Introduction
Squamous cell carcinoma (SCC) is more common in
Eastern countries, including Japan. Despite the pathological
difference, esophageal cancer has a poor prognosis due to early
metastasis and direct invasion. Multimodal therapies including
surgery, chemotherapy, and radiotherapy are necessary; however, the
response rate to chemoradiation therapy is low; in one Japanese
study it was found to be 68% in patients with advanced SCC in the
thoracic esophagus (1). Although
chemo or radiosensitivity is closely related to prognosis, it is
currently impossible to predict the therapeutic effect of
therapies. Therefore, detecting resistant genes and mechanisms is
essential for tailoring treatment to improve the prognosis.
Many studies have shown numerous genes or microRNA
related to radioresistance in esophageal cancer cells (2). A few comprehensive gene analyses by
microarrays have also detected many candidate radioresistant genes
(3,4).
However, most radioresistant genes are not yet known and the
mechanisms underlying their radioresistance remain unclear.
Comprehensive gene screening with transposons is a
novel procedure for systematically identifying chemoresistant
genes, developed by Chen et al in 2013 (5). A transposon is a mobile genetic element
which transports at random to other locations in the genome. By
inserting a cytomegalovirus (CMV) promotor as a transcriptional
activator in the transposon, the following gene in the new location
will be overexpressed and the gene located at the transposon
insertion site will be downregulated. We inserted transposons into
tumor cells to form a library of ‘transposon-tagged cells.’ Each
transposon-tagged cell has randomly activated or inactivated genes.
After drug treatment is administered to the cell library, the
surviving cells can either have overexpressed chemoresistant genes
or downregulated chemosensitive genes due to the transposons with
the CMV promoter. By detecting the location of the transposon in
surviving cells, it is possible to pick up candidate
chemotherapy-resistant genes. What makes this screening method
distinct is that it is a gain-of-function genetic screening, unlike
past genetic interrogation approaches for genomes. Thus, this
screening can be used to survey untranscribed regions (5).
Using this technique, we have already succeeded in
generating cell libraries and identifying candidate genes for
cisplatin resistance in esophageal squamous cell carcinoma
(6). The aim of the present study is
to use this method to identify candidate radioresistant genes in
esophageal squamous cell carcinoma.
Materials and methods
Cell cultures and treatments
We established transposon-tagged cells in TE4 and
TE15, human well-differentiated esophageal squamous cell carcinoma
cell lines provided by the Cell Resource Center for Biomedical
Research Institute of Development, Aging and Cancer, Tohoku
University (Sendai, Japan) (6).
Puromycin-resistant genes in transposons were also inserted into
the transposon-tagged cells for puromycin selection. The cell lines
were cultured in Dulbecco's modified Eagles medium supplemented
with 10% fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and
penicillin-streptomycin solution (penicillin 10,000 U/ml,
streptomycin 10,000 µg/ml, puromycin 0.5 µg/ml) (all from Nacalai
Tesque, Inc., Kyoto, Japan). In all the experiments, the cells were
cultured at 37°C in a humidified atmosphere consisting of 5%
CO2 in the air. The parent cell lines, TE4, and TE15,
were cultured without puromycin.
Radiation treatment
Around 250,000 transposon-tagged cells were plated
on 100-mm tissue culture dishes for radiation treatment. Dishes
were irradiated by 9–11 Gy using an X-ray cell irradiator, CellRad
(Faxitron Bioptics, Tucson, AZ, USA). Irradiated cells were
cultured for more than 14 days until radioresistant cells (RRCs)
formed colonies.
Comprehensive gene screening with
transposon
The base sequences of the transposons' insertion
sites were detected using Splinkerette polymerase chain reaction
(PCR), TOPO cloning, and Sanger sequencing performed in accordance
with the protocol of our previous study (6). Insertion sites were aligned using the
BLAST function of the National Library of Medicine (http://blast.ncbi.nlm.nih.gov/Blast.cgi). We included
a maximum of 15 genes that had somewhat similar sequences (blastn)
as candidate genes for each colony.
Real-time quantitative PCR
analysis
The cells were harvested, and total RNA was
extracted with an RNeasy kit (Qiagen, Valencia, CA, USA). The
concentration of total RNA was determined using a NanoDrop 2000c
(NanoDrop Technologies, San Diego, CA, USA). cDNA was synthesized
from total RNA using a High Capacity RNA-to-cDNA kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in accordance with the
manufacturer's protocol. PCR reaction mixes were prepared using the
template cDNA samples and Fast SYBR-Green Master Mix (Thermo Fisher
Scientific, Inc.), while the expressions of human cytochrome
c oxidase 1 (MT-CO1), cytochrome c oxidase
subunits 2 (MT-CO2), and 3 (MT-CO3), ND2, and
GAPDH were analyzed using a ViiA7 Real-Time PCR system
(Thermo Fisher Scientific, Inc). The thermal cycling reaction
included incubation at 95°C for 20 sec, 40 cycles of 95°C for 3
sec, and 60°C for 30 sec. Data were collected using analytical
software (Applied Biosystems, Foster City, CA, USA). The expression
level of each gene was determined using the ΔΔCt method.
MTT assay
Survival rates after irradiation were measured by an
MTT assay. One day before 7-Gy irradiation, 5×102/well
of TE4 cells and MT-CO1-downregulated cells were seeded on a
96-well plate. TE4 cells and MT-CO1-downregulated cells with
and without irradiation were incubated with MTT (3-(4,
5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide, a yellow
tetrazole) for 3 h and incubated with dimethyl sulfoxide for 20
min. The survival rate after irradiation was measured as the ratio
of absorbance at 550 nm of irradiated cells to non-irradiated
cells.
Cytochrome c oxidase assay
A cell pellet of 10 million cells was resuspended in
200 µl of a cell lysis buffer containing 250 mM sucrose, 20 mM
HEPES-KOH (pH 7.4), 10 mM KCl, 1.5 mM Na-EGTA, 1.5 mM Na-EDTA, 1 mM
MgCl2, and a protease inhibitor cocktail. After a 20-min
incubation on ice, the cells were disrupted with a Dounce
homogenizer (50 strokes with a loose glass pestle and 50 strokes
with a tight glass pestle). The resulting homogenates were
immediately used in the cytochrome c oxidase assay.
Cytochrome c oxidase activity was determined
by using a Cytochrome c Oxidase Assay kit (Sigma-Aldrich,
St. Louis, MO, USA). Absorbance at 550 nm of ferrocytochrome
c was measured using NanoDrop 2000c (NanoDrop Technologies).
Cytochrome c oxidase activity was measured as the reduction
in absorbance in the sample at 550 nm.
To get reduced cytochrome c, 2.7 mg of
cytochrome c, dissolved in 1 ml water, was incubated with 5
µl of 0.1 M 1.4-dithiothreitol (DTT) for 15 min on ice. The
reduction in absorbance at 550 nm for 10 min was measured
immediately after adding 50 µl of reduced cytochrome c
solution to 950 µl of the homogenate (100 µg of total protein).
Caspase-3 activity
Caspase-3 activity was determined by using an
APOPCYTO™ Caspase-3 Colorimetric Assay kit (Medical &
Biological Laboratories, Co., Ltd., Nagoya, Japan). After 5-Gy
irradiation, a 3×105/well of TE4 cells and
MT-CO1-downregulated cells were seeded on a 96-well plate.
Caspase-3 activity was measured by NanoDrop 2000c as the absorbance
at 405 nm after incubation with a colorimetric substrate (DEVD-pNA)
with cell lysate (90 µg of total protein in 50 µl of lysing buffer)
at 37°C for 24 h.
Statistical analysis
Univariate analyses were performed using Student's
t-test or the Mann-Whitney U test. IBM SPSS Statistics version 21
(IBM Corp., Armonk, NY, USA) was used for statistical analyses. A
p-value <0.05 was considered statistically significant.
Results
Detection of candidate radioresistant
genes
After irradiation of the TE4 and TE15
transposon-tagged cells, 108 radioresistant colonies were picked
up. The nucleotide sequences of the transposons' insertion sites in
each colony were amplified by Splinkerette PCR and TOPO cloning.
Fifty nucleotide sequences were thus, detected. Insertion sites
were searched for by aligning the detected nucleotide sequences on
the BLAST site. The genes located up to 25 kbp downstream from the
insertion site were identified as candidate radioresistant genes.
Eleven such genes were detected; these are listed in Table I.
 | Table I.List of the 11 candidate
radioresistant genes detected by cyclopedic gene screening with
transposons. |
Table I.
List of the 11 candidate
radioresistant genes detected by cyclopedic gene screening with
transposons.
| Gene |
Overexpression/downregulation | Number of
detection |
|---|
| Sorting nexin 3
(SNX3) | Downregulation | 3 |
| Cytochrome c
oxidase 1 (MT-CO1) | Downregulation | 3 |
| mediator of RNA
polymerase II transcription subunit 13 (MED13) | Downregulation | 2 |
| Yae1 domain
containing protein 1 isoform 2 (YAE1D1) | Overexpression | 1 |
| centromere protein
V | Downregulation | 1 |
| Signal-induced
proliferation associated 1 like protein2 (SIPA1L2) | Downregulation | 1 |
| beta crystallin A4
(CRYBA4) | Overexpression | 1 |
| RNA-binding motif
single-stranded interacting protein 3 (RBMS3) | Downregulation | 1 |
| Transcription
factor 4 isoform 1, isoform 4 | Downregulation | 1 |
| protein FAM178B
isoform A (FAM178B) | Downregulation | 1 |
| Nck-associated
protein 5 (NCKAP5) | Downregulation | 1 |
Nucleotide sequences of the transposon's insertion
site matched a part of the gene sequence of nine genes which may
have been downregulated by the inserted transposons. Two genes were
detected downstream of the transposon's insertion site and may have
been overexpressed by the CMV promoter in the transposon. Three
genes were detected in two or three different radioresistant
colonies.
MT-CO1 was one of the detected candidate
radioresistant genes, found in three radioresistant colonies. One
hundred ninety-one nucleotide bases, detected by sequence, matched
the base sequence of MT-CO1 coded in mitochondrial DNA
(mtDNA) with a 99% concordance rate. Cytochrome c oxidase is
known to be an enzyme associated with apoptosis by activating the
caspase cascade. Thus, we conducted further experiments on the
hypothesis that downregulation of MT-CO1 induced
radioresistance by blocking activation of the caspase cascade in
esophageal cancer cells.
Downregulation of MT-CO1 in RRCs
In RRCs in which transposon was inserted into the
MT-CO1, MT-CO1 may be downregulated by the inserted
transposon; and MT-CO2 and MT-CO3, coded downstream
of MT-CO1, could possibly have been overexpressed by the CMV
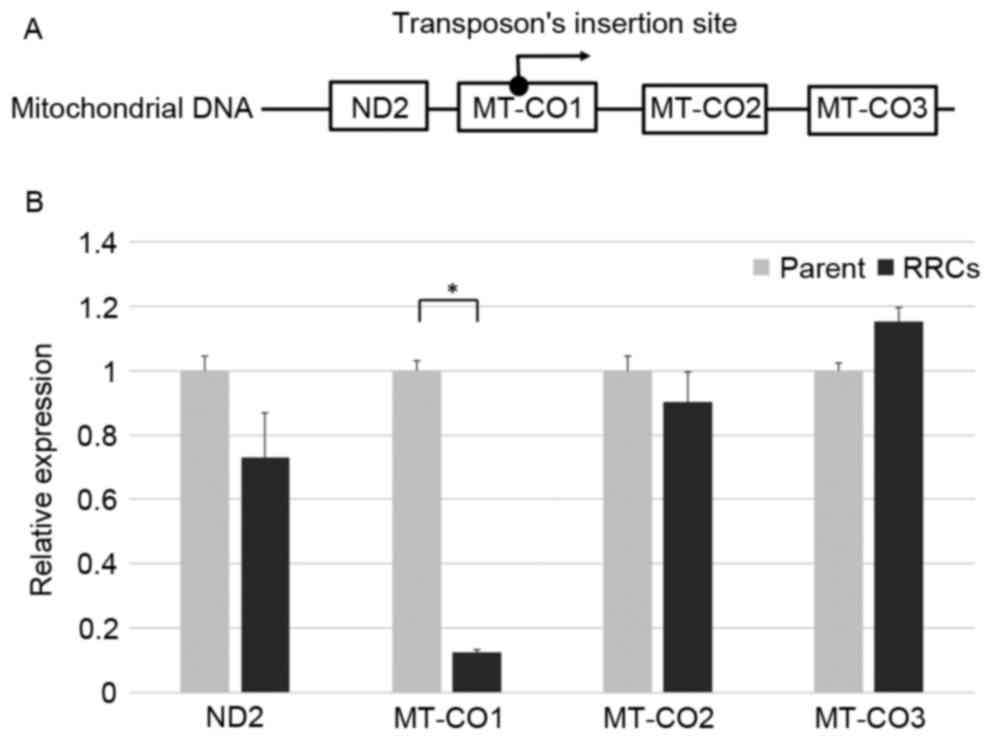
promotor in the transposon (Fig. 1A).
The relative expression levels of MT-CO1, MT-CO2, and
MT-CO3 were 0.12, 0.90, and 1.15, respectively, when the
expression level of parent TE4 cell was 1. MT-CO1 was
significantly downregulated in these cells (P<0.001) (Fig. 1B).
Radioresistance and downregulation of
MT-CO1
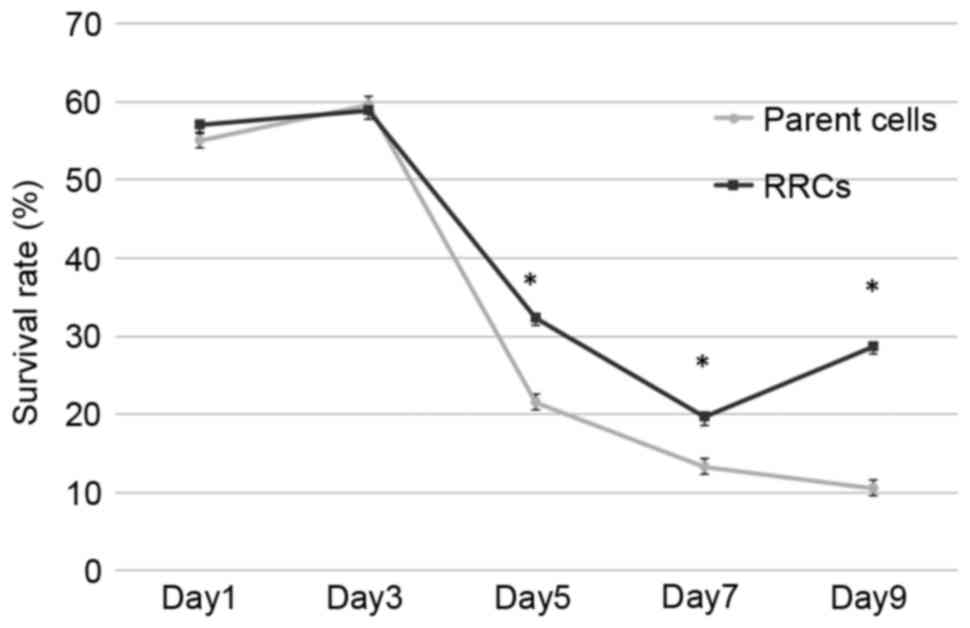
To establish whether the RRCs (in which
MT-CO1 was inactivated) showed radioresistance, survival
rates after irradiation were measured by an MTT assay (Fig. 2). The relative survival rate became
significantly higher compared to non-irradiated cells in the
MT-CO1-downregulated cells 5 days after 7-Gy irradiation
(P<0.001). Although the survival rate kept decreasing in the
parent (TE4) cells, it increased in the RRCs
(MT-CO1-downregulated cells) at day 9. The difference
between the parent cells and RRCs increased, with survival rates at
day 9 of 10.5 and 28.7%, respectively (P<0.001).
The activation of cytochrome c oxidase
was inhibited in RRCs
The cytochrome c oxidase changed reduced
cytochrome c (ferrochrome c) into oxidized cytochrome
c (ferrichrome c). Cytochrome c has a sharp
absorption band at 550 nm in the reduced state. Activation of
cytochrome c oxidase was measured as a decrease in
absorbance at 550 nm after adding cytochrome c reduced by
DTT.
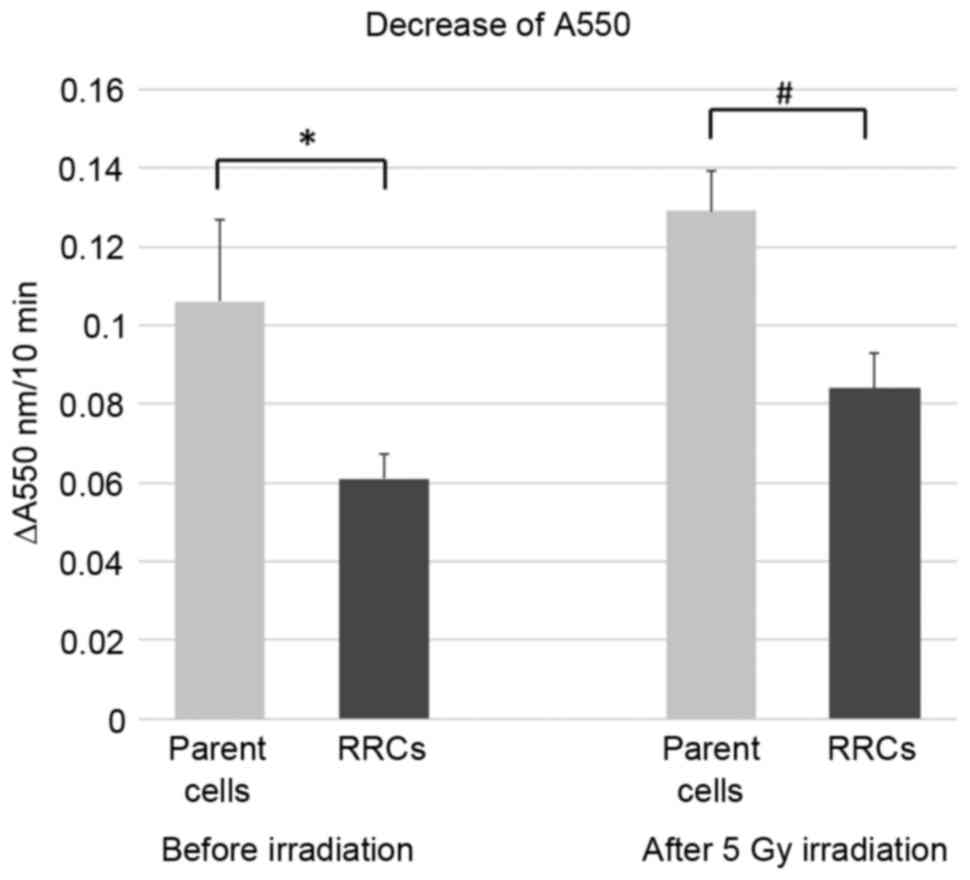
Fig. 3 shows the
decrease in the absorbance at 550 nm, which was 0.106 in the parent
(TE4) cells and 0.061 in the RRCs before irradiation. Six h after
5-Gy irradiation, the decrease in absorbance was 0.129 in the
parent cells and 0.084 in the RRCs. Activation of cytochrome
c oxidase was significantly lower in RRCs with MT-CO1
downregulation both before and after irradiation (P=0.023 and
P=0.005, respectively).
The activation of caspase-3 was
inhibited in RRCs after irradiation
To show that activation of the caspase cascade
induced by oxidized cytochrome c was suppressed in RRCs
after irradiation, we measured the activity of caspase-3.
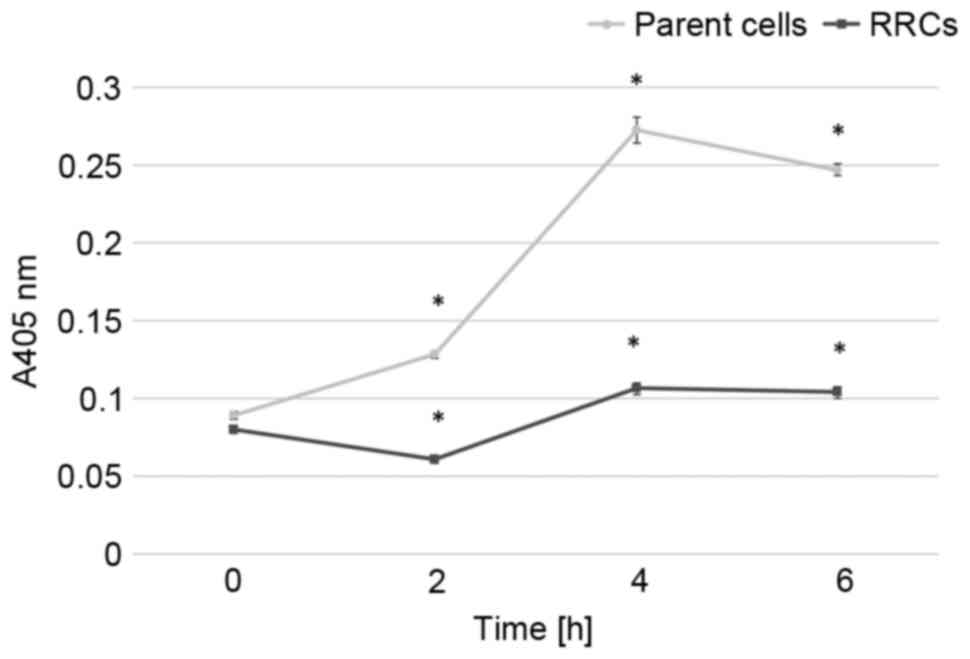
Caspase-3 activity after 5-Gy irradiation is shown
in Fig. 4. In the parent cells,
caspase-3 activity rapidly increased after irradiation and reached
its peak at 4 h after the irradiation. Caspase-3 activity gradually
increased after irradiation in the RRCs, although it remained
significantly lower than in the parent cells (P<0.001).
Discussion
We detected MT-CO1 as a candidate
radioresistant gene from radioresistant transposon-tagged
esophageal squamous cancer cells. Downregulation of MT-CO1
was verified by real-time quantitative PCR analysis of RRCs. RRCs
with the downregulation of MT-CO1 showed higher survival
rates than parent cells after irradiation. We also showed that
inhibiting apoptosis by the activation of caspase cascade caused
radioresistance because the activity of cytochrome c and
caspase-3 after irradiation was significantly lower in the
MT-CO1-downregulated RRCs.
MT-CO, or complex IV, is the terminal complex
of the electron transport chain on the mitochondrial membrane. It
is composed of three catalytic subunits encoded in mtDNA (subunits
1 to 3) and 10 accessory subunits (subunits 4 to 13) encoded in
nuclear DNA (7,8).
It is known that cytochrome c oxidase is
related to apoptosis (9). Cytochrome
c, released from mitochondria into the cytosol by various
apoptotic stimuli, binds to Apaf-1 and forms the apoptosome
which, in turn, activates pro-caspase-9 leading to apoptosis
(10). The redox state of cytochrome
c is important in this caspase-dependent apoptosis. Only
oxidized cytochrome c can activate the apoptosome, whereas
reduced cytochrome c cannot (11–13). The
most potent enzyme that oxidizes cytochrome c is cytochrome
c oxidase (13). Multiple
caspase-dependent and -independent cell death pathways induce
apoptosis after DNA damage (14). Our
results revealed that downregulation of the MT-CO1 gene could not
activate casepase-3 in a few hours after irradiation. It seemed to
affect radioresistant, high survival rate in 5 to 9 days after
irradiation, in our study.
The relationship between cytochrome c oxidase
and chemo or radioresistance has been reported in various tumor
cell lines (15–19). Comprehensive gene analysis on a
microarray in cervical carcinoma cells (squamous cell carcinoma)
showed that MT-CO1 was increased by a factor greater than
two in RRCs compared with radiosensitive cells (15). Also, in human acute myelogenic
leukemia cells, MT-CO3 was upregulated in chemoresistant
cells compared with in chemosensitive cells (16). However, these study did not verify
that overexpression of MT-CO1 actually induces
radioresistance. Because the technique for transfection into
mitochondrial DNA has not yet been established, it is impossible to
overexpress or downregulate a mitochondrial gene (17). Aichler et al (18) reported that knockdown of
MT-CO7A2 with siRNA increased chemosensitivity after
chemotherapy with cisplatin and 5-flurouracil in esophageal
adenocarcinoma cells. Oliva et al (19,20)
revealed that pharmacologic and genetic manipulation of cytochrome
c oxidase restored chemosensitivity in chemoresistant glioma
cells. From these and our results, cytochrome c oxidase
seems to be a significant gene which induces chemo or
radioresistance, and it may be a target gene for treatment to
improve chemo or radiosensitivity.
A question raised by our study is about the
homogeneity of mtDNA. Each cell contains hundreds to thousands of
copies of mtDNA, so if a transposon transports into the
MT-CO1 gene in one mtDNA, it should not affect the
properties of the whole cell. However, in this study MT-CO1
was actually downregulated in RRCs. It is known that, in general,
all copies of mtDNA are identical within a cell, a genetic state
known as homoplasmy (21). In cell
proliferation, only one mtDNA is copied by rolling circle DNA
replication (22). Although this
mechanism has not yet been completely clarified, homoplasmy of
mtDNA could relate to the downregulation of MT-CO1.
It was unclear whether MT-CO1 gene mutation
was actually related to radioresistance in patients with esophageal
cancer. Multiple gene mutations with complicated interactions could
induce radioresistance. Although our comprehensive gene screening
is ongoing, it is proving to be a useful approach for detecting
radioresistant genes. Verification of the relationships between all
the detected radioresistant genes and the clinical responses to
radiation therapy is essential for clinical application.
In conclusion, eleven candidate radioresistant genes
in esophageal squamous cancer were detected by comprehensive gene
screening with transposons. The mechanism for radioresistance
induced by MT-CO1 downregulation was also revealed. The
candidate genes were detected under conditions more similar to a
clinical setting than was the case in previous forms of gene
screening. By detecting all radioresistant genes and revealing the
underlying mechanisms for this, we can select the appropriate
therapy for patients or enhance the effect of the therapy by
treating target genes.
References
|
1
|
Ishida K, Ando N, Yamamoto S, Ide H and
Shinoda M: Phase II study of cisplatin and 5-fluorouracil with
concurrent radiotherapy in advanced squamous cell carcinoma of the
esophagus: A Japan Esophageal Oncology Group (JEOG)/Japan Clinical
Oncology Group trial (JCOG9516). Jpn J Clin Oncol. 34:615–619.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guo W and Jiang YG: Current gene
expression studies in esophageal carcinoma. Curr Genomics.
10:534–539. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fukuda K, Sakakura C, Miyagawa K, Kuriu Y,
Kin S, Nakase Y, Hagiwara A, Mitsufuji S, Okazaki Y, Hayashizaki Y
and Yamagishi H: Differential gene expression profiles of
radioresistant oesophageal cancer cell lines established by
continuous fractionated irradiation. Br J Cancer. 91:1543–1550.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ogawa R, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Mori Y, Mori R, Tomoda K, Katada T, Harada K and Fujii Y:
Identification of candidate genes involved in the radiosensitivity
of esophageal cancer cells by microarray analysis. Dis Esophagus.
21:288–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Stuart L, Ohsumi TK, Burgess S,
Varshney GK, Dastur A, Borowsky M, Benes C, Lacy-Hulbert A and
Schmidt EV: Transposon activation mutagenesis as a screening tool
for identifying resistance to cancer therapeutics. BMC Cancer.
13:932013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsutsui M, Kawakubo H, Hayashida T, Fukuda
K, Nakamura R, Takahashi T, Wada N, Saikawa Y, Omori T, Takeuchi H
and Kitagawa Y: Comprehensive screening of genes resistant to an
anticancer drug in esophageal squamous cell carcinoma. Int J Oncol.
47:867–874. 2015.PubMed/NCBI
|
|
7
|
Kadenbach B, Jarausch J, Hartmann R and
Merle P: Separation of mammalian cytochrome c oxidase into
13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic
procedure. Anal Biochem. 129:517–521. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weishaupt A and Kadenbach B: Selective
removal of subunit VIb increases the activity of cytochrome
c oxidase. Biochemistry. 31:11477–11481. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pradelli LA, Bénéteau M and Ricci JE:
Mitochondrial control of caspase-dependent and -independent cell
death. Cell Mol Life Sci. 67:1589–1597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brown GC and Borutaite V: Regulation of
apoptosis by the redox state of cytochrome c. Biochim
Biophys Acta. 1777:877–881. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan Z, Voehringer DW and Meyn RE: Analysis
of redox regulation of cytochrome c-induced apoptosis in a
cell-free system. Cell Death Differ. 6:683–688. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suto D, Sato K, Ohba Y, Yoshimura T and
Fujii J: Suppression of the pro-apoptotic function of cytochrome
c by singlet oxygen via a haem redox state-independent
mechanism. Biochem J. 392:399–406. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Borutaite V and Brown GC: Mitochondrial
regulation of caspase activation by cytochrome oxidase and
tetramethylphenylenediamine via cytosolic cytochrome c redox
state. J Biol Chem. 282:31124–31130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim R, Emi M and Tanabe K:
Caspase-dependent and -independent cell death pathways after DNA
damage (Review). Oncol Rep. 14:595–599. 2005.PubMed/NCBI
|
|
15
|
Achary MP, Jaggernauth W, Gross E, Alfieri
A, Klinger HP and Vikram B: Cell lines from the same cervical
carcinoma but with different radiosensitivities exhibit different
cDNA microarray patterns of gene expression. Cytogenet Cell Genet.
91:39–43. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang TS, Myklebust LM, Kjarland E,
Gjertsen BT, Pendino F, Bruserud Ø, Døskeland SO and Lillehaug JR:
LEDGF/p75 has increased expression in blasts from
chemotherapy-resistant human acute myelogenic leukemia patients and
protects leukemia cells from apoptosis in vitro. Mol Cancer.
6:312007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Inoue K, Nakada K, Ogura A, Isobe K, Goto
Y, Nonaka I and Hayashi JI: Generation of mice with mitochondrial
dysfunction by introducing mouse mtDNA carrying a deletion into
zygotes. Nat Genet. 26:176–181. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aichler M, Elsner M, Ludyga N, Feuchtinger
A, Zangen V, Maier SK, Balluff B, Schöne C, Hierber L, Braselmann
H, et al: Clinical response to chemotherapy in oesophageal
adenocarcinoma patients is linked to defects in mitochondria. J
Pathol. 230:410–419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Oliva CR, Nozell SE, Diers A, McClugage SG
III, Sarkaria JN, Markert JM, Darley-Usmar VM, Bailey SM, Gillespie
GY, Landar A and Griguer CE: Acquisition of temozolomide
chemoresistance in gliomas leads to remodeling of mitochondrial
electron transport chain. J Biol Chem. 285:39759–39767. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oliva CR, Moellering DR, Gillespie GY and
Griguer CE: Acquisition of chemoresistance in gliomas is associated
with increased mitochondrial coupling and decreased ROS production.
PLoS One. 6:e246652011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lightowlers RN, Chinnery PF, Turnbull DM
and Howell N: Mammalian mitochondrial genetics: Heredity,
heteroplasmy and disease. Trends Genet. 13:450–455. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shibata T and Ling F: DNA recombination
protein-dependent mechanism of homoplasmy and its proposed
functions. Mitochondrion. 7:17–23. 2007. View Article : Google Scholar : PubMed/NCBI
|