Introduction
Esophageal cancer is one of most common malignant
tumors in the clinic. The incidence of esophageal cancer is the 8th
highest in malignant tumors and the mortality rate ranks the 6th
highest worldwide (1). In China, the
morbidity of esophageal cancer, particularly esophageal squamous
cell carcinoma, is very high (2). In
the early stage no specific clinical manifestations in patients of
esophageal cancer are evident (3).
Approximately 80% patients are in advanced or late stage when
diagnosed, and the majority of patients cannot be treated with
surgery (4). Invasion and metastasis
of esophageal cancer in patients is an important factor affecting
treatment efficacy and induced poor prognosis (4). It is important to investigate the
molecular mechanisms of proliferation-, invasion- and
metastasis-associated genes in esophageal cancer, which may provide
evidence to prevent and cure esophageal cancer.
Protease activated receptor-2 (PAR-2) is one type of
receptor on the cell surface of numerous cells and it belongs to
the superfamily of G protein-coupled protease-activated phase
receptor (5). Trypsin, tryptase and
coagulation factors are the natural agonists for PAR-2, and
activated PAR-2 is involved in a series of biological behaviors,
including cell proliferation, invasion and metastasis in tumors
(6,7).
Our previous study found that PAR-2 performed important roles in
growth, invasion and metastasis of the esophageal cancer EC109 cell
line (8). The present study aimed to
investigate whether PAR-2 effects cell characteristics of EC109
through RNA interference technology. A PAR-2 targeted short hairpin
RNA (shRNA) vector was constructed and transfected into the EC109
cell line. The silencing effect of PAR-2 shRNA on its target gene
was then observed.
Materials and methods
Cell line and plasmid
The esophageal cancer EC109 cell line, E.
coli top 10 strain and pGFP-V-RS plasmid were provided by the
Department of Cell Biology in Logistics University of People's
Armed Police Force (Tianjin, China). The DH5α competent cell was
bought from Transgene Biotech Co., Ltd. (Beijing, China).
Construction of vector
The sequence of human PAR-2 mRNA (gene ID, 55065)
was retrieved from the GeneBank database (https://www.ncbi.nlm.nih.gov/gene/55065). Subsequent
to selecting a suitable target site, two oligonucleotide sequences
were synthesized: Sequence 1, 5′-TTCCTAACTCTGGCCTTGGTGTTGGCAAT-3′;
sequence 2, 5′-GTGTTCTCATATGTGAAGGTGGCTGCAAG-3′, while another one
non-specific sequence (5′-GCCTGTTGTACCTCTAATGTCACTTTCCT-3′) was
synthesized, all of these sequences were supplied by OriGene
Technologies, Inc. (Rockville, MD, USA). The shRNA stem-loop
structure was TCAAGAG, and restriction sites of BamHI and HindIII
were introduced at the 5′ and 3′ ends, respectively. The sequences
were then connected to the pGFP-V-RS plasmid vector. The
recombinant plasmids were transformed to DH5α competent cells.
Briefly, the competent cells (100 µl) were incubated with pGFP-V-RS
plasmid vector (5 µl) on ice for 30 min, followed by 42°C for 45s
and ice bathed for 2 min. Subsequently, 500 µl SOC medium
(Zhongaobio Company, Tianjin, China) was added and mixed. The
mixture was centrifuged at 37°C and 40 × g for 1 h. The product
then was transferred to a Kana resistant (30 µg/ml) preloaded
lysogeny broth (LB) medium plate and incubated overnight at 37°C.
Three monoclonal colonies were selected on each dish and were
inoculated with 3 ml of Kana resistant (final concentration 30
µg/ml) LB medium (Zhongaobio Company) at 37°C overnight. The
plasmid DNA was extracted according the manufacturer's instructions
(Transgene company, Beijing, China). Plasmid DNA was quantificated.
Briefly, 2 µl of plasmid DNA and 98 µl of TE buffer (Tianjin
Chemical Reagent Factory, Tianjin, China) were mixed. A nucleic
acid quantifier (GeneQuant 80–2114-98, Cambridge, UK) was used to
determine the concentration and record the absorbance ratio at 260
and 280 nm to assess the purity. Subsequent to detecting the
concentration and purity, the plasmids were termed PAR-2 shRNA-1,
PAR-2 shRNA-2 and non-specific sequences. The positive bacterium
solution was sent to Shanghai Shangon Co., Ltd. (Shanghai, China)
for identification by sequence technology.
Cell culture and transfection
EC109 cells were cultured with RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS; Minhai Biotech, Beijing,
China) in a 5% CO2 incubator at 37°C, and EC109 cells
were digested with trypsin for passage following growth 80–90%.
PAR-2 shRNA-1, PAR-2 shRNA-2 and non-specific sequences were
transfected into cells in the logarithmic growth phase. Briefly,
EC109 cells at logarithm phase were digested by trypsin and seeded
into six-well plate (5×104 per well). The cells were
transfected when the growth reached 80% confluence. The cells and
the transfected plasmids were incubated at 37°C for 24 h in the
incubator. The blank control of EC109 cells received no
transfection. The pGFP-V-RS plasmid with green fluorescent protein
was observed using a Microscope Digital Camera DP27 (Olympus,
Tokyo, Japan). The transfection efficiency was calculated based on
the expression of green fluorescent protein (GFP) at 24 h
post-transfection. The stably transfected cells were selected using
1.0 mg/ml puromycin by several passages. The EC109 cells without
transfection were regarded as the blank control group. Other groups
were named based on the transfection: PAR-2 shRNA-1 group; PAR-2
shRNA-2 group; and non-specific sequence group.
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA from cells at logarithm phase was isolated
in each group using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
cDNA was synthesized from total RNA by reverse transcription with a
Reverse Transcription System (Biometra UNO II Thermoblock; Biometra
GmbH, Göttingen, German) and stored at −20°C. The reverse
transcription conditions were 30°C for 10 min, 42°C for 30 min,
99°C for 5 min and 5°C for 5 min. The primers of PAR-2 and internal
reference β-actin were designed by Omiga 2.0 software (Accelrys,
San Diego, CA, USA) as follows: PAR-2 forward,
5′-AGAAGCCTTATTGGTAAGGTT-3′ and reverse,
5′-AACATCATGACAGGTCGTGAT-3′, with amplification length 582 bp; and
β-actin forward, 5′-TGTTTGAGACCTTCAACACCC-3′ and reverse,
5′-AGCACTGTGTTGGCGTACAGG-3′, with amplification length 540 bp. The
reaction conditions of RT-PCR for PAR-2 used the RT-qPCR kit
purchased from Beijing Transgen Biotech Co., Ltd. (Beijing, China),
and the reaction included 35 cycles at 94°C for 45 sec, 51°C for 45
sec and 75°C for 1 min. The reaction conditions for β-actin were 35
cycles at 94°C for 45 sec, 55°C for 60 sec and 72°C for 45 sec. The
PCR products were separated by 2% agarose gel electrophoresis and
scanned using a GDS 8000 gel documentation system (UVP LLC, Upland,
CA, USA). The relative expression of PAR-2 was calculated based on
the expression of β-actin. The experiments were repeated three
times.
Western blot analysis
The experimental cells (PAR-2 shRNA-1 EC109 cells,
PAR-2 shRNA-2 EC109 cells, nonspecific sequence transfection EC109
cells and EC109 cells) were lysed with RIPA lysate (Beyotime
Institute of Biotechnology, Haimen, China) in each group and
centrifuged at 3,000 × g and 4°C for 10 min to obtain supernatant.
The protein concentration was detected by the bicinchoninic acid
method using a BCA kit (Beyotime Institute of Biotechnology). Total
proteins (80 µg) were loaded onto each well (six wells in total) of
10% SDS-PAGE and then transferred to polyvinylidene fluoride
membranes. The membrane was washed with TBST and blocked with 5%
skimmed milk at 37°C for 2 h. The primary antibody (rabbit mAb;
cat. no. 6976; Cell Signaling Technology, Inc., Danvers, MA, USA;
dilution, 1:1,000) was incubated at 37°C for 2 h, followed by
incubation at 4°C overnight. This was then washed with TBST for 15
min four times. The secondary antibody (goat anti-rabbit; dilution,
1:1,000, Beijing Zhong Shan-Golden Bridge Biological Technology
Co., Ltd., Beijing, China) was then added and incubated at 4°C for
2 h, and then washed with TBST for 15 min four times. Finally, the
membrane was developed by enhanced chemiluminescence plus reagent
(Beijing Dingguo Changshen Biotechnology co., Ltd., Beijing,
China). The developed film was scanned and analyzed by Quantity One
4.62 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
β-actin was used as an internal control to calculate the relative
expression of PAR-2.
MTT assay
In the MTT assay, three groups of EC109 cells were
included: Negative control group (transfected with non-specific
sequence vector); transfection reagent group (just adding
transfection reagents); and PAR-2 shRNA group (PAR-2 downregulated
group). The EC109 cells in different groups were cultured in
RPMI-1640 medium with 10% FBS at 37°C with 5% CO2 for 24
h, and the cells in logarithmic growth phase were used for MTT
assay. Cells in each group were seeded onto 96-well plates
(2×103 per well). The 20 µl MTT solution with 5 mg/ml
concentration was added to each well at 24, 48 and 72 h. Following
culture for 4 h at 37°C, 200 µl dimethyl sulfoxide was added to end
the reaction, The samples were analyzed using an ELISA analyzer
(Biomad, Sacremento, CA, USA) and the absorbance of each well was
measured at a wavelength of 490 nm. The ratio of inhibited growth
was calculated by the formula: Ratio (%)=[absorbance (A) control-A
experiment]/(A control-A blank) ×100. The ratio of inhibited growth
was used to draw cell growth inhibition curves.
Cell cycle analysis by flow
cytometry
The cells in logarithmic phase were seeded into a
culture flask at a density of 1×105/ml for 24 h at 37°C
with 5% CO2. The medium was then replaced with medium
without FBS to culture for another 24 h at 37°C with 5%
CO2. The cells were collected by centrifugation 300 × g
for 3 min at room temperature and washed twice with pre-cooled PBS.
The 70% pre-cooled ethanol was added to fix at 4°C overnight. The
cells were reserved at 4°C and subjected to propidium iodide
staining and then the cell cycle was detected using BD flow
cytometry (BD Biosciences, San Jose, CA, USA), and data was
analyzed using ModFit LT for Windows Version 3.2 (Verity Software
House, Topsham, ME, USA).
Transwell migration and Matrigel
invasion assays
The migration and invasion assays were performed
using Transwell chambers (EMD Millipore, Billerica, MA, USA).
Matrigel was thawed at 4°C and mixed with serum-free medium with
1:4 ratio as an artificial basement membrane. The Transwell chamber
with an 8 µm microporous membrane was placed in 24-well culture
plates, and the upper chamber was uniformly covered with 40 µl
artificial basement membrane (BD Matrigel™ Basement Membrane
Matrix; BD Biosciences) mixed with RPMI-1640 at the ratio of 1:4.
Subsequent to agglutination for 1 at 37°C h in an incubator, the
moisture was absorbed from Matrigel to form a matrix barrier.
The cells were seeded into the upper chambers
(2×105 cell/well), while 600 µl of medium containing 10%
FBS was added to the bottom of the chamber. Each well had 3
replicates. Following culture at 37°C with 5% CO2 for 28
h, cells were fixed using 10% formaldehyde at room temperature for
30 min, washed with PBS, and stained with eosin for 5 min at room
temperature. The cells that could not pass through the membrane
were wiped. Subsequent to natural drying, the membrane was then
dried and moved to the slide and sealed by neutral balsam. Finally,
images of the cells were captured under a light microscope (Olympus
BX41; Olympus Corporation) with 5 random views. The number of
migrated/invaded cells in the bottom chamber was counted and the
average of 3 replicates was used. The experimental steps in the
migration assay did not include use of Matrigel. The density of
cell suspension was 1×105/ml.
Statistical analysis
Data are presented as the mean ± standard deviation,
and were analyzed by SPSS 13.0 software (SPSS, Inc., Chicago, IL,
USA). The differences between two groups were determined by
Student's t-test. Multi-group analysis was performed by one way
analysis of variance, the post hoc test was the least significant
difference test. P<0.05 was considered to indicate a
statistically significant difference.
Results
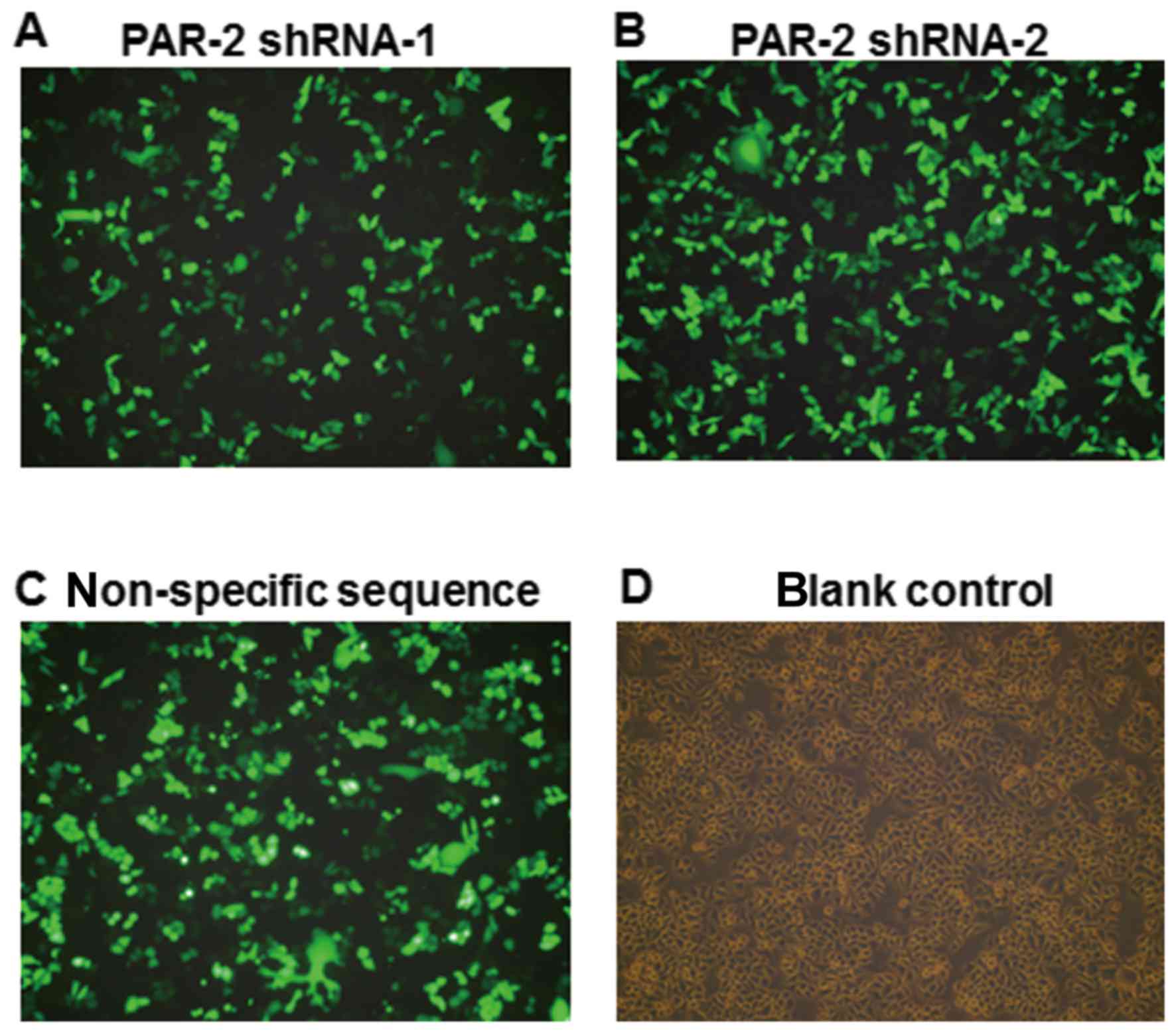
Efficiency of cell transfection
The sequences of the PAR gene in vector were
completely consistent with the PAR-2 target sequences. Following
transfection for 24 h, the expression of green fluorescence was
observed by inverted fluorescence microscopy, as shown in Fig. 1. The transfection efficiency was
calculated by the percentage of the number of cells expressing
green fluorescence among the total number of cells. The mean
transfection efficiency of PAR-2 shRNA-1, PAR-2 shRNA-2 and
non-specific sequence group was 67.6%, while no green fluorescence
was observed in the blank control group.
PAR-2 expression in different EC109
cell lines by RT-PCR and western blot analysis
To detect PAR-2 mRNA expression following
transfection, RT-PCR was applied to different groups. As shown in
Fig. 2A, the band intensities were
0.35±0.03, 0.43±0.04, 0.44±0.04 and 0.45±0.41 in PAR-2 shRNA-1
group, PAR-2 shRNA-2 group, non-specific sequence group and blank
control group, respectively. Compared with the other three groups,
PAR-2 was significantly downregulated in the PAR-2 shRNA-1 group
(P<0.05). Among the PAR-2 shRNA-2 group, non-specific sequence
group and blank control group, PAR-2 mRNA expression had no
significant difference (P>0.05). The results indicated that the
constructed PAR-2 shRNA-1 recombinant vector efficiently silenced
PAR-2 mRNA.
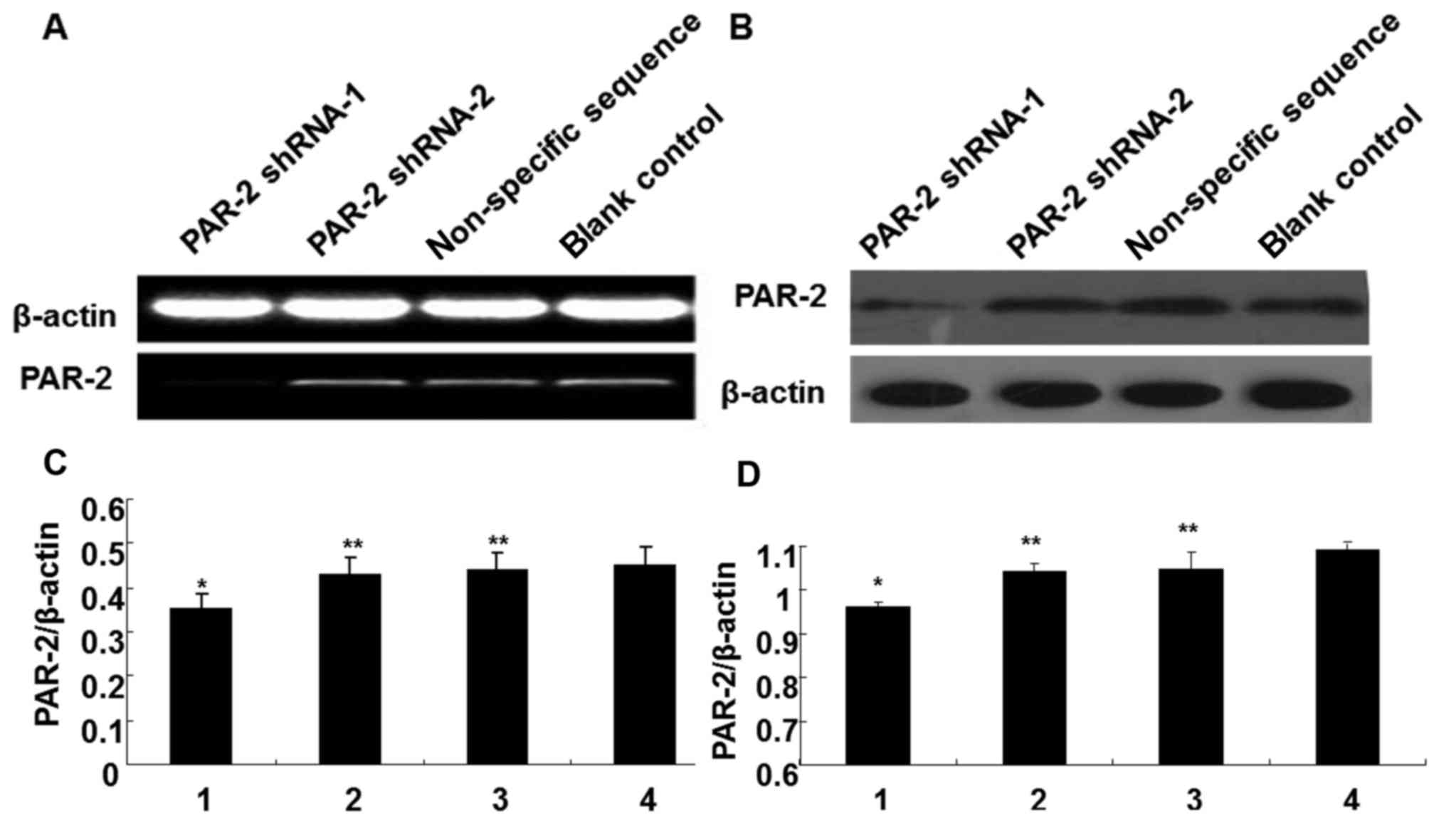 | Figure 2.Detection of PAR-2 expression in
different EC109 cell lines by RT-PCR and western blot. (A) RT-PCR
method to detect mRNA expression of PAR-2 in different EC109 cell
lines. (B) Western blot analysis to detect protein expression of
PAR-2 in different EC109 cell lines. (C) RT-PCR analysis of PAR-2
mRNA expression levels in EC109 cells following transfection. 1,
PAR-2 shRNA-1 group; 2, PAR-2 shRNA-2 group; 3, nonspecific
sequence transfection group; 4, control group. (D) Western blot
analysis of PAR-2 protein expression levels in EC109 cells
following transfection. 1, PAR-2 shRNA-1 group; 2, PAR-2 shRNA-2
group; 3, nonspecific sequence transfection group; 4, control
group. *P<0.05 vs. control group, **P>0.05 vs. control group.
RT-PCR, reverse transcription-polymerase chain reaction; PAR-2,
protease-activated receptor 2; shRNA, small hairpin RNA. |
To detect PAR-2 protein expression subsequent to
transfection with recombinant vectors, western blot analysis was
performed in different groups. As shown in Fig. 2B, the grey intensities were 0.96±0.01,
1.04±0.02, 1.05±0.04 and 1.09±0.04 in the PAR-2 shRNA-1, PAR-2
shRNA-2, non-specific sequence and blank control groups,
respectively. Compared with the other three groups, PAR-2 was
significantly decreased in the PAR-2 shRNA-1 group (P<0.05).
Among the PAR-2 shRNA-2 group, non-specific sequence group and
blank control group, PAR-2 protein expression had no significant
difference (P>0.05). The results indicated that the constructed
PAR-2 shRNA-1 recombinant vector could efficiently silence PAR-2
expression, which was consistent with PAR-2 mRNA expression.
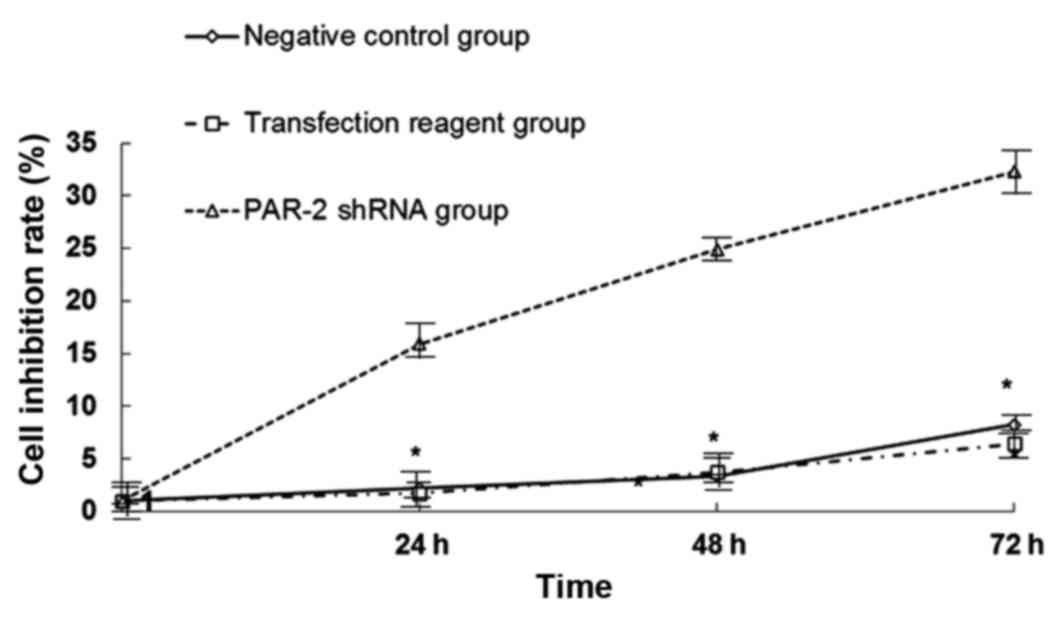
EC109 cell viability by MTT assay
For studying the effect of PAR-2 on cell viability,
a MTT assay was used to detect the changes of proliferation
following transfection. As shown in Fig.
3, EC109 cell proliferation in the PAR-2 shRNA group was
significantly lower than the negative control group (P<0.05),
and the growth inhibition ratios were 5.92, 24.89 and 32.28% at 24,
48 and 72 h post-transfection, respectively. Between the
transfection reagent group and the negative control group, no
significant difference was observed (P>0.05). The results
indicated that silenced PAR-2 can repress cellular proliferation in
EC109 cell lines, while the transfection reagent and negative
plasmid have no significant inhibition roles in EC109 cell
viability.
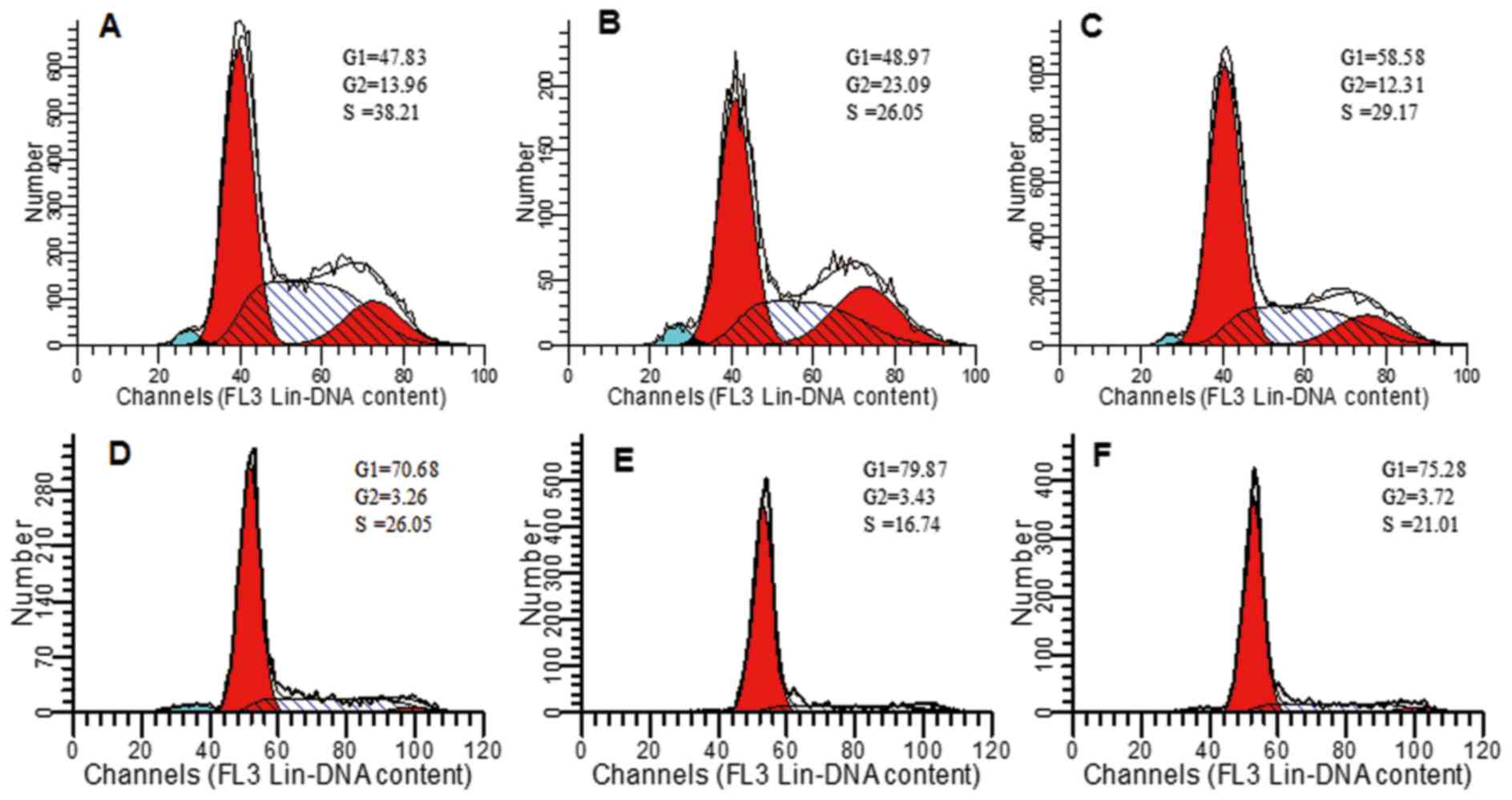
PAR-2-induced changes of cell cycle by
flow cytometry
To investigate the effect of silenced PAR-2 on cell
cycles of EC109 cells, flow cytometry was used to detect the
changes of cell cycle. As shown in Fig.
4, S phase arrest was observed in EC109 cells with PAR-2 shRNA
transfection. The percentages of cells in the S phase in PAR-2
shRNA-transfected cells and normal cultured cells were 19.37±2.19,
16.93±2.56 and 18.74±2.92% and 32.79±4.06, 26.54±1.37 and
33.45±2.46% after 24, 48 and 72 h, respectively. The ratio of S
phase cells was significantly lower in EC109 cells transfected with
PAR-2 shRNA than in EC109 cells (P<0.05), which indicated that
silenced PAR-2 inhibited cell cycle of EC109 cells.
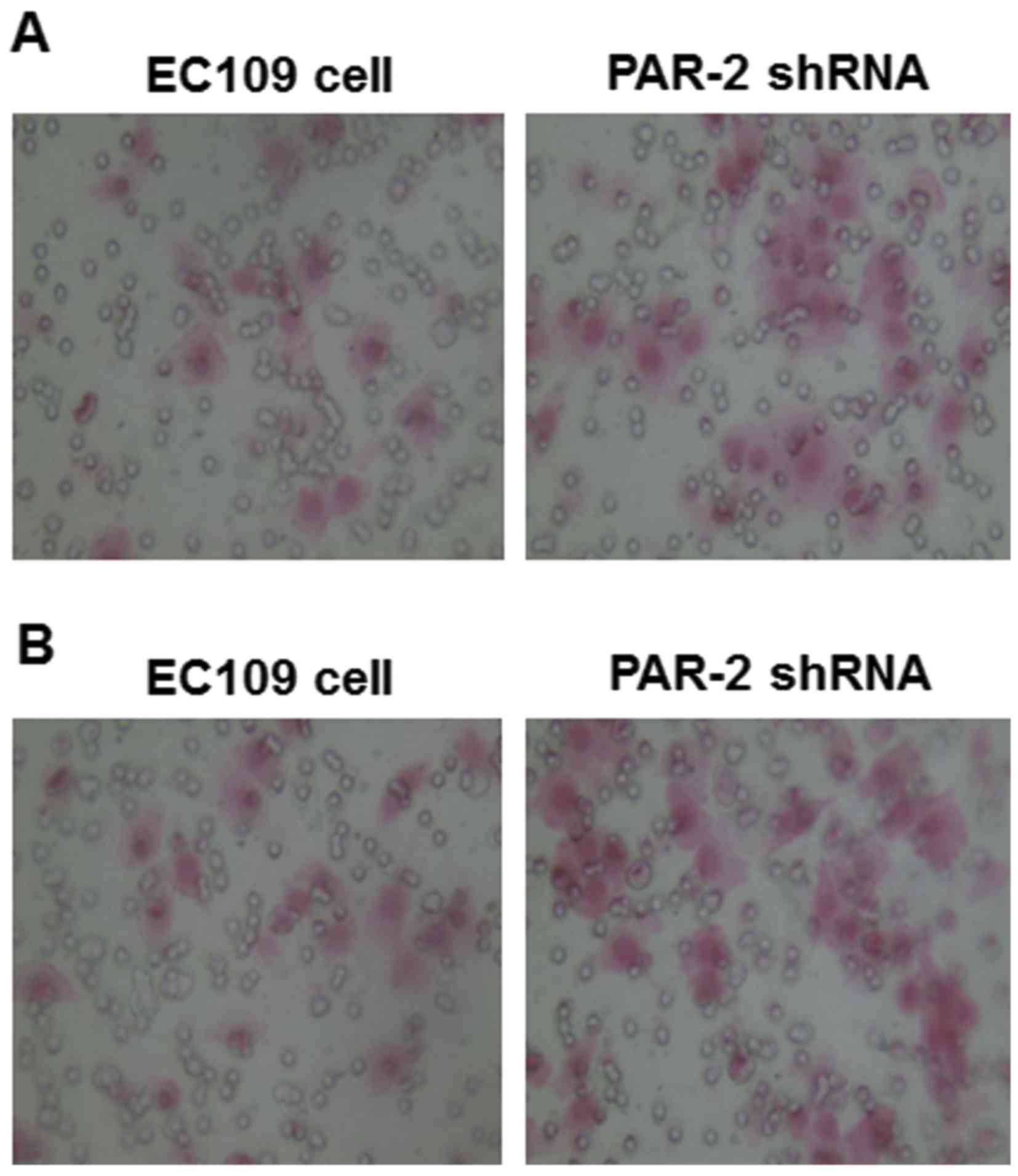
Effect of PAR-2 shRNA on EC109 cell
invasion and migration
To study the changes of EC109 cell behaviors induced
by PAR-2 silence, Matrigel and Transwell assays were applied to
detect the cell invasion and migration of EC109 cells. As shown in
Fig. 5A, numbers of EC109 cells that
passed through Matrigel were 19.6±2.11 and 30.1±2.64 in the PAR-2
shRNA and control groups, respectively, and the difference was
statistically significant, as presented in Table I (P<0.05). The results indicated
that PAR-2 silence repressed the invasion capability of EC109
cells.
 | Table I.Effects of PAR-2 on invasion and
migration of silenced and normal EC109 cells. |
Table I.
Effects of PAR-2 on invasion and
migration of silenced and normal EC109 cells.
|
| EC109 |
|---|
|
|
|
|---|
| Cell number of
Transwells | Invasion | Migration |
|---|
| Silenced |
19.6±2.11a |
24.2±2.82a |
| Control | 30.1±2.64 | 43.8±2.14 |
As shown in Fig. 5B,
under the microscope (magnification, ×200), the number of EC109
cells that moved through the artificial basement membrane by
deformation was significantly lower in the PAR-2 shRNA group
(24.2±2.82) compared with the control group (43.8±2.14; Table I, P<0.01). The results indicated
that PAR-2 silence repressed the invasion capability of EC109
cells.
Discussion
Protease-activated receptors (PARs) belong to a G
protein-coupled receptor superfamily with seven transmembrane
domains, which includes four subtypes: PAR-1, PAR-2, PAR-3 and
PAR-4 (5). All the subtypes contain
N-terminus, extracellular loop, seven transmembrane helix,
intracellular loop and C-terminal regions (5). The serine cleavage site is hidden in the
N-terminus of the PARs, which serves important roles in activated
processes (9). In 1994, Nystedt et
al (10) identified the DNA
sequence of PAR-2 in mice, which was named PAR-2 as it shared a
similar structure and activating mechanism to thrombin receptor.
PAR-2 is widely expressed in various tissues and organs in the
body, and is highly expressed in a variety of gastrointestinal
cancer cells, including esophageal, liver, stomach, pancreatic and
colon cancers, which are associated with the morbidity, progression
and prognosis of tumors (11,12). It was revealed that PAR-2 can be
activated by a variety of molecules, including trypsin, human
airway trypsin-like protease, sperm acrosin, cathepsin G, tissue
factor Xa and activated coagulation factor VIIa and Xa in
vivo (8,13,14), and
trypsin is one of most effective activators (9). Among tumors in the digestive system,
activated PAR-2 can promote invasion and metastasis of tumor cells,
regulate tumor cell proliferation, adhesion and angiogenesis
(15,16).
RNA interference (RNAi) refers to
post-transcriptional silencing processes induced by specific
sequences, which can inhibit mRNA expression mediated by small
interfering RNA (siRNA) (17). When
viral or exogenous genes were randomly integrated into the host
genome, double-stranded RNA (dsRNA) was produced, and these dsRNA
can be cut into small interfering RNA fragments (siRNAs) 21–23 bp
in length by Dicer (18). Under RNA
helicase processing, siRNA antisense can form into an RNA-induced
silencing complex (RISC) with different enzymes (19). Based on complementary binding, RISC
can combine with target mRNA sequences, then the mRNA was cut at
position with 12 bp of siRNA 3′ end. The cut mRNA was degraded,
which induced post-transcriptional gene silencing (18–20). RNAi
is one of most efficient and specific gene silencing mechanisms,
which is becoming an effective targeting gene therapy for tumors
(21). In the present study, a
pGFP-V-RS vector containing PAR-2 shRNA was constructed and
transfected into EC109 cells. Following selection by puromycin,
stably-transfected EC109 cells were validated by RT-PCR and western
blot analysis. PAR-2 was downregulated in mRNA level and protein
level, which indicated that recombinant siRNA vector was
successfully constructed. It was revealed that activated PAR-2
induces DNA synthesis and cell division (22). MTT assay was applied to detect the
proliferation of EC109 cells subsequent to silencing PAR-2. It was
revealed that silenced PAR-2 repressed cell proliferation. Through
flow cytometry, S phase arrest was observed in PAR-2 silencing
cells. Additional studies are required to investigate whether
repression of cell proliferation induced by PAR-2 silencing was
associated with regulators of the cell cycle.
Besides local proliferation and infiltration, tumor
cells will invade to other distant tissues (8). Cell invasion and metastasis in tumors is
a complex and continuous process, among which extracellular matrix
degradation is a key step (23).
Matrix metalloproteinases (MMPs) are a class of Zn+
dependent proteolytic enzymes, which can be expressed and secreted
in tumor cells and stromal cells, resulting in tumor invasion and
metastasis (24). In previous years,
it was shown that PAR-2 can promote tumor growth, invasion and
metastasis through activation of MMP pathways (24). Through promoting the release of
transforming growth factor-α, activating epithelial growth factor
receptor and kinase extracellular signal-regulated kinase 1/2,
PAR-2 can promote cellular proliferation, invasion and metastasis
in colorectal and gastric cancers (24,25). Our
previous study revealed that PAR-2 can promote invasion and
metastasis of HepG2 cells through degrading extracellular matrix by
the PAR-2-focal adhesion kinase-MMP-2/9 pathway (8). By RT-PCR, immunohistochemistry and
zymography methods, Zhou et al (8) found that PAR-2 receptor expressed in
EC109 cells, and appropriate concentrations of SLIGKV and trypsin
can activate PAR-2 receptor to increase the expression level.
Therefore, this promotes cell invasion and metastasis of EC109
cells by promoting MMP-9 secretion and degradation of the
extracellular matrix. In the present study, Matrigel and Transwell
assays were applied to detect the changes of cell invasion and
metastasis of EC109 cells following PAR-2 silencing. It was
indicated that the capabilities of cell invasion and metastasis
were decreased subsequent to PAR-2 gene silencing. The molecular
mechanisms and associated signaling transduction pathways require
additional exploration.
In summary, PAR-2 can be downregulated in EC109
cells through RNAi technology. The capabilities of cellular
proliferation, invasion and metastasis were decreased following the
downregulation of PAR-2 expression. Although it remains unclear
whether PAR-2 is involved in repressing the capabilities of cell
proliferation, invasion and metastasis in EC109 cells subsequent to
gene silencing, PAR-2 target silence by RNAi technology may become
a new candidate for treatment in esophageal cancers.
Acknowledgements
The present study was supported by the
Hospital-Level Seed Fund Project of the Affiliated Hospital of
Medical School of the Armed Police (grant no. FYM201115).
References
|
1
|
Montgomery EA: Oesophageal cancerWorld
Cancer Report 2014. Stewart BW and Wild CP: IARC; Lyon: pp.
374–382. 2014
|
|
2
|
Hao J and Shao K: The epidemiology,
current status of management, challenge and future strategy for
esophageal cancer. Chin Oncol. 21:501–504. 2011.
|
|
3
|
Ciocirlan M, Lapalus MG, Hervieu V,
Souquet JC, Napoléon B, Scoazec JY, Lefort C, Saurin JC and Ponchon
T: Endoscopic mucosal resection for squamous premalignant and early
malignant lesions of the esophagus. Endoscopy. 39:24–29. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim T, Grobmyer SR, Smith R, Ben-David K,
Ang D, Vogel SB and Hochwald SN: Esophageal cancer-the five year
survivors. J Surg Oncol. 103:179–183. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ossovskaya VS and Bunnett NW:
Protease-activated receptors: Contribution to physiology and
disease. Physiol Rev. 84:579–621. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caruso R, Pallone F, Fina D, Gioia V,
Peluso I, Caprioli F, Stolfi C, Perfetti A, Spagnoli LG, Palmieri
G, et al: Protease-activated receptor-2 activation in gastric
cancer cells promotes epidermal growth factor receptor
transactivation and proliferation. Am J Pathol. 169:268–278. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shimamoto R, Sawada T, Uchima Y, Inoue M,
Kimura K, Yamashita Y, Yamada N, Nishihara T, Ohira M and Hirakawa
K: A role for protease-activated receptor-2 in pancreatic cancer
cell proliferation. Int J Oncol. 24:1401–1406. 2004.PubMed/NCBI
|
|
8
|
Zhou J, Xie LQ, Li X, Chen X, Chen L,
Zheng Y and Li F: Protease-activated receptor-2 agonists promote
cell invasion and metastasis in human esophageal cancer cell line
EC109. World Chin J Digestol. 13:1313–1319. 2010.(In Chinese).
|
|
9
|
Macfarlane SR, Seatter MJ, Kanke T, Hunter
GD and Plevin R: Proteinase-activated receptors. Pharmacol Rev.
53:245–282. 2001.PubMed/NCBI
|
|
10
|
Nystedt S, Emilsson K, Wahlestedt C and
Sundelin J: Molecular cloning of a potential proteinase activated
receptor. Proc Natl Acad Sci USA. 91:9208–9212. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Inci K, Edebo A, Olbe L and Casselbrant A:
Expression of protease-activated-receptor 2 (PAR-2) in human
esophageal mucosa. Scand J Gastroenterol. 44:664–671. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Uchima Y, Sawada T and Hirakawa K: Action
of antiproteases on pancreatic cancer cells. JOP. 8:(4 Suppl).
S479–S487. 2007.
|
|
13
|
Uusitalo-Jarvinen H, Kurokawa T, Mueller
BM, Andrade-Gordon P, Friedlander M and Ruf W: Role of protease
activated receptor 1 and 2 signaling in hypoxia induced
angiogenesis. Arterioscler Thromb Vasc Biol. 27:1456–1462. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Awasthi V, Mandal SK, Papanna V, Rao LV
and Pendurthi UR: Modulation of tissue factor VIIa signaling by
lipid rafts and caveolae. Arterioscler Thromb Vasc Biol.
27:1447–1455. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fujimoto D, Hirono Y, Goi T, Katayama K,
Hirose K and Yamaguchi A: Expression of protease activated
receptor-2 (PAR-2) in gastric cancer. J Surg Oncol. 93:139–144.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ribeiro FS, Simão TA, Amoêdo ND, Andreollo
NA, Lopes LR, Acatauassu R, Rumjanek FD, Albano RM, Pinto LF and
Monteiro RQ: Evidence for increased expression of tissue factor and
protease-activated receptor-1 in human esophageal cancer. Oncol
Rep. 21:1599–1604. 2009.PubMed/NCBI
|
|
17
|
Hannon GJ: RNA interference. Nature.
418:244–251. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brusselmans K, De Schrijver E, Verhoeven G
and Swinnen JV: RNA interference-mediated silencing of the
acetyl-CoA-carboxylase-alpha gene induces growth inhibition and
apoptosis of prostate cancer cells. Cancer Res. 65:6719–6725. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Filipowicz W: RNAI: The nuts and bolts of
the RISC machine. Cell. 122:17–20. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mette MF, Aufsatz W, Kanno T, Daxinger L,
Rovina P, Matzke M and Matzke AJ: Analysis of double-strand RNA and
small RNAs involved in RNA-mediated transcriptional gene silencing.
Methods Mol Biol. 309:61–82. 2005.PubMed/NCBI
|
|
21
|
Zhang H, Zheng X, Chen K, et al: SiRNA
inhibit the expression of VEGF-C in patients with esophageal
carcinoma. Cancer Res Prev Treatment. 37:132–135. 2010.(In
Chinese).
|
|
22
|
Zheng YM, Xie LQ, Zhao JY, Li X, Chen XY,
Chen L, Hai O and Zhou J: Effects of proteinase activated
receptor-2 agonists on proliferation of hepatoma cells. Zhonghua
Gan Zang Bing Za Zhi. 17:701–702. 2009.(In Chinese). PubMed/NCBI
|
|
23
|
Zeng ZS, Shu WP, Cohen AM and Guillem JG:
Matrix metalloproteinase-7 expression in colorectal cancer liver
metastases: Evidence for involvement of MMP-7 activation in human
cancer metastases. Clin Cancer Res. 8:144–148. 2002.PubMed/NCBI
|
|
24
|
Caruso R, Pallone F, Fina D, Gioia V,
Peluso I, Caprioli F, Stolfi C, Perfetti A, Spagnoli LG, Palmieri
G, et al: Protease-activated receptor-2 activation in gastric
cancer cells promotes epidermal growth factor receptor
trans-activation and proliferation. Am J Pathol. 169:268–278. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Darmoul D, Gratio V, Devaud H and Laburthe
M: Protease-activated receptor 2 in colon cancer: Trypsin-induced
MAPK phosphorylation and cell proliferation are mediated by
epidermal growth factor receptor transactivation. J Biol Chem.
279:20927–20934. 2004. View Article : Google Scholar : PubMed/NCBI
|