated melanomas. Our work is thus of significance for the
development of accurate personalized medicine to treat
melanoma.
Acknowledgements
This work was supported by grants from the Major
State Basic Research Development Program of China (2013CB911004),
National Natural Science Foundation of China (81301984, 81402264,
81672696), Beijing Municipal Natural Science Foundation (7152033,
7154187), Beijing Talents Fund (2014000021223ZK26,
2016000021223ZK18), Beijing Baiqianwan Talents Project, Beijing
Municipal Administration of Hospitals Clinical Medicine Development
of special funding support (ZYLX201603), and Beijing Municipal
Science and Technology Commission (Z151100003915074). An English
language service by Editage (http://www.editage.cn/english-editing) was used to
help prepare the paper.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wada T and Penninger JM: Mitogen-activated
protein kinases in apoptosis regulation. Oncogene. 23:2838–2849.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gray-Schopfer V, Wellbrock C and Marais R:
Melanoma biology and new targeted therapy. Nature. 445:851–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Si L, Kong Y, Xu X, Flaherty KT, Sheng X,
Cui C, Chi Z, Li S, Mao L and Guo J: Prevalence of BRAF V600E
mutation in Chinese melanoma patients: Large scale analysis of BRAF
and NRAS mutations in a 432-case cohort. Eur J Cancer. 48:94–100.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Flaherty KT, Puzanov I, Kim KB, Ribas A,
McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K and
Chapman PB: Inhibition of mutated, activated BRAF in metastatic
melanoma. N Engl J Med. 363:809–819. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hauschild A, Grob JJ, Demidov LV, Jouary
T, Gutzmer R, Millward M, Rutkowski P, Blank CU, Miller WH Jr,
Kaempgen E, et al: Dabrafenib in BRAF-mutated metastatic melanoma:
A multicentre, open-label, phase 3 randomised controlled trial.
Lancet. 380:358–365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ascierto PA, McArthur GA, Dréno B,
Atkinson V, Liszkay G, Di Giacomo AM, Mandalà M, Demidov L,
Stroyakovskiy D, Thomas L, et al: Cobimetinib combined with
vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM):
Updated efficacy results from a randomised, double-blind, phase 3
trial. Lancet Oncol. 17:1248–1260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Robert C, Karaszewska B, Schachter J,
Rutkowski P, Mackiewicz A, Stroiakovski D, Lichinitser M, Dummer R,
Grange F, Mortier L, et al: Improved overall survival in melanoma
with combined dabrafenib and trametinib. N Engl J Med. 372:30–39.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng G, Tseng LH, Chen G, Haley L, Illei
P, Gocke CD, Eshleman JR and Lin MT: Clinical detection and
categorization of uncommon and concomitant mutations involving
BRAF. BMC Cancer. 15:7792015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heidorn SJ, Milagre C, Whittaker S, Nourry
A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer
CJ, Pritchard C, et al: Kinase-dead BRAF and oncogenic RAS
cooperate to drive tumor progression through CRAF. Cell.
140:209–221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wan PT, Garnett MJ, Roe SM, Lee S,
Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ,
Barford D, et al: Mechanism of activation of the RAF-ERK signaling
pathway by oncogenic mutations of B-RAF. Cell. 116:855–867. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Larkin J, Chiarion-Sileni V, Gonzalez R,
Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M,
Rutkowski P, et al: Combined nivolumab and ipilimumab or
monotherapy in untreated melanoma. N Engl J Med. 373:23–34. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smalley KS, Xiao M, Villanueva J, Nguyen
TK, Flaherty KT, Letrero R, Van Belle P, Elder DE, Wang Y,
Nathanson KL and Herlyn M: CRAF inhibition induces apoptosis in
melanoma cells with non-V600E BRAF mutations. Oncogene. 28:85–94.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Curtin JA, Fridlyand J, Kageshita T, Patel
HN, Busam KJ, Kutzner H, Cho KH, Aiba S, Bröcker EB, LeBoit PE, et
al: Distinct sets of genetic alterations in melanoma. N Engl J Med.
353:2135–2147. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ikenoue T, Hikiba Y, Kanai F, Tanaka Y,
Imamura J, Imamura T, Ohta M, Ijichi H, Tateishi K, Kawakami T, et
al: Functional analysis of mutations within the kinase activation
segment of B-Raf in human colorectal tumors. Cancer Res.
63:8132–8137. 2003.PubMed/NCBI
|
|
20
|
Bauer J, Büttner P, Murali R, Okamoto I,
Kolaitis NA, Landi MT, Scolyer RA and Bastian BC: BRAF mutations in
cutaneous melanoma are independently associated with age, anatomic
site of the primary tumor, and the degree of solar elastosis at the
primary tumor site. Pigment Cell Melanoma Res. 24:345–351. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cremolini C, Di Bartolomeo M, Amatu A,
Antoniotti C, Moretto R, Berenato R, Perrone F, Tamborini E, Aprile
G, Lonardi S, et al: BRAF codons 594 and 596 mutations identify a
new molecular subtype of metastatic colorectal cancer at favorable
prognosis. Ann Oncol. 26:2092–2097. 2015. View Article : Google Scholar : PubMed/NCBI
|