Introduction
Like every major malignant tumor threatening the
health of women, breast cancer, accounting for 7–10% of the
systemic tumors, has a high mortality rate world-wide (1). Locally advanced breast cancer, the major
cause of postoperative recurrence and death, can rarely be
completely removed by surgical resection. Neoadjuvant chemotherapy
can narrow lesions, decrease tumor clinical stage, reduce
micro-metastases, increase breast conservation rate and improve the
quality of life (2). There remains no
unified neoadjuvant chemotherapy regimen, but TAC scheme
(docetaxel, pirarubicin and cyclophosphamide) is proved to have
better safety and efficacy (3).
Appropriate indexes with better sensitivity and accuracy for
predicting the clinical outcome are of great value in improving the
efficacy and decreasing the side effects of chemotherapy. Several
studies (4,5) have confirmed that downregulation or
deficiency in expression of tissue and serum Ras association domain
family 1A (RASSF1A) gene and the Wnt inhibitory factor (WIF)-1 gene
is closely related with the occurrence and metastasis of breast
cancer, in which the abnormal methylation of promoter is one of the
main mechanisms (6) for the decrease
of gene activity. Our study aimed at analyzing the methylation of
RASSF1A and WIF-1 and their value in prediction of clinical outcome
of neoadjuvant chemotherapy in patients with locally advanced
breast cancer.
Patients and methods
Patient profile
We continuously selected 126 female patients,
diagnosed as locally advanced breast cancer in the Affiliated Union
Hospital of Fujian Medical University from January 2013 to January
2016.
Inclusion criteria
a) Patients who were confirmed with no distant
metastasis by molybdenum target radiography, CT, MRI and ultrasound
and whole body bone imaging; b) patients with no contraindications
of surgery and anesthesia; c) patients who were diagnosed as
locally advanced breast cancer for the first time with no therapy
history such as surgeries, chemotherapy, radiotherapy or endocrine
therapy; and d) patients with Karnofsky Performance Status (KPS)
score >90 points.
Exclusion criteria
a) Patients with metastatic breast tumor; b)
patients with the primary malignant tumor in other parts; c)
patients who were unable to accomplish four cycles of chemotherapy;
d) patients whose clinical data were incomplete. The study was
approved by the Ethics Committee of the Affiliated Union Hospital
of Fujian Medical University, and patients or their family provided
signed written informed consent.
Methods
Patients agreed to accept TAC scheme,
cyclophosphamide 600 mg/m2, intravenous drip, the first
day; pirarubicin 50 mg/m2, intravenous drip, the first
day; docetaxel 75 mg/m2, intravenous drip, the first
day; one cycle consisted of 21 days, and at least 4 cycles were
conducted. During the execution of TAC scheme, the liver and kidney
function, blood routine and coagulation function were regularly
monitored and the symptomatic treatment was conducted for patients
with minor complication; for patients with severe complications,
drug withdrawal and close observation were performed. We recorded
the total effective rate, incidence of complications,
progression-free survival and survival rate. The tumor diameter was
measured with mammary gland molybdenum target in two dimensions,
and the efficacy was determined by UICC solid tumor response
criterion which was divided into complete response (CR), partial
response (PR), stable disease (SD) and disease progression (PD).
The total effective rate = (CR + PR + SD)/total number of ×
100%.
Tumor tissues and peripheral blood samples collected
in this study was used to detect methylation positive rate of
RASSF1A and WIF-1 by methylation-specific PCR (MSP) method and the
relative expression level of RASSF1A and WIF-1 mRNA by reverse
transcription PCR (RT-PCR) method.
MSP process
DNA extraction process
Tissue DNA extraction process: 50 mg of cancer
tissue was ground and later procedures were performed according to
the tissue DNA extraction kit (Beyotime Institute of Biotechnology,
Jiangsu, China) instructions; 500 µl of tissue lysate buffer was
added and heated at 50°C water bath for 1 h. Then the mixture was
diluted with proteinase K until the concentration reached 100
µg/ml. After water bath for 3 h at 50°C, extraction was performed
using equal volume of saturated phenol, phenol-chloroform (volume
ratio, 1:1) and chloroform-isoamyl alcohol (volume ratio, 24:1),
respectively. Then 1/10 volume of sodium ethoxide and 2 times the
volume of absolute ethanol was added to precipitate the DNA which
was later dissolved by a certain volume of TE solution. Then the
DNA concentration and purity were detected using ultraviolet
spectrophotometer (Applied Biosystems Life Technologies, Foster
City, CA, USA).
Peripheral blood DNA extraction process: 5 ml venous
blood was collected and centrifuged at 2,500 × g for 20 min; the
supernatant was then preserved at −80°C; 200 µl of the supernatant
was for later procedures according to the instruction of plasma DNA
extraction kit (Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.,
Beijing, China) using paramagnetic particle method.
Sulfite modification
Relevant procedures were performed according to the
instruction of DNA methylation modification kit (Sigma, St. Louis,
MO, USA); 1 µg of DNA was dissolved in 45 µl of TE solution. Then
it was diluted by 5 µl of NaOH (3 mol/l) until the concentration
reached 0.3 mol/l. After 20 min of denaturation at 37°C, 30 µl of
hydroquinone (10 nmol/l) and 520 µl of sodium bisulfite (pH 5.0, 3
mol/l) were added and heated in 50°C water bath for 16 h. For
purification by columns, DNA purification agent was added in 45 µl
of deionized water. Then it was diluted by 5 µl of NaOH (3 mol/l)
until the concentration reached 0.3 mol/l. After desulfurization as
well as precipitation by ethanol, it was dissolved in the 20 µl of
TE solution and preserved at −20°C. DNA of the lymphocyte in human
umbilical cord blood treated by bisulphite was set as the
non-methylation positive control, DNA of the lymphocyte in human
umbilical cord blood modified by SssI methyltransferase and treated
by bisulphite as methylation positive control, and the distilled
water as negative control; the DNA of the same sample was amplified
by non-methylated primers and methylation-specific primers.
MSP
SYBR-Green I method was applied. Primers were
synthesized by Invitrogen (Carlsbad, CA, USA) and the primer
sequences are shown in Table I. The
reaction system included: 10X buffer 2.5 µl + Mg2 + l
(25 mol/l) 1.5 µl + dNTP (2.5 mol/l) 1.8 µl + DNA 50 ng + upstream
and downstream primers (10 µmol/l) 1 µl + Taq polymerase (5 U/µl)
0.25 µl. The reaction system was diluted by water to the total
volume of 25 µl. The reaction was performed for 40 cycles under the
conditions of 95°C for 5 min, 95°C for 1 min, 56°C for 1 min, and
72°C for 1 min and the product was extended at 72°C for 10 min; 2%
agarose gel electrophoresis and analysis by gel imaging analysis
system (Media Cybernetics, Inc., Rockville, MD, USA) were performed
on the MSP product. The production with results of positive
methylation were selected for control to calculate the copies of
DNA. The production treated by 10 times gradient dilution was
chosen for standard substance to perform PCR amplification.
Specimens to be detected with more than 500 copies/ml were
considered as positive gene methylated.
 | Table I.Primer sequences of genes. |
Table I.
Primer sequences of genes.
| Genes | Sequences | Length (bp) |
|---|
| RASSF1A (U) | F:
5′-GGGGGTTTTGTGAGAGTGTGTTT-3′ |
|
|
| R:
5′-CCCAATTAAACCCATACTTCACTAA-3′ | 204 |
| RASSF1A (M) | F:
5′-CGAGAGCGCGTTTAGTTTCGTT-3′ |
|
|
| R:
5′-CGATTAAACCCGTACTTCGCTAA-3′ | 192 |
| WIF-1 (U) | F:
5′-GGGTGTTTTATTGGGTGTATTGT-3′ |
|
|
| R:
5′-AAAAAAACTAACACAAACAAAATACAAAC-3′ | 154 |
| WIF-1 (M) | F:
5′-CGCTCCACTGGGCGCACCGC-3′ |
|
|
| R:
5′-TCGCACCTCGCTCGCGCCAGC-3′ | 145 |
| RASSF1A | F:
5′-CAGATTGCAAGTTCACCTGCCACTA-3′ |
|
|
| R:
5′-GATGAAGCCTGTGTAAGAACCGTCCT-3′ | 249 |
| WIF-1 | F:
5′-GTCTAAACGGGAACAGCCCT-3′ |
|
|
| R:
5′-GCTGGCATTCTCTGTTGTGC-3′ | 354 |
| β-actin | F:
5′-AAAGACCTGTACGCCAACAC-3′ |
|
|
| R:
5′-GTCATACTCCTGCTTGCTGAT-3′ | 219 |
RT-PCR process
DNA was extracted using conventional TRIzol method,
concentration and purity of RNA were determined by ultraviolet
spectrophotometric method, and cDNA was synthesized by reverse
transcription kit. Primers were synthesized and designed by Takara
Bio, Inc. (Otsu, Japan) as shown in Table
I. Reaction system: 5X 2.5 µl buffer l + 1.5 µl
MgCl2 l + 0.5 µl dNTP + GAP-43 and upstream and
downstream primers for internal reference each 1 + 0.3 µl Taq
polymerase + 2 µl cDNA template. The reaction system was diluted by
water to the total volume of 25 µl. Reactions were performed for 35
cycles under the conditions of 95°C for 5 min, 95°C 30 sec, 62°C,
30 sec, 30 sec 72°C for 30 sec and the product was extended at 72°C
for 10 min. We composed the dissolution curve and used
2−ΔΔCq method (7) to
calculate the relative expression level of mRNA.
Statistical analysis
We used SPSS 20.0 software (IBM SPSS, Armonk, NY,
USA) for statistical analysis. Measurement data are presented as
mean ± standard deviation. Independent sample t-test was performed
in intergroup comparison. Countable data are presented as the cases
or (%). Chi-square (χ2) test was performed in intergroup
comparison; the specificity of clinical outcome prediction using
RASSF1A and WIF-1 gene was analyzed by receiver operating
characteristic (ROC) curve, and the accuracy was presented in the
area under the curve (AUC). P<0.05 was considered to indicate a
statistically significant difference.
Results
Chemotherapy outcome analysis
The 126 studied cases constituted 18 cases of CR, 32
cases of PR, 50 cases of SD and 26 cases of PD. The total effective
rate was 79.37%. There was no difference in comparison of baseline
data between effective group and ineffective group (P>0.05)
(Table II).
 | Table II.Baseline data comparison. |
Table II.
Baseline data comparison.
| Groups | Effective group
(n=100) | Ineffective group
(n=26) | t/χ2 | P-value |
|---|
| Age (years) | 56.7±14.5 | 55.9±16.3 | 0.152 | 0.932 |
| Invasive ductal
carcinoma, case (%) | 68 | 19 | 0.249 | 0.618 |
| Invasive lobular
carcinoma | 32 | 7 |
|
|
| Maximum diameter of
tumor (cm) | 3.8±1.2 | 3.9±1.4 | 0.163 | 0.879 |
| Phase II, case
(%) | 46 | 12 | 0.000 | 0.989 |
| Phase III | 54 | 14 | 0.002 | 0.966 |
| Lymphatic metastasis,
case (%) | 38 | 10 |
|
|
| Chemotherapy
cycle | 4.9±0.8 | 4.6±0.5 | 0.232 | 0.824 |
| Follow-up time
(months) | 15.5±3.4 | 13.6±3.8 | 0.356 | 0.763 |
The major adverse reactions, including
gastrointestinal reaction, bone marrow suppression, cardiac
toxicity, and hair loss, were divided into 0-IV levels, according
to the World Health Organization (WHO) classification standard for
adverse reaction of chemotherapy. Comparison on adverse reactions
occurrence between the two groups showed no difference (P>0.05);
compared with the ineffective group, the effective group had longer
progress-free survival and higher survival rate and the differences
between the groups were statistically significant (P<0.05)
(Table III).
 | Table III.Survival outcome analysis. |
Table III.
Survival outcome analysis.
| Groups | Effective group
(n=100) | Ineffective group
(n=26) | χ2 | P-value |
|---|
| Gastrointestinal
reaction, case (%) |
| I–II | 50 | 11 | 0.489 | 0.783 |
|
III–IV | 20 | 6 |
|
|
| Bone marrow
suppression, case (%) |
| I–II | 30 | 8 | 0.134 | 0.935 |
|
III–IV | 10 | 2 |
|
|
| Cardiotoxicity, case
(%) |
| I–II | 15 | 5 | 0.405 | 0.817 |
|
III–IV | 6 | 2 |
|
|
| Hair loss, case
(%) |
| I–II | 35 | 9 | 0.101 | 0.951 |
|
III–IV | 13 | 4 |
|
|
| Progression-free
survival time (months) | 6.5±1.2 | 3.4±0.9 | 6.532 | 0.013 |
| Survival rate, case
(%) | 78 (78.0) | 12 (46.2) | 8.064 | 0.005 |
Comparison on methylation positive
rates of RASSF1A and WIF-1 of tissue and serum
In comparison on methylation positive rates of
RASSF1A and WIF-1 of tissue and serum, the effective group was
significantly lower than the ineffective group (P<0.05)
(Table IV).
 | Table IV.Comparison on methylation positive
rates of RASSF1A and WIF-1 of tissue and serum, case (%). |
Table IV.
Comparison on methylation positive
rates of RASSF1A and WIF-1 of tissue and serum, case (%).
| Groups | Effective group
(n=100) | Ineffective group
(n=26) | χ2 | P-value |
|---|
| Tissue RASSF1A | 33 (33.00) | 20 (76.92) | 16.335 | <0.001 |
| Tissue WIF-1 | 24 (24.00) | 15 (57.69) | 10.960 | 0.001 |
| Serum RASSF1A | 15 (15.00) | 11 (42.31) | 9.396 | 0.002 |
| Serum WIF-1 | 6 (6.00) | 8
(30.77) | 10.433 | 0.001 |
Comparison on relative expression
level of RASSF1A and WIF-1 mRNA in tissue and serum
The relative expression level of RASSF1A and WIF-1
mRNA in tissue and serum of the effective group was obviously
higher than the ineffective group (P<0.05) (Table V).
 | Table V.Comparison on relative expression
level of RASSF1A and WIF-1 mRNA in tissue and serum. |
Table V.
Comparison on relative expression
level of RASSF1A and WIF-1 mRNA in tissue and serum.
| Groups | Effective group
(n=100) | Ineffective group
(n=26) | t-test | P-value |
|---|
| Tissue RASSF1A | 0.6358±0.1236 | 0.3256±0.1124 | 7.532 | <0.001 |
| Tissue WIF-1 | 0.5427±0.1247 | 0.2142±0.1032 | 7.123 | <0.001 |
| Serum RASSF1A | 0.4326±0.1326 | 0.1213±0.0639 | 5.629 |
0.008 |
| Serum WIF-1 | 0.3598±0.1528 | 0.0659±0.0124 | 6.124 | <0.001 |
Analysis of the prediction for clinical outcome
using the methylation positive rate of RASSF1A and WIF-1 in tissue
and serum. The sensitivity of clinical outcome prediction using
tissue RASSF1A methylation was 67.0%, the specificity 15.4%,
positive predictive value 69.0% and negative predictive value
31.0%. The above mentioned indexes of tissue WIF-1 were 76.0, 31.4,
72.2 and 27.8%, respectively. These indexes of serum RASSF1A were
85.0, 50.0, 76.2 and 23.8%, respectively, and indexes of serum
WIF-1 were 94.0, 75.0, 81.0 and 19.0%, respectively.
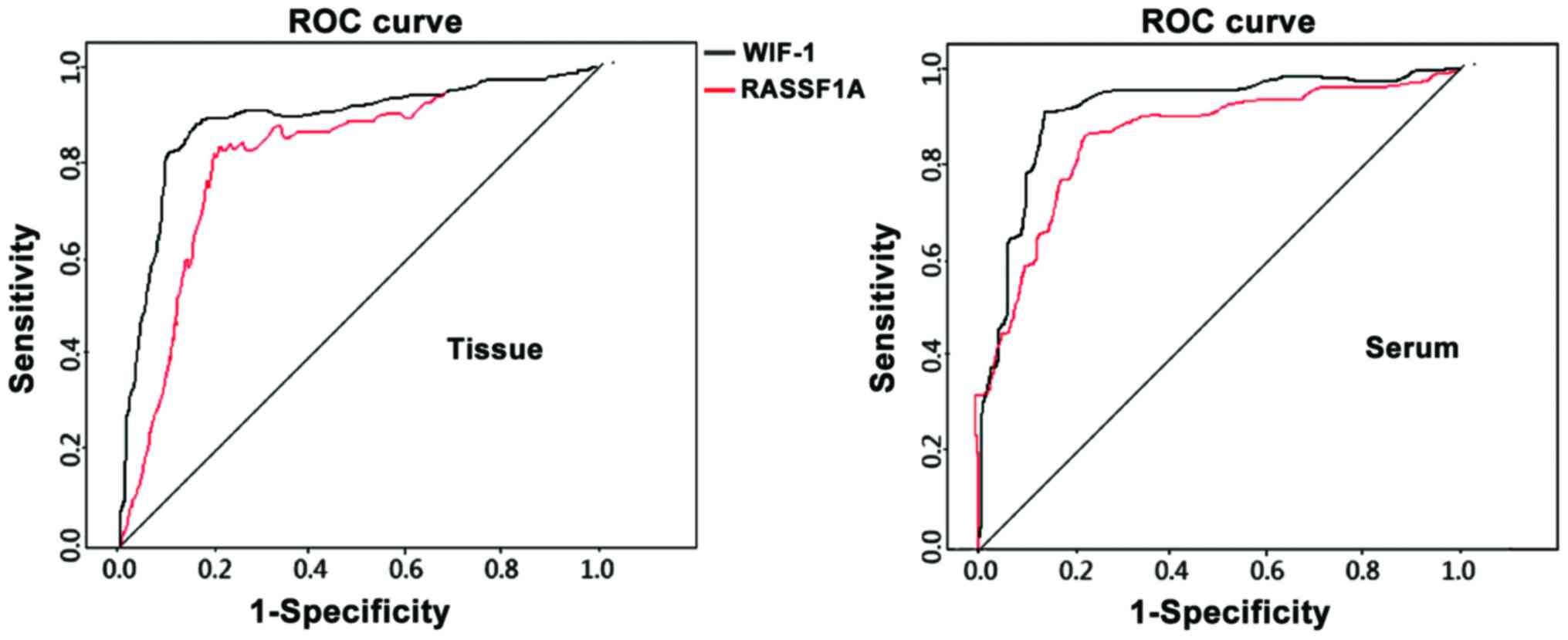
ROC analysis of tissue and serum
RASSF1A mRNA level in clinical outcome prediction
The tissue and serum RASSF1A and WIF-1 mRNA levels
served as the diagnostic index and the clinical outcome as the
diagnosis, which were substituted into the ROC for analysis and
suggested: The accuracy of clinical outcome prediction using tissue
RASSF1A mRNA level was 0.812 (CI=0.756–0.932, P=0.008). The
sensitivity 85.2%, the specificity 76.3% and the critical value
0.4256. These indexes of tissue WIF-1 were 0.833 (95%
CI=0.721–0.948, P=0.010), 86.7%, 75.4% and 0.3562, respectively.
These indexes of serum RASSF1A were 0.864 (CI=0.737–0.964,
P=0.003), 88.3%, 77.4% and 0.2564, respectively. These indexes of
serum WIF-1 were 0.882 (95% CI=0.727–0.986, P=0.005), 89.4%, 73.5%
and 0.1562, respectively (Fig.
1).
Discussion
RASSF1A is highly expressed in almost every normal
tissue, through inhibiting tumor formation by regulating the cell
cycle, apoptosis and genome stability, and the deficiency of its
expression is closely related to tumor occurrence and development
(8). It has been proved (9,10) that the
deficiency of its expression is observed in more than 20 tumor
types, including the lung, breast, ovarian and prostate cancer, ans
its expression could be activated by antitumor drugs to have its
biological activity recovered. Abnormal methylation of gene
promoter is an important cause for deactivation. Malignantly cloned
DNA in the tumor tissues can be continuously released into the
peripheral circulating blood and the circulating tumor cells as
well as micro-metastases can also be delivered into the blood for
detection (11). High levels of
methylation of RASSF1A and low level of mRNA expression are closely
related with clinical stages, differentiation degree, sensitivity
to chemotherapy and survival rate of breast cancer (12). Although levels of RASSF1A methylation
and mRNA have been widely applied in the early diagnosis of breast
cancer (13), there are few studies
assessing the clinical efficacy of neoadjuvant chemotherapy on
locally advanced breast cancer. In the present study, we found that
the total effective rate of TAC scheme was 79.37% with lower
incidence and slight symptoms of adverse reactions, suggesting
better safety and efficacy of TAC scheme. The progression-free
survival of the effective group was ~6.5 months, and the survival
rate was 78.0%. Rates of positive methylation of tissue, and serum
RASSF1A and WIF-1 in the effective group were remarkably lower than
those in the ineffective group; but, with levels of mRNA
significantly higher than those in the ineffective group.
Wnt/β-catenin signaling pathway plays a key role in biological
behavior of breast cancer occurrence, differentiation,
proliferation and invasion (14).
C-myc and cyclin D1, as two important downstream target genes,
regulate the cell division cycle (15). Not only is E-cadherin an important
effector molecular in this signal pathway, but also a key factor in
the invasion, metastasis and recurrence of breast cancer (16). As a major tumor suppressor gene of Wnt
pathways, WIF-1 which is highly methylated or expresses a low level
of mRNA can significantly affect the occurrence and development of
breast cancer (17).
Further study found that the sensitivity of clinical
outcome prediction using tissue RASSF1A and WIF-1 methylation
positive rate was 67.0–76.0%, positive predictive value was
69.0–72.2%, and the specificity and negative predictive value were
relatively low. The sensitivity of clinical outcome prediction
using the rate of serum RASSF1A and WIF-1 positive methylation was
85.0–94.0%, positive predictive value was 76.2–81.0%, specificity
was 50.0–75.0%, and the negative predictive value was relatively
low. For tumor suppression genes such as RASSF1A and WIF-1, higher
efficacy could be achieved by lowering the methylation positive
rate of these two genes; besides, some abnormally methylated genes
may be involved in the sensitivity to chemotherapy. Through the
analysis of ROC, we found that the mRNA levels of RASSF1A and WIF-1
in tissues and serum show better accuracy, sensitivity and
specificity when applied to predict the clinical outcomes. In
conclusion, the detection of methylation and mRNA expression levels
of RASSF1A and WIF-1 genes in tissues and serum of the patient with
locally advanced breast cancer is of great significance in
predicting clinical efficacy of the neoadjuvant chemotherapy TAC
scheme.
Acknowledgements
This study was supported by the Youth Project of
Fujian Provincial Health and Family Planning Commission (grant no.
20130233).
References
|
1
|
Yung KW, Yung TT, Chung CY, Tong GT, Liu
Y, Henderson J, Welbeck D and Oseni S: Principles of cancer
staging. Asian Pac J Surg Oncol. 1:1–16. 2015.
|
|
2
|
Lee R, Yeung AW, Hong SE, Brose MS and
Michels DL: Principles of medical oncology. Asian Pac J Surg Oncol.
1:39–46. 2015.
|
|
3
|
Lupichuk S, Tilley D, Kostaras X and Joy
AA: Real-world adjuvant TAC or FEC-D for HER2-negative
node-positive breast cancer in women less than 50 years of age.
Curr Oncol. 23:164–170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ji Y, Jin HH, Wang MD, Cao WX and Bao JL:
Methylation of the RASSFIA promoter in breast cancer. Genet Mol
Res. 15:156–157. 2016. View Article : Google Scholar
|
|
5
|
Trifa F, Karray-Chouayekh S, Jmal E, Jmaa
ZB, Khabir A, Sellami-Boudawara T, Frikha M, Daoud J and
Mokdad-Gargouri R: Loss of WIF-1 and Wnt5a expression is related to
aggressiveness of sporadic breast cancer in Tunisian patients.
Tumour Biol. 34:1625–1633. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takahashi H, Kagara N, Tanei T, Naoi Y,
Shimoda M, Shimomura A, Shimazu K, Kim SJ and Noguchi S:
Correlation of methylated circulating tumor DNA with response to
neoadjuvant chemotherapy in breast cancer patients. Clin Breast
Cancer. Jun 16–2016.(Epub ahead of print). PubMed/NCBI
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative geneexpression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kristiansen S, Nielsen D and Sölétormos G:
Detection and monitoring of hypermethylated RASSF1A in serum from
patients with metastatic breast cancer. Clin Epigenetics. 8:352016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Spitzwieser M, Holzweber E, Pfeiler G,
Hacker S and Cichna-Markl M: Applicability of HIN-1, MGMT and
RASSF1A promoter methylation as biomarkers for detecting field
cancerization in breast cancer. Breast Cancer Res. 17:1252015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han JC, Xu F, Chen N, Qi GB, Wei YJ, Li
HB, Zhang YJ, Li JH, Wang XL, Xu W, et al: Promoter methylations of
RASSF1A and p16 is associated with clinicopathological features in
lung cancers. J Cancer Res Ther. 12:340–349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Groot JS, Moelans CB, Elias SG, Fackler
Jo M, van Domselaar R, Suijkerbuijk KP, Witkamp AJ, Sukumar S, van
Diest PJ and van der Wall E: DNA promoter hypermethylation in
nipple fluid: a potential tool for early breast cancer detection.
Oncotarget. 7:24778–24791. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rauscher GH, Kresovich JK, Poulin M, Yan
L, Macias V, Mahmoud AM, Al-Alem U, Kajdacsy-Balla A, Wiley EL,
Tonetti D, et al: Exploring DNA methylation changes in promoter,
intragenic, and intergenic regions as early and late events in
breast cancer formation. BMC Cancer. 15:8162015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pirouzpanah S, Taleban FA, Mehdipour P and
Atri M: Association of folate and other one-carbon related
nutrients with hypermethylation status and expression of RARB,
BRCA1, and RASSF1A genes in breast cancer patients. J Mol Med
(Berl). 93:917–934. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu D, Wong P, Li W, Vogel CF and Matsumura
F: Suppression of WIF-1 through promoter hypermethylation causes
accelerated proliferation of the aryl hydrocarbon receptor (AHR)
overexpressing MCF10AT1 breast cancer cells. Toxicology.
285:97–103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xiao C, Wu CH and Hu HZ: LncRNA UCA1
promotes epithelial-mesenchymal transition (EMT) of breast cancer
cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med
Pharmacol Sci. 20:2819–2824. 2016.PubMed/NCBI
|
|
16
|
Li M, Cai H, Yang Y, Zhang J, Sun K, Yan
Y, Qu H, Wang W, Wang J and Duan X: Perichondrium mesenchymal stem
cells inhibit the growth of breast cancer cells via the
DKK-1/Wnt/β-catenin signaling pathway. Oncol Rep. 36:936–944. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jacobsen A, Heijmans N, Verkaar F, Smit
MJ, Heringa J, van Amerongen R and Feenstra KA: Construction and
experimental validation of a Petri net model of Wnt/β-catenin
signaling. PLoS One. 11:e01557432016. View Article : Google Scholar : PubMed/NCBI
|