Introduction
Colorectal carcinoma (CRC) is the one of the most
common types of cancer, and is the fifth leading cause of
cancer-associated mortalities worldwide (1). Despite recent advancements in the
surgical techniques, radiotherapy and chemotherapy available to
patients diagnosed with CRC over the past several decades, the
overall survival rate has not significantly improved. The existence
of numerous known carcinogens and varying genetic backgrounds makes
it difficult to determine which factors are most important in the
development of CRC. Thus, there is a requirement to further
understand the underlying molecular mechanisms and identify novel
prognostic biomarkers and therapeutic targets to provide improved
treatment strategies for CRC.
Bactericidal or permeability-increasing protein
fold-containing family A member 1 (BPIFA1) is a protein-coding gene
specifically expressed in the upper airways and nasopharyngeal
regions. A number of previous studies have demonstrated that BPIFA1
is involved in various physiological and pathological processes
(2–19). It is considered to be involved in
inflammatory responses to irritants in the upper airway (2–7). BPIFA1
may decrease mycoplasma pneumonia expression levels and inhibit
interleukin 8 (8). BPIFA1 protein was
also revealed to have antibacterial activity against gram-negative
bacteria (9,10). The anti-inflammatory function has been
associated with the regulation of macrophagic activity (10), particularly cellular responses to
lipopolysaccharide (11). Previous
studies have demonstrated that the expression levels of BPIFA1 are
upregulated in lung cancer (12–14),
gastric cancer (15) and head and
neck neoplasms, such as mucoepidermoid carcinoma and nasopharyngeal
carcinoma (16–19). BPIFA1 (LUNX) was identified to be a
potential marker for the micro-metastasis of non-small cell lung
cancer (NSCLC) (14). BPIFA1 was also
revealed to be a novel marker able to distinguish gastric hepatoid
adenocarcinoma from primary hepatocellular carcinoma (15). Recently, it was demonstrated that
anti-LUNX antibody slowed tumor growth and metastasis and improved
the survival time of mice bearing lung cancer xenografts (20). However, little is known about BPIFA1
in CRC.
In the present study, the mRNA and protein
expression levels of BPIFA1 in clinically resected human CRC and
adjacent noncancerous tissues were examined by quantitative
polymerase chain reaction (qPCR) and immunohistochemistry (IHC),
and the association between BPIFA1 protein expression levels and
the prognosis of patients with CRC was analyzed.
Materials and methods
Tissue specimens
Fresh formalin-fixed and paraffin-embedded CRC tumor
tissue samples were obtained from patients with a diagnosis of
primary CRC, who underwent surgical resection at Nanfang Hospital,
Southern Medical University (Guangzhou, China) from February 2000
to November 2010. The use of tissues for this study was approved by
the Ethics Committee of Nanfang Hospital, Southern Medical
University. Written informed consent was obtained from all patients
prior to enrollment in the present study. A total of 36 cases of
fresh CRC tissue (20 males and 16 females) of a mean age of
52.08±9.16 (range 35–67) were snap-frozen in liquid nitrogen and
stored at −80°C until further use. A total of 118 cases of archived
CRC tissue samples and 73 adjacent non-tumors tissues were from
Nanfang Hospital were used for immunohistochemistry to investigate
the expression of BPIFA1 protein. None of the patients received
pre-operative chemotherapy or radiotherapy. The patients included
76 males and 42 females, ranging in age from 24–88 years (mean,
57.83±1.35 years). During the follow-up period, 56 patients
succumbed to disease between months 1 and 122 months of (median, 56
months).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 50–100 mg of tissue
using TRIzol Reagent (Takara Biotechnology Co., Ltd., Dalian,
China). cDNA was synthesized using the PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd.). RT-qPCR was performed to detect
the expression of BPIFA1 using the One-Step SYBR PrimeScript RT-PCR
kit (Takara Biotechnology Co., Ltd.), using the following thermal
cycling profile: 95°C for 5 min, followed by 40 cycles of
amplification (95°C for 40 sec, 56°C for 40 sec and 72°C for 40
sec), followed by dissociation curve analysis to validate the
amplification of the product, normalized to the expression of
β-actin. The primers were as follows: β-actin forward,
5′-TAAGGAGAAGCTGTGCTACG-3′; reverse, 5′-GACTCGTCATACTCCTGCTT-3′;
BPIFA1 forward, 5′-GTGGGGGAGAGAGAG-GAGAC-3′, reverse,
5′-GTCAAGCTTCCTGCAAGACC-3′. The assay was performed in triplicate
for each case to allow for the assessment of technical
variability.
IHC
IHC staining of tissue samples was performed
according to a previously described protocol (21) using a Dako EnVision System (Dako;
Agilent Technologies, Inc., Santa Clara, CA, USA), in order to
evaluate BPIFA1 protein expression levels in 118 human CRC tissue
samples. Briefly, the sections were incubated with primary
antibodies against BPIFA1 (cat. no. LS-B3549; dilution 1:400,
LifeSpan BioSciences, Inc., Seattle, WA, USA) for 1 h at room
temperature. Following incubation with the peroxidase-conjugated
secondary antibody from the Dako EnVision System (ready-to-use
dilution), expression patterns were visualized using the substrate
diaminobenzidine to generate a stained product. For negative
controls, the antibodies were replaced with normal goat serum
(Maixin Biotech, Fuzhou, China) and incubated under the same
conditions as the BPIFA1 antibody.
Evaluation of staining for BPIFA1
To eliminate inter-observer bias, the expression
levels of BPIFA1 were reviewed and scored by two independent
pathologists who were blinded to the patients' clinicopathological
data. Staining for BPIFA1 was assessed using a method previously
described (22,23). On a scale of 0–3, the staining
intensity was scored as follows: Negative (no staining, 0), weak
(light yellow, 1), medium (yellow-brown, 2) or strong (brown, 3).
The extent of the staining was defined as the percentage of
positively stained areas of tumor cells or normal mucosal
epithelial cells relative to the whole tumor area or to the entire
section for the normal tissue samples. The extent of staining was
scored on a scale of 0–4 as follows: 0, 0%; 1, >0-≤25%; 2,
>25-≤50%; 3, >50-≤75% and 4, >75-<100%. The sum of the
staining-intensity and staining-extent scores was used as the final
staining score for BPIFA1. For statistical analysis, a final
staining score of ≥3 was regarded to represent high expression
levels.
Statistical analysis
The SPSS software package (version 16.0; SPSS Inc.,
Chicago, IL, USA) was used for all statistical analysis.
Differences in BPIFA1 expression levels in fresh tissue samples
were evaluated using the paired Student's t-test. Differences
between variables were determined using the χ2 test.
Survival curves for the patients with various expression levels of
BPIFA1 protein were constructed using the Kaplan-Meier method and
compared using the log-rank test. Cox proportional hazards
regression analysis was performed for univariate and multivariate
analyses of the prognostic values. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of BPIFA1 mRNA expression
level in CRC tissues and adjacent noncancerous tissues
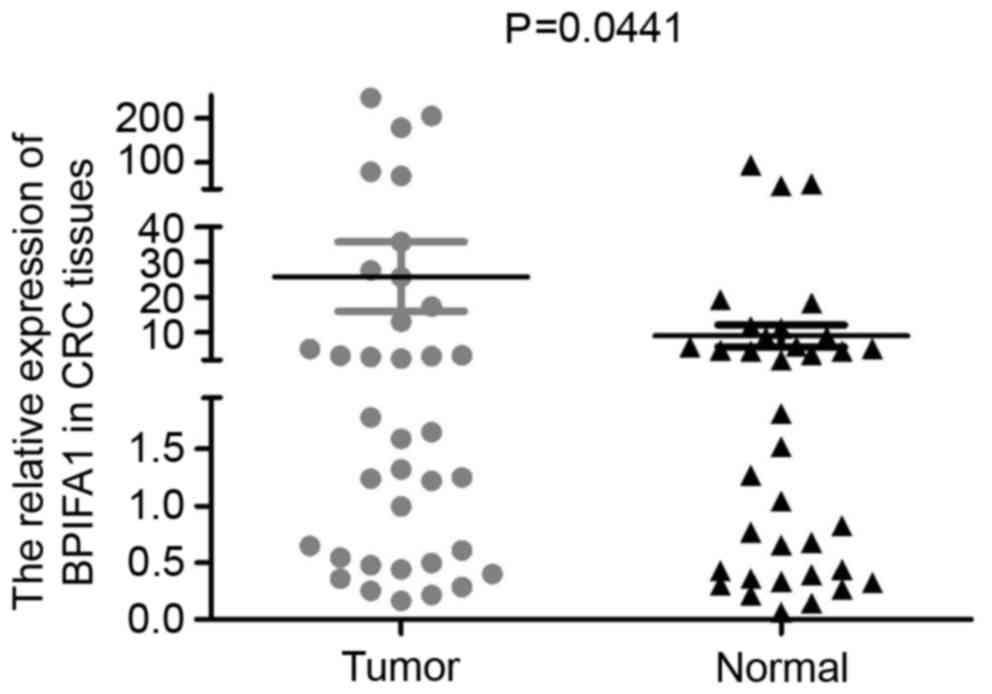
The expression levels of BPIFA1 mRNA in 36 paired
human CRC tissues and adjacent noncancerous tissues were quantified
by qPCR. The results of the present study indicated that there was
variability in the expression levels of BPIFA1 across the tissue
samples. Notably, BPIFA1 mRNA was upregulated in CRC tissues, when
compared with in adjacent noncancerous tissues (Fig. 1; P=0.0441), and the majority of CRC
tissues exhibited a >2-fold increase in BPIFA1 expression
levels, as compared with normal tissues.
Expression levels of BPIFA1 protein in
CRC tissues
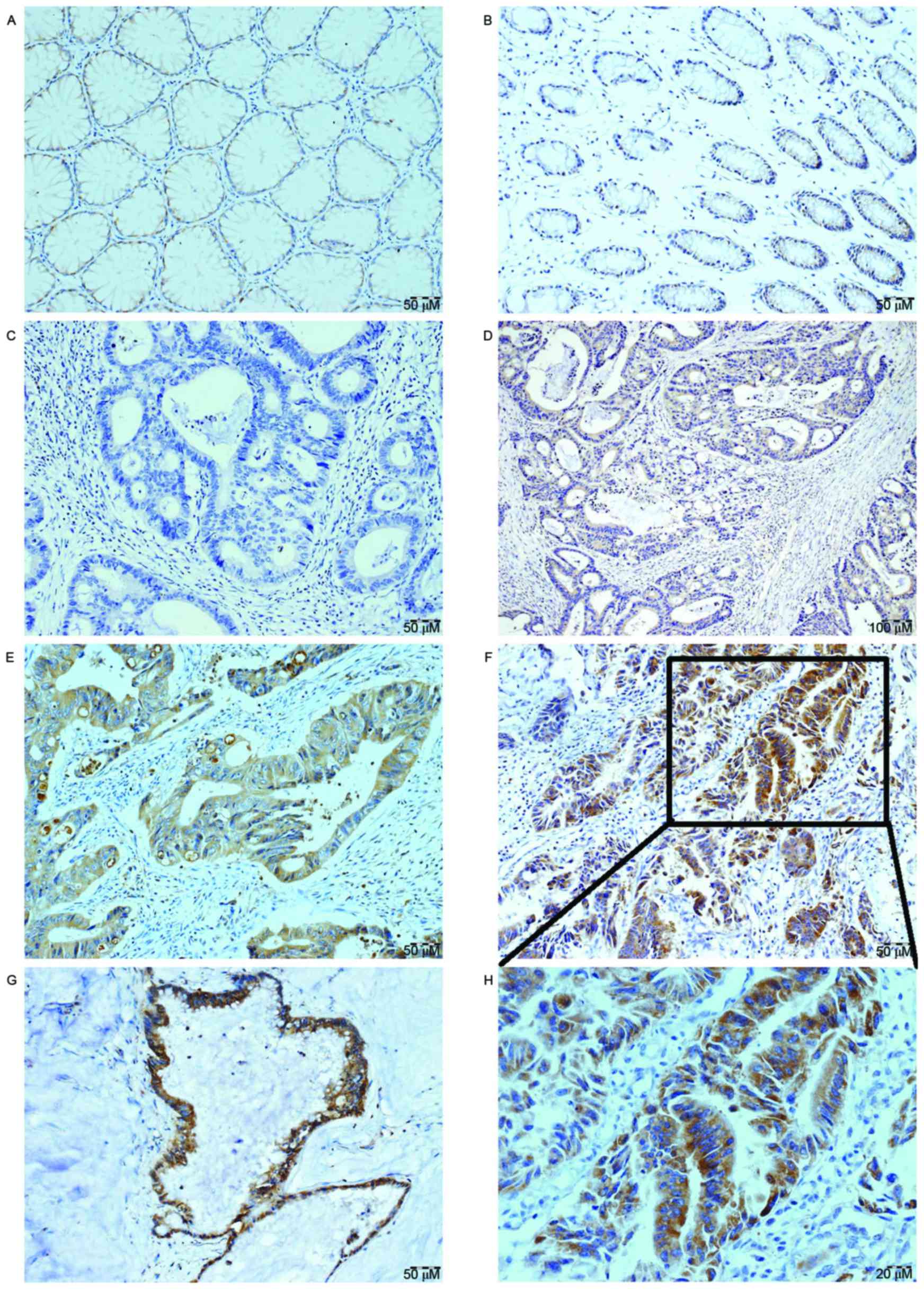
The present study evaluated the protein expression
levels of BPIFA1 in archived paraffin-embedded primary CRC tissues
and normal colon tissue samples by performing immunohistochemical
staining. The results revealed that BPIFA1 protein was expressed in
the cytoplasm of benign and malignant epithelial cells. It was
observed that BPIFA1 protein was expressed in 23/73 (31.51%) normal
colon mucosa samples. In comparison, BPIFA1 was expressed in
110/118 (93.22%) CRC tumor tissue samples. The expression level of
BPIFA1 was markedly upregulated in CRC tissues compared with in
normal mucosa tissues (P<0.001). According to the aforementioned
reclassification guidelines, the present study determined there was
high BPIFA1 expression in 70/118 (59.32%) of the CRC tissue samples
(Fig. 2).
Association between
clinicopathological characteristics and BPIFA1 expression level in
patients with CRC
The association analysis between clinicopathological
characteristics and BPIFA1 protein expression level was evaluated
in individuals. The data is presented in Table I. The upregulation of BPIFA1 was
significantly associated with invasion depth (P=0.040), positive
regional lymph node metastasis (P=0.035) and distant metastasis
(P=0.010); conversely, there was no association with age, sex,
tumor site, tumor size and differentiation grade (P>0.05).
 | Table I.Associations between the
clinicopathological features and expression levels of BPIFA1. |
Table I.
Associations between the
clinicopathological features and expression levels of BPIFA1.
|
|
| BPIFA1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristics | n | Low (%) | High (%) | P-value | χ2 |
|---|
| Sex |
|
|
|
|
|
| Male | 76 | 32 (42.11) | 44 (57.89) |
|
Female | 42 | 16 (38.10) | 26 (61.90) | 0.671 | 0.180 |
| Age (years) |
|
|
|
|
|
| ≤57 | 59 | 21 (35.59) | 38 (64.41) |
|
|
|
>57 | 59 | 27 (45.76) | 32 (54.24) | 0.261 | 1.264 |
| Tumor site |
|
|
|
|
|
| Proximal
colon | 37 | 15 (41.57) | 22 (58.43) |
|
|
| Distant
colon | 26 | 13 (50.00) | 13 (50.00) |
|
|
|
Rectum | 55 | 20 (36.36) | 35 (63.64) | 0.506 | 1.361 |
| Tumor size (diameter
in cm) |
|
|
|
|
|
|
<5 | 60 | 25 (41.67) | 35 (58.33) |
| ≥5 | 58 | 23 (39.65) | 35 (60.35) | 0.879 | 0.023 |
| Tumor
differentiation |
|
|
|
|
|
| Good | 51 | 26 (50.98) | 25 (49.02) |
|
|
|
Moderate | 45 | 16 (35.56) | 29 (64.44) |
|
|
|
Poor | 22 | 6 (27.27) | 16 (72.73) | 0.112 | 4.371 |
| T-stage |
|
|
|
|
|
|
1–2 | 31 | 18 (58.06) | 13 (51.94) |
|
|
| 3 | 73 | 27 (36.99) | 46 (63.01) |
|
|
| 4 | 14 | 3 (21.43) | 11 (78.57) | 0.040a | 6.445 |
| N-stage |
|
|
|
|
|
|
1–2 | 48 | 14 (29.17) | 34 (70.83) |
|
|
| 0 | 70 | 34 (48.57) | 36 (51.43) | 0.035a | 4.443 |
| Distant
metastasis |
|
|
|
|
|
| 1 | 13 | 1 (7.69) | 12 (92.31) |
|
|
| 0 | 105 | 47 (44.34) | 58 (55.66) | 0.010a | 6.587 |
Association between BPIFA1 expression
level and patient survival
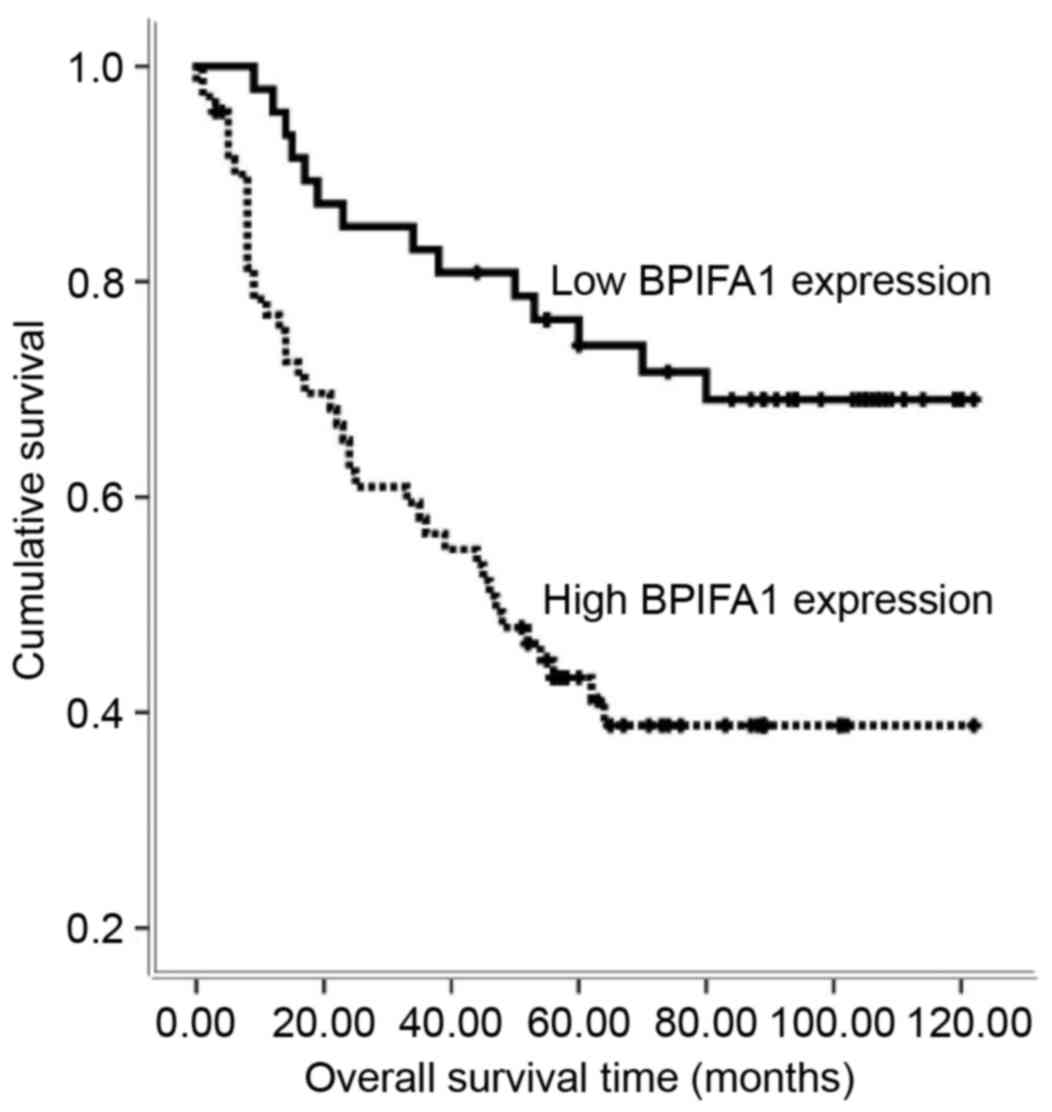
The present study used Kaplan-Meier analysis as a
first step to assess the prognostic value of BIPFA1 in CRC. It was
observed that the expression level of BPIFA1 was significantly
associated with overall survival (log-rank test statistic=11.898;
P=0.001; Fig. 3). The high expression
level of BPIFA1 was associated with a shorter survival time for
patients with CRC (high BPIFA1 expression group, 62.63±6.08 months;
low BPIFA1 expression group, 95.57±6.12 months).
Univariate and multivariate analyses
of prognostic variables in patients with CRC
In order to evaluate the expression levels of BPIFA1
as an independent prognostic factor for patients with CRC,
univariate and multivariate analyses were preformed to determine
the prognostic value of clinicopathological factors, including sex,
age, tumor size, differentiation grade, T-stage, N-stage and
distant metastasis. The results revealed that high expression
levels of BPIFA1 protein are an independent prognostic factor of
disease outcome in patients with CRC (Table II; P=0.049).
 | Table II.Summary of overall survival analyses
by univariate and multivariate Cox regression analysis. |
Table II.
Summary of overall survival analyses
by univariate and multivariate Cox regression analysis.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| Sex | 0.4410 | 0.8010 | 0.456–1.408 |
|
|
|
| Age | 0.4770 | 1.2540 | 0.672–2.340 |
|
|
|
| Tumor site | 0.4560 | 1.1240 | 0.827–1.528 |
|
|
|
| Tumor size | 0.6090 | 0.8710 | 0.513–1.479 |
|
|
|
| Tumor
differentiation | 0.0620 | 1.3970 | 0.983–1.985 |
|
|
|
| T-stage | 0.0020 | 2.0130 | 1.305–3.104 | 0.093 | 1.465 | 0.938–2.286 |
| N-stage | <0.001 | 2.5990 | 1.525–4.428 | 0.002 | 2.430 | 1.402–4.213 |
| M-stage | <0.001 | 7.5280 | 3.860–14.683 | <0.001 | 6.657 | 3.258–13.604 |
| BPIFA1 | 0.0010 | 2.7930 | 1.515–5.146 | 0.049 | 1.903 | 1.002–3.615 |
Discussion
The palate, lung and nasal epithelium clone (PLUNC)
protein was first described in the epithelium, trachea and bronchus
of mouse embryos (24); subsequently,
a family with ten members of human equivalents was recorded
(9). Based on their predicted
structure of being homologous to one or both domains of the
bactericidal or permeability-increasing protein (BPI), PLUNCs can
be subdivided in two groups: Short (SPLUNC) and long (LPLUNC)
proteins (11). Recently, PLUNC
family members have been included in the BPI fold-containing
superfamily, leading to a novel nomenclature whereby SPLUNC1
protein has the designation BPIFA1 (25,26).
Previously, various antimicrobial activities have
been attributed to BPIFA1, in addition to evidence that it may
function as a host defense protein (4,6,7). However, the specific function of BPIFA1
has not yet been well defined. In prior studies, its expression was
detected in lung cancer (12–14,20),
gastric cancer (15), salivary gland
tumors (16–19) and nasopharyngeal carcinoma (19). However, to the best of our knowledge,
there have been no reports investigating the role of BPIFA1 in
CRC.
Therefore, the current study presents the first
evidence that BPIFA1 expression is upregulated at the
transcriptional and translational levels in CRC tissues, compared
with in normal mucosa tissues. These finding suggest that the
upregulation of BPIFA1 in CRC may be associated with the
carcinogenesis of CRC. BPIFA1 was overexpressed in CRC and weakly
expressed in normal colon mucosa tissue, which suggested that
BPIFA1 may be a potentially superior diagnostic marker for CRC.
Notably, the upregulation of BPIFA1 was
significantly associated with tumor invasion depth (T-stage),
positive regional lymph node metastasis (N-stage) and distant
metastasis (M-stage) of patients with CRC. Local invasion is the
initial step in tumor metastasis. Metastatic colonization at a
distant tissue is a key step in the metastatic cascade (27). The overexpression of BPIFA1 was
significantly associated with invasion and migration of CRC cells,
suggesting that BPIFA1 may serve a critical role in the metastatic
cascade of CRC. The results of the present study are consistent
with those of certain previous studies; Iwao et al (14) identified BPIFA1 to be a marker of
micrometastasis in NSCLC and Zheng et al (20) observed that BPIFA1 promotes lung
cancer cell migration and proliferation by targeting 14–3-3ζ and
14–3-3θ proteins. The metastasis of tumor cells to vital organs is
responsible for the majority of cancer-associated mortalities. The
present results indicated that targeting BPIFA1 may exert an
anti-metastasis effect and prolong the survival time of patients
with CRC.
The present study also demonstrated an association
between BPIFA1 expression and the prognosis of patients with CRC.
The results indicated that BPIFA1 protein expression level is
inversely associated with overall survival. Patients with CRC and a
high level of BPIFA1 expression experienced a shorter survival
time. In univariate and multivariate analyses, a high expression
level of BPIFA1 protein was associated with an increased risk of
mortality from CRC, indicating that a high expression level of
BPIFA1 may be an independent factor for poor prognosis for patients
with CRC. These findings are consistent with those of a previous
study investigating gastric cancer (15) and lung cancer (20), wherein the upregulation of BPIFA1 was
revealed to be associated with advanced disease stage and/or poor
prognosis. Therefore, these results suggested that overexpression
of BPIFA1 may be a promising predictor of prognosis and a potential
therapeutic target for CRC.
In conclusion, the results of the present study have
extended previous findings regarding the role of BPIFA1 in cancer
progression. The present study revealed that BPIFA1 expression was
upregulated in CRC tissues and, for the first time, highlighted the
clinical and prognostic significance of BPIFA1 in CRC. The elevated
BPIFA1 expression level was associated with tumor invasion and
metastasis, which is significantly associated with tumor
progression in patients, leading to a poor clinical outcome. These
results indicated that BPIFA1 may have potential as a clinical
predictor for aggressive phenotypes and as a prognostic predictor
for patients with CRC. However, further studies are required in
order to verify the molecular mechanisms underlying the function of
BPIFA1, and to illustrate the therapeutic value of BPIFA1 for the
treatment of CRC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81172242 and
8147238).
Glossary
Abbreviations
Abbreviations:
|
BPIFA1
|
BPI fold-containing family A member
1
|
|
CRC
|
colorectal carcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
PLUNC
|
palate, lung and nasal epithelium
clone protein
|
|
SPLUNC1
|
short palate, lung and nasal
epithelium clone protein 1
|
|
qPCR
|
quantitative polymerase chain
reaction
|
References
|
1
|
Bernard W: Stewart CPW: World cancer
report 2014: IARC. 2014.
|
|
2
|
Lukinskiene L, Liu Y, Reynolds SD, Steele
C, Stripp BR, Leikauf GD, Kolls JK and Di YP: Antimicrobial
activity of PLUNC protects against Pseudomonas aeruginosa
infection. J Immunol. 187:382–390. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gally F, Di YP, Smith SK, Minor MN, Liu Y,
Bratton DL, Frasch SC, Michels NM, Case SR and Chu HW: SPLUNC1
promotes lung innate defense against Mycoplasma pneumoniae
infection in mice. Am J Pathol. 178:2159–2167. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ghafouri B, Kihlström E, Ståhlbom B,
Tagesson C and Lindahl M: PLUNC (palate, lung and nasal epithelial
clone) proteins in human nasal lavage fluid. Biochem Soc Trans.
31:810–814. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sayeed S, Nistico L, St Croix C and Di YP:
Multifunctional role of human SPLUNC1 in Pseudomonas
aeruginosa infection. Infect Immun. 81:285–291. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bingle L, Barnes FA, Cross SS, Rassl D,
Wallace WA, Campos MA and Bingle CD: Differential epithelial
expression of the putative innate immune molecule SPLUNC1 in cystic
fibrosis. Respir Res. 8:792007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di YP, Harper R, Zhao Y, Pahlavan N,
Finkbeiner W and Wu R: Molecular cloning and characterization of
spurt, a human novel gene that is retinoic acid-inducible and
encodes a secretory protein specific in upper respiratory tracts. J
Biol Chem. 278:1165–1173. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chu HW, Thaikoottathil J, Rino JG, Zhang
G, Wu Q, Moss T, Refaeli Y, Bowler R, Wenzel SE, Chen Z, et al:
Function and regulation of SPLUNC1 protein in Mycoplasma infection
and allergic inflammation. J Immunol. 179:3995–4002. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bingle CD and Bingle L: Characterisation
of the human plunc gene, a gene product with an upper airways and
nasopharyngeal restricted expression pattern. Biochim Biophys Acta.
1493:363–367. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bingle CD and Gorr SU: Host defense in
oral and airway epithelia: Chromosome 20 contributes a new protein
family. Int J Biochem Cell Biol. 36:2144–2152. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bingle CD and Craven CJ: PLUNC: A novel
family of candidate host defence proteins expressed in the upper
airways and nasopharynx. Hum Mol Genet. 11:937–943. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bingle L, Cross SS, High AS, Wallace WA,
Devine DA, Havard S, Campos MA and Bingle CD: SPLUNC1 (PLUNC) is
expressed in glandular tissues of the respiratory tract and in lung
tumours with a glandular phenotype. J Pathol. 205:491–497. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cheng M, Chen Y, Yu X, Tian Z and Wei H:
Diagnostic utility of LunX mRNA in peripheral blood and pleural
fluid in patients with primary non-small cell lung cancer. BMC
Cancer. 8:1562008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Iwao K, Watanabe T, Fujiwara Y, Takami K,
Kodama K, Higashiyama M, Yokouchi H, Ozaki K, Monden M and Tanigami
A: Isolation of a novel human lung-specific gene, LUNX, a potential
molecular marker for detection of micrometastasis in non-small-cell
lung cancer. Int J Cancer. 91:433–437. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sentani K, Oue N, Sakamoto N, Arihiro K,
Aoyagi K, Sasaki H and Yasui W: Gene expression profiling with
microarray and SAGE identifies PLUNC as a marker for hepatoid
adenocarcinoma of the stomach. Mod Pathol. 21:464–475. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vargas PA, Speight PM, Bingle CD, Barrett
AW and Bingle L: Expression of PLUNC family members in benign and
malignant salivary gland tumours. Oral Dis. 14:613–619. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gonzalez-Arriagada WA, Santos-Silva AR,
Ito FA, Vargas PA, Speight PM, Bingle L and Lopes MA: Expression
pattern of PLUNC proteins as an auxiliary tool for the diagnosis of
high-grade mucoepidermoid carcinoma of the salivary gland. J Oral
Pathol Med. 41:589–597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He Y, Zhou G, Zhai Y, Dong X, Lv L, He F
and Yao K: Association of PLUNC gene polymorphisms with
susceptibility to nasopharyngeal carcinoma in a Chinese population.
J Med Genet. 42:172–176. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang B, Nie X, Xiao B, Xiang J, Shen S,
Gong J, Zhou M, Zhu S, Zhou J, Qian J, et al: Identification of
tissue-specific genes in nasopharyngeal epithelial tissue and
differentially expressed genes in nasopharyngeal carcinoma by
suppression subtractive hybridization and cDNA microarray. Genes
Chromosomes Cancer. 38:80–90. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng X, Cheng M, Fu B, Fan X, Wang Q, Yu
X, Sun R, Tian Z and Wei H: Targeting LUNX inhibits non-small cell
lung cancer growth and metastasis. Cancer Res. 75:1080–1090. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pietruszewska W, Bojanowska-Poźniak K and
Kobos J: Matrix metalloproteinases MMP1, MMP2, MMP9 and their
tissue inhibitors TIMP1, TIMP2, TIMP3 in head and neck cancer: An
immunohistochemical study. Otolaryngol Pol. 70:32–43. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang S, Zhou J, Wang XY, Hao JM, Chen JZ,
Zhang XM, Jin H, Liu L, Zhang YF, Liu J, et al: Down-regulated
expression of SATB2 is associated with metastasis and poor
prognosis in colorectal cancer. J Pathol. 219:114–122. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang S, Yang MH, Wang XY, Lin J and Ding
YQ: Increased expression of miRNA-182 in colorectal carcinoma: An
independent and tissue-specific prognostic factor. Int J Clin Exp
Pathol. 7:3498–3503. 2014.PubMed/NCBI
|
|
24
|
Weston WM, LeClair EE, Trzyna W, McHugh
KM, Nugent P, Lafferty CM, Ma L, Tuan RS and Greene RM:
Differential display identification of plunc, a novel gene
expressed in embryonic palate, nasal epithelium, and adult lung. J
Biol Chem. 274:13698–13703. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bingle L and Bingle CD: Distribution of
human PLUNC/BIP fold-containing (BPIF) proteins. Biochem Soc Trans.
39:1023–1027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bingle CD, Seal RL and Craven CJ:
Systematic nomenclature for the PLUNC/PSP/BSP30/SMGB proteins as a
subfamily of the BPI fold-containing superfamily. Biochem Soc
Trans. 39:977–983. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar : PubMed/NCBI
|