Introduction
Gastric cancer is one of the most common malignant
tumors, and although chemotherapy and neoadjuvant chemotherapy have
been widely used in the treatment of gastric cancer, the prognosis
of gastric cancer is poor, especially for patients with clinical
metastasis and tumor recurrence (1,2).
Epidermal-type fatty acid binding protein-5 (FABP-5) gene is
a fatty acid binding protein found in epidermal cells. It is a key
molecule in tumor development. In a variety of tumors, the
tumor-associated epithelial cell adhesion molecule upregulates the
expression of FABP-5 gene. Tumor cells with a high
expression of FABP-5 gene can affect cell signal
transduction function by fatty acid metabolites (3–6). At
present, there is no relevant report regarding the role of
FABP-5 gene in gastric cancer. In order to further study the
mechanism of FABP-5 gene in gastric cancer, this study
employed RNA interference technology to silence FABP-5 gene
in human gastric SGC-7901 cancer cells, and the effect on tumor
cell proliferation, apoptosis and invasiveness was observed.
Materials and methods
Materials
The human gastric cancer cell line (SGC-7901) was
purchased from Shanghai Ji Kai Gene Technology Co., Ltd. (Shanghai,
China). shRNA target designed recombinant FABP-5 gene
silencing lentiviral particles LV-shRNA-FABP-5 and the control
empty vector lentiviral particles (LV-shRNA-NC) were provided by
the Shanghai Bio-engineering Co., Ltd. (Shanghai, China). DMEM,
phosphate buffer (phosphatic buffered saline, PBS), and fetal
bovine serum were purchased from Hyclone; GE Healthcare (Logan, UT,
USA). TRIzol was purchased from Invitrogen; Thermo Fisher
Scientific (Waltham, MA, USA) and the reverse transcription kit was
purchased from Fermentas; Thermo Fisher Scientific (Waltham, MA,
USA). A fluorescence quantitative PCR kit was purchased from Takara
Biotechnology Co., Ltd. (Dalian, China); DNA marker was purchased
from Guangzhou Dongsheng Biotech Co., Ltd. (Guangzhou, China);
western blot analysis and IP cell lysate, phenylmethylsulfonyl
(phenylmethanesulfonyl fluoride, PMSF), loading buffer (5X) on
SDS-PAGE protein, BCA protein concentration of the test kit
(Enhanced), 20X TBS buffer were purchased from Jiangsu Pik days
Biotechnology Research Institute (Nanjing, China); PVDF membrane
was purchased from Merck Millipore (Billerica, MA, USA); propidium
iodide (PI) and RNase enzymes were purchased from Fermentas; Thermo
Fisher Scientific; flow cytometry was purchased from BD Biosciences
(Franklin Lakes, NJ, USA); fluorescence microscope was purchased
from Olympus Corp. (Tokyo, Japan); apoptosis kit was purchased from
eBioscience company (Vienna, Austria); and the cell invasion assay
was purchased from cell Invasion Assay Kit (Chemicon International
(Billerica, MA, USA); cat. no. ECM550). The primers for the FABP-5
and GAPDH gene sequence were produced and verified by Shanghai
Bio-engineering Co. Ltd., the same to a previous report (5). Rabbit anti-human FABP-5 monoclonal
antibody was purchased from Abcam (Cambridge, UK); and mouse
anti-human GAPDH monoclonal antibodies were purchased from Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing,
China).
Methods
Cell culture and transfection. Gastric cancer
SGC-7901 cells were cultured with DMEM medium containing 10% fetal
calf serum and 100 U/ml levofloxacin in a sealed incubator (37°C,
5% CO2). Cells in the logarithmic growth phase were
randomly selected: the FABP-5-shRNA group was treated with
Lv-shRNA-FABP-5, the negative control group was treated with
(LV-shRNA-NC), and the control group was routinely cultured. At 12
h before transfection, human gastric cancer SGC-7901 cells in the
logarithmic phase were digested with trypsin and prepared into cell
suspension. The cells were seeded in 6-well plates
(5×104) and cultured in a 37°C, 50 ml/l CO2
incubator until cell confluence was up to 20–30%. Transfection was
performed, and polybrene and infection enhancement solution were
added to each well, with multiplicity of infection (MOI) being 10.
Three replicate wells were set for each group. The LV-shRNA-FABP-5
target sequence was 5-TGGGAAGGAAAGCACAATA-3, while the target
sequence for the NC (negative control) was 5-TTCTCCGAACGTGTCACGT-3.
The cells were harvested 3 days after transfection. The same amount
of medium instead of the transfection system was used for the blank
control group. Cells continued to be cultured in the 37°C, 5%
CO2 humidified incubator.
RT-PCR to detect FABP-5 mRNA
expression
At 72 h after transfection, total RNA of SGC-7901
cells (4×105 cells) was extracted using the TRIzol kit.
Quantitation cDNA was then synthesized according to the reverse
transcription kit protocol. Primer 5.0 software was used to design
primers as follows: FABP-5 upstream primer:
5-TGAAGGAGCTAGGAGTGGGAA-3, downstream primer:
5-TGCACCATCTGTAAAGTTGCAG-3, amplified fragment 212 bp; internal
reference GAPDH upstream primer: 5-TGACTTCAACAGCGACACCCA-3
downstream primers: 5-CACCCTGTTGCTGTAGCCAAA-3, amplified fragment
121 bp. Total RNA of five groups of cells was extracted using
TRIzol reagent and reverse transcribed to cDNA, after which three
replicate wells were set for each group. PCR primers were designed
and synthesized by Shanghai Sangon Biological Engineering
Technology & Services Co., Ltd. (Shanghai, China), and three
replicate wells were set for each group. Each experiment was
repeated three times.
The RT-PCR reaction conditions used were: 95°C
denaturation for 5 min, 95°C denaturation for 30 sec, 60°C
annealing for 30 sec, 72°C extension for 60 sec, a total of 40
cycles. PCR products were subjected to 1.2% agarose gel
electrophoresis, and a gel imager was used to observe results. The
UVI gel imaging system was used to capture pictures. Image-Pro Plus
7.0 software was used to analyze the gray values, with the
FABP-5/GAPDH ratio as the FABP-5 mRNA relative expression
level.
Western blot analysis to detect FABP-5
protein expression
At 72 h after transfection, the total protein of
cells was extracted in each experimental group and the protein
concentration was determined using the BCA kit. Total protein (50
µg) was subjected to 8% SDS-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred to PVDF membranes by the wet transfer
method. After film transfer, the membrane was blocked with 10%
non-fat dry milk at room temperature for 2 h. Subsequently, rabbit
polyclonal FABP-5 antibody (dilution, 1/500; cat. no. ab37267) and
rabbit polyclonal GAPDH antiboody (dilution, 1:500, cat. no.
ab37168) purchased from Abcam (Cambridge, MA, USA) were added. They
were placed on a shaker and incubated overnight at 4°C. The next
day, the membrane was washed by (triethanolamine buffered saline
solution) TBS-T three times for 10 min each time. Then secondary
goat anti-rabbit (HRP) IgG antibody (dilution, 1:2,000 (Abcam);
cat. no. ab6721) was added after washing the membrane. The
membranes were then exposed in the dark. Ultra-sensitive ECL
chemiluminescence was used to detect protein bands, images were
captured and striped gray value analyzed. The relative expression
of the target protein was calculated as: gray value of target
protein bands/gray value of internal reference protein bands.
CCK8 assay to detect SGC-7901 cell
proliferation
SGC-7901 cells were seeded in 96-well culture plates
at 4×103/well and cultured with 200 µl DMEM medium
containing 10% fetal calf serum. Each group set five wells and
separate blank wells were set as the control. CCK-8 (20 µl) was
added into each well. After incubation for 4 h, the absorbance at
490 nm was detected by a microplate reader, the average of five
wells was calculated, and the growth curve was drawn.
Transwell chamber invasion assay
The polycarbonate membrane filter was capped with 50
µg Matrigel per well in a well-polymerized lower chamber, after
which 10% fetal bovine serum was added as conditioned medium. Then,
100 µl SGC-7901 cell suspension of the three groups
(3×105/l) was added in the upper chamber, placed in an
incubator for 24 h and fixed with 4% paraformaldehyde for 10 min
prior to staining with hematoxylin for 20 min. The cells on the
lower surface of membrane were counted under a light microscope
(BX-42, Olympus Corp.). Penetrating cells in five random fields
were counted for each film, and the average was calculated. Each
group set three chambers in parallel. Experiments were repeated
three times. The cell invasion rate (%) was calculated as the total
number of penetrating cells/total number of cells seeded in the
upper chamber × 100%.
Cell cycle detection
Three groups of cells were trypsinized and washed
with PBS twice and fixed with 1 ml pre-chilled 70% ethanol at 4°C
overnight. The cells were washed with PBS twice and measured for
1.0×105/ml cell suspension. After mixing, an appropriate
amount of PI solution (cell suspension, PI=1:1) was added. The
cells were incubated for 30 min at 4°C in the dark. A 300 mesh
screen filter was used to filter the cell suspension and remove
adhesion cells. Flow cytometry was used to analyze DNA content, and
the software was used to analyze the cell number in G0/G1, S, G2/M
phases and the proportion.
Cell apoptosis detected by flow
cytometry
At 72 h after transfection, the cells in each group
were collected and digested by EDTA-free trypsin. Then they were
transferred into a 1.5 ml sterile Eppendorf tube. The
centrifugation was performed at 4°C, 1,650 × g for 5 min. The
supernatant was discarded and the cells were washed with PBS twice,
and 500 µl binding buffer was added to re-suspend cells. Annexin
V-FITC (5 µl) and PI (5 µl) were added at room temperature in the
dark. After reacting for 5–15 min, the cells were detected by flow
cytometry within 1 h. The excitation wavelength (Ex) was 488 nm,
and the emission wavelength (Em) was 530 nm.
TUNEL staining
At 72 h after grouping, the cells in each group were
collected and MGC-803 glass slides were prepared. After aspirating
the cultured medium, the cells were air dried, fixed with 4%
paraformaldehyde, treated with fresh 3% H202 at room temperature.
Then, 0.1% Triton X-100 (dissolved in 0.1% sodium citrate solution)
was used for drilling. According to the TUNEL kit instructions, DAB
staining and hematoxylin re-staining were performed. This was
followed by gradient ethanol dehydration, xylene transparency and
neutral gum sealing, after which the samples were observed under
the microscope. Three horizons were observed in each slice, and 300
consecutive cells were counted in each field. The percentage of
apoptotic cells was the apoptosis index (m), and was calculated as:
AI (%) = number of apoptotic cells/total number of cells ×
100%.
Statistical analysis
SPSS 16.0 statistical software (Chicago, IL, USA)
was used for analysis. Apoptosis data of the two groups were
compared using the t-test, and one-way ANOVA was used to determine
the remaining measurement data among groups. Experimental data were
presented as mean ± SD. P<0.05 was considered statistically
significant.
Results
shRNA downregulated FABP-5 mRNA
expression level in SGC-7901 cells
After RNA interference, RT-PCR results showed the
FABP-5 mRNA level in SGC-7901 cells (Fig.
1A and B). The relative expression level (0.12±0.03) was
significantly reduced in the FABP-5-shRNA group compared with the
negative control group (0.62±0.08%) and blank control group
(0.57±0.11%) (P<0.01) and the inhibition rate was (70.27±1.38%).
There was no significant difference in FABP-5 mRNA expression
between the negative and blank control groups, suggesting that the
interference sequence designed and produced in this study can
specifically inhibit the expression of FABP-5 gene (Fig. 1).
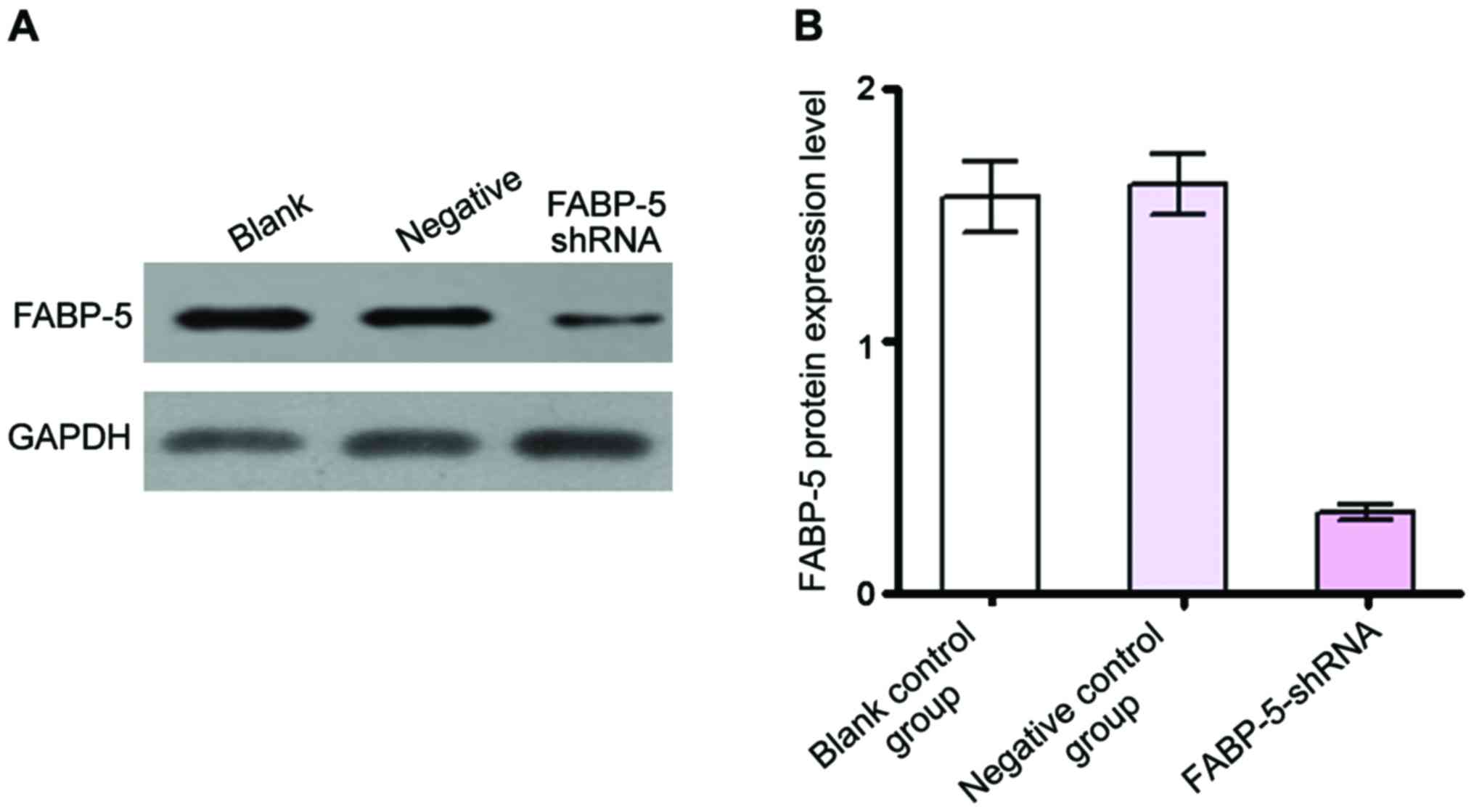
shRNA down-regulated FABP-5 protein
expression level in SGC-7901 cells
Western blot analysis revealed, after RNA
interference, the FABP-5 protein level in SGC-7901 cells (Fig. 2A and B). The relative expression level
(0.32±0.03) was significantly reduced in the FABP-5-shRNA group
compared with the negative control group (1.62±0.12) and blank
control group (1.57±0.14) (P<0.01). The software Image J was
used to analyze the gray values of bands and calculate the
inhibition rate, which was (72.56±1.24%). By contrast, there was no
significant difference in FABP-5 protein expression between the
negative and blank control groups, indicating that the interference
sequence designed and produced in this study can specifically
inhibit the expression of FABP-5 protein (Fig. 2).
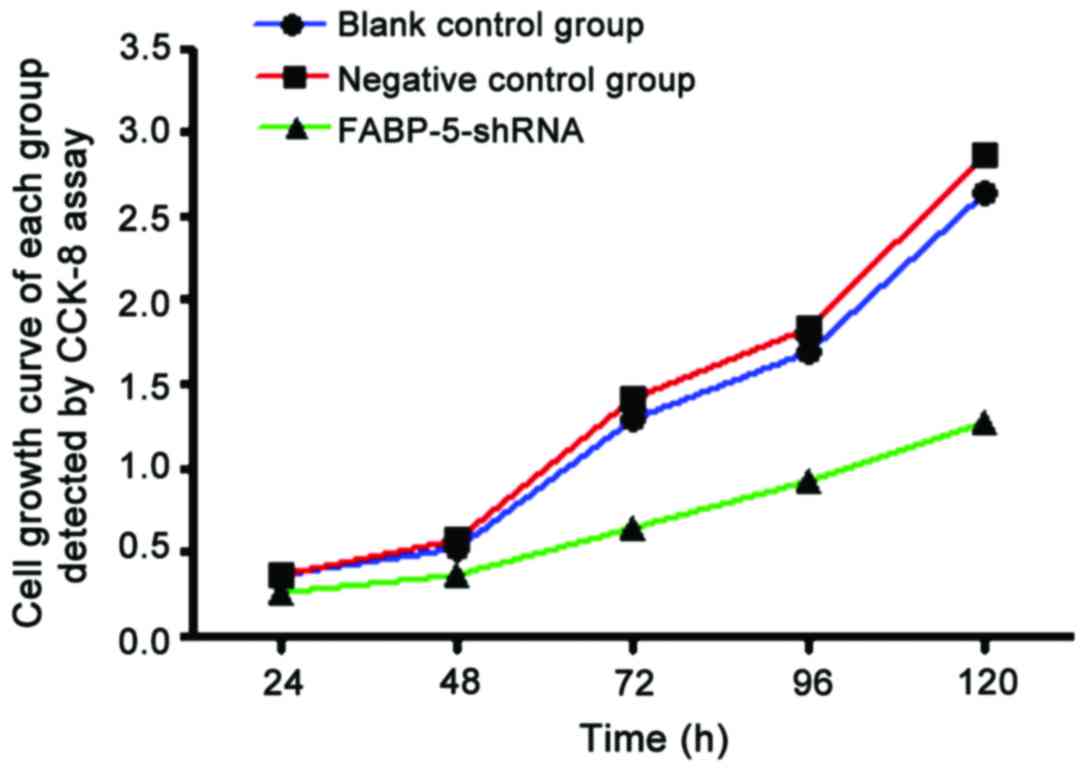
CCK-8 assay to detect reduced SGC-7901
cell proliferation in FABP-5-shRNA group
CCK-8 test results showed that compared with the
blank and negative control groups, A values at 490 nm in the
FABP-5-shRNA group were lower at 24, 48, 72, 96 and 120 h after
transfection, and the differences were statistically significant
(P<0.05). The growth curve showed that, the curve of
FABP-5-shRNA group was significantly lower than that of the blank
and negative control groups, and the difference was statistically
significant (P<0.05). It indicated that the cell proliferation
in the FABP-5-shRNA group was significantly inhibited (Fig. 3; Table
I).
 | Table I.Comparison of cell viability of three
groups of cells at different time pointsa. |
Table I.
Comparison of cell viability of three
groups of cells at different time pointsa.
| Groups | 24 h | 48 h | 72 h | 96 h | 120 h |
|---|
| Blank control
group | 0.36±0.03 | 0.55±0.04 | 1.36±0.11 | 1.79±0.18 | 2.74±0.21 |
| Negative control
group | 0.35±0.02 | 0.56±0.06 | 1.40±0.13 | 1.81±0.16 | 2.75±0.19 |
| FABP-5-shRNA
group | 0.26±0.02 | 0.36±0.04 | 0.64±0.09 | 0.92±0.12 | 1.28±0.17 |
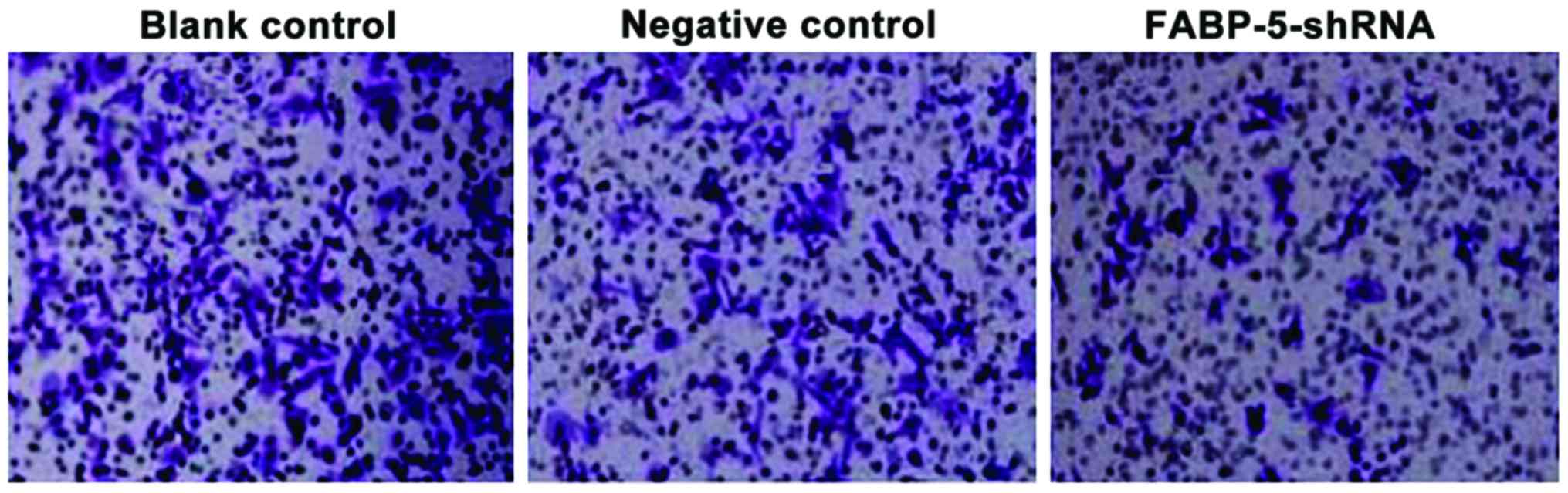
Cell invasiveness was reduced in the
FABP5-siRNA group
As shown in Fig. 4,
the number of cells identified across the membrane in the blank and
negative control groups were higher [(59.22±6.32) and
(61.27±7.36)], the number of cells across the membrane in the
FABP-5-shRNA group was significantly reduced (28.46±4.58), and the
difference was statistically significant (P<0.01). The results
showed that specifically interfering with FABP-5 gene
expression effectively reduces the invasiveness of SGC-7901 cells
(Fig. 4).
Flow cytometry to detect cell
cycle
The proportion of cells in G1 phase in the
FABP-5-shRNA group was reduced compared to the blank and negative
control groups (P<0.05). Proportions of S-phase cells and
G2/M-phase cells in the FABP-5-shRNA group increased compared with
the blank and negative control groups (all P<0.05). In the
FABP-5-shRNA group, cells in G1 phase decreased, and cells in
S-phase and G2/M phase increased in the negative and blank control
groups, and the difference was not statistically significant
(P>0.05) (Table II; Fig. 5).
 | Table II.Cell cycle distribution and apoptosis
rate (%). |
Table II.
Cell cycle distribution and apoptosis
rate (%).
| Groups | G1 | S | G2/M |
|---|
| Blank control
group | 62.83±0.84 | 30.56±0.64 | 7.53±0.56 |
| Negative control
group | 63.46±0.79 | 29.17±0.46 | 7.49±0.73 |
| FABP-5-shRNA
group | 32.75±0.56 | 48.34±0.96 | 18.84±0.57 |
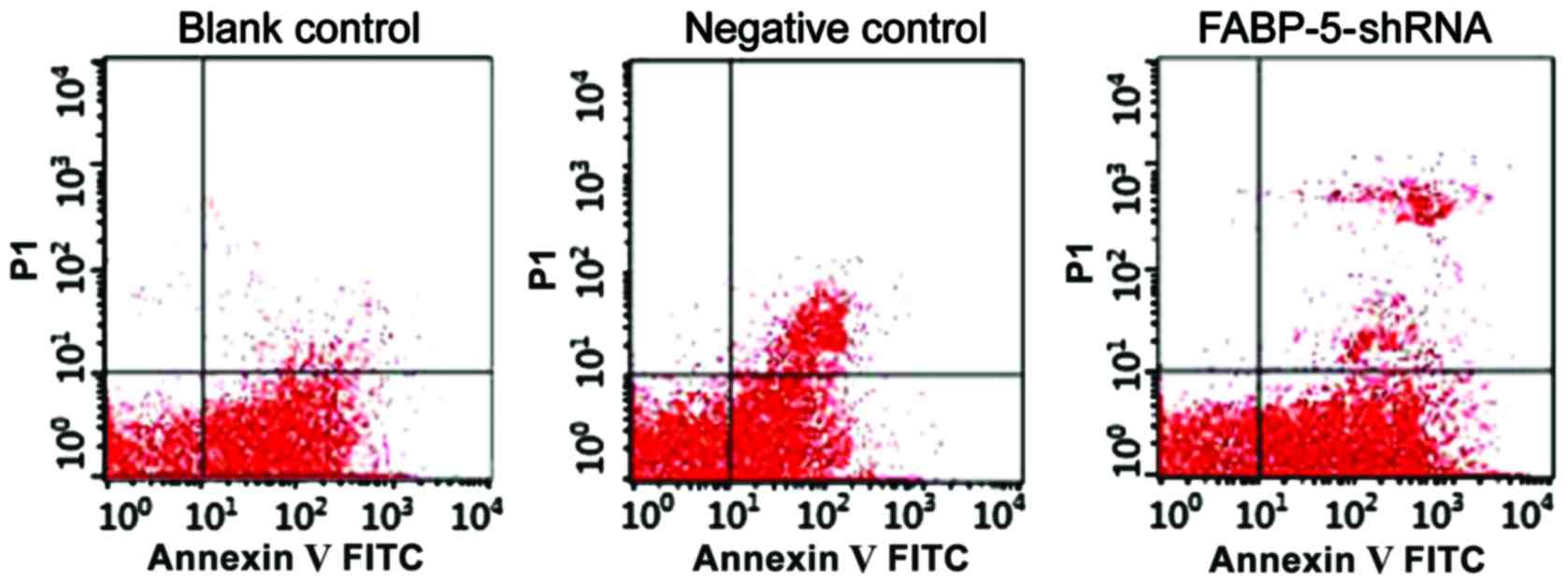
Apoptotic cells detected by flow
cytometry
In the DNA histogram of flow cytometry the SGC-7901
cell apoptosis rate of the FABP-5-shRNA group (4.76±0.16%) was
significantly higher than that of the blank control group
(2.13±0.36%) and the negative control group (2.13±0.36%), and the
difference was statistically significant (P<0.05) (Fig. 6). RNA interference to silence
FABP-5 gene can significantly promote apoptosis of SGC-7901
cells.
TUNEL staining to observe
apoptosis
In the FABP-5-shRNA group, there were apoptotic
cells with brown-stained nuclei (Fig.
7, arrow). A comparison of the negative and blank control
groups showed the results for the apoptotic index were
(5.86±1.23%), (6.26±1.75%) and (38.64±6.84%), and the difference
was significant (P<0.01).
Discussion
FABPs are a group of small and highly-conserved
cytoplasmic proteins, widely expressed in a variety of mammal
cells, and which can bind to long-chain fatty acid cytoplasmic
protein, playing an important role in the uptake, transport and
metabolism of long-chain fatty acids (7). FABPs are tissue-specific, named by the
tissue from which they were initially isolated or identified.
Currently, there are nine types of FABP, including the
epidermal-relevant type FABP and myocardium-type FABP. The most
basic function of FABPs is to be involved in the intake and
intracellular transport of fatty acids (8). Expression of FABPs is increased in many
types of cancer. FABPs affect tumor growth and progression by
combining with transported fatty acids and their derivatives,
hormones, steroids, carcinogens and other ligands (9,10).
Recently, an increasing number of studies have shown that FABPs are
expressed in different degrees in most malignancies, including
breast, prostate, liver, lung and bladder epithelium cancer, and
are associated with the incidence, metastasis, invasion, poor
prognosis and resistance of malignant tumors (11–15).
FABP-5 is epidermal-relevant type, and the current
study found that FABP-5 expressed by cells can combine with
long-chain fatty acids, provide energy and raw materials for cell
growth and participate in tumor growth-associated signal
transduction. Fatty acid binding proteins are closely associated
with tumors and a variety of other diseases (9,16). In
breast cancer (17) and lung squamous
cell carcinoma (18), FABP-5
gene was significantly upregulated to promote tumorigenesis. In
primary NSCLC tissues, FABP-5 expression was associated with tumor
grade and metastasis. The larger the tumor volume and the higher
the tumor grade, the higher the expression of FABP-5, including
patients with metastasis (19). Celis
et al found that the FABP-5 expression level was positively
correlated with the degree of differentiation of bladder cancer
(20). Additionally, the expression
level of FABP-5 decreased as the degree of differentiation of
bladder cancer decreased (20). In
prostate cancer, in vivo experiments confirmed that FABP5
downregulation can reduce tumor cell metastasis and inhibit tumor
growth (21,22). In intrahepatic bile duct cell
carcinoma and squamous cell carcinoma, FABP-5 can promote the
proliferation of tumor cells and enhance the invasion ability of
cells (23,24). Zhou et al transfected
FABP-5-shRNA expression vector into human HepG2 cells and found
that FABP-5-shRNA can significantly promote tumor cell apoptosis,
arrest the cell cycle in G2/M phase to inhibit the proliferation of
liver cancer cells, and reduce the invasiveness of liver cancer
cells (6). In addition, head and neck
cancer (25), endometrial cancer
(26) and melanoma (27) are closely associated with the
expression of FABP-5. A previous study on pathological tissues of
esophageal cancer showed that FABP-5 gene expression was
significantly increased, suggesting that the upregulation of
FABP-5 gene expression may contribute to the development of
esophageal cancer (28).
In the present study, we transfected FABP-5-shRNA
expression vector into human gastric SGC-7901 cancer cells, and
found that the relative expression levels of FABP-5 gene and
protein in the FABP-5-shRNA group were significantly lower than
those in the negative and blank control groups, indicating that the
interference sequence designed and synthesized in this study can
specifically inhibit the expression of FABP-5 gene. CCK-8
detection results showed that compared to the blank and negative
control groups, cell proliferation in the FABP-5-shRNA group was
significantly inhibited. Flow cytometry and TUNEL staining showed
that FABP-5 gene silencing can significantly promote
SGC-7901 cell apoptosis. Flow cytometry showed that after FABP-5
gene silencing, the SGC-7901 cell cycle was arrested in G2/M phase,
and the proliferation of SGC-7901 cells was inhibited. The cell
invasion chamber assay showed that cell invasiveness in the
FABP-5-shRNA group was significantly lower than that in the blank
and negative control groups, suggesting that FABP-5 gene
silencing reduced the invasiveness of gastric cancer cells.
In summary, using the lentivirus RNA interference to
knockout FABP-5 gene can influence the proliferation of
gastric cancer cells and induce apoptosis, and can inhibit the
invasiveness of gastric cancer cells. These indicated that
FABP-5 gene may be directly or indirectly involved in the
cell cycle regulation and apoptosis of gastric cancer cells. These
changes of the gene expression levels were closely associated with
tumor cell invasiveness. Therefore, FABP-5 gene may become a
target for the treatment of gastric cancer.
References
|
1
|
Zhu YL, Yang L, Sui ZQ, Liu L and Du JF:
Clinicopathological features and prognosis of Borrmann type IV
gastric cancer. J BUON. 21:1471–1475. 2016.PubMed/NCBI
|
|
2
|
Yildiz B, Etiz D, Dal P, Junushova B,
Pasaoglu O, Yilmaz E, Erkasap S and Dincer M: Tumor deposits:
Prognostic significance in gastric cancer patients. J BUON.
21:1476–1481. 2016.PubMed/NCBI
|
|
3
|
Veerkamp JH, Maatman RG and Prinsen CF:
Fatty acid-binding proteins: Structural and functional diversity.
Biochem Soc Trans. 20:801–805. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Münz M, Zeidler R and Gires O: The
tumour-associated antigen EpCAM upregulates the fatty acid binding
protein E-FABP. Cancer Lett. 225:151–157. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Uma RS, Naresh KN, D'Cruz AK, Mulherkar R
and Borges AM: Metastasis of squamous cell carcinoma of the oral
tongue is associated with down-regulation of epidermal fatty acid
binding protein (E-FABP). Oral Oncol. 43:27–32. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu RZ, Graham K, Glubrecht DD, Germain
DR, Mackey JR and Godbout R: Association of FABP5 expression with
poor survival in triple-negative breast cancer: implication for
retinoic acid therapy. Am J Pathol. 178:997–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith S, Witkowski A and Joshi AK:
Structural and functional organization of the animal fatty acid
synthase. Prog Lipid Res. 42:289–317. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chmurzyńska A: The multigene family of
fatty acid-binding proteins (FABPs): Function, structure and
polymorphism. J Appl Genet. 47:39–48. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thumser AE, Moore JB and Plant NJ: Fatty
acid binding proteins: Tissue-specific functions in health and
disease. Curr Opin Clin Nutr Metab Care. 17:124–129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zimmerman AW, van Moerkerk HT and Veerkamp
JH: Ligand specificity and conformational stability of human fatty
acid-binding proteins. Int J Biochem Cell Biol. 33:865–876. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawamura T, Kanno R, Fujii H and Suzuki T:
Expression of liver-type fatty-acid-binding protein, fatty acid
synthase and vascular endothelial growth factor in human lung
carcinoma. Pathobiology. 72:233–240. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lawrie LC, Dundas SR, Curran S and Murray
GI: Liver fatty acid binding protein expression in colorectal
neoplasia. Br J Cancer. 90:1955–1960. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hammamieh R, Chakraborty N, Barmada M, Das
R and Jett M: Expression patterns of fatty acid binding proteins in
breast cancer cells. J Exp Ther Oncol. 5:133–143. 2005.PubMed/NCBI
|
|
14
|
Tölle A, Suhail S, Jung M, Jung K and
Stephan C: Fatty acid binding proteins (FABPs) in prostate, bladder
and kidney cancer cell lines and the use of IL-FABP as survival
predictor in patients with renal cell carcinoma. BMC Cancer.
11:3022011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hashimoto T, Kusakabe T, Watanabe K,
Sugino T, Fukuda T, Nashimoto A, Honma K, Sato Y, Kimura H, Fujii
H, et al: Liver-type fatty acid-binding protein is highly expressed
in intestinal metaplasia and in a subset of carcinomas of the
stomach without association with the fatty acid synthase status in
the carcinoma. Pathobiology. 71:115–122. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Armstrong EH, Goswami D, Griffin PR, Noy N
and Ortlund EA: Structural basis for ligand regulation of the fatty
acid-binding protein 5, peroxisome proliferator-activated receptor
β/δ (FABP5-PPARβ/δ) signaling pathway. J Biol Chem.
289:14941–14954. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Levi L, Lobo G, Doud MK, von Lintig J,
Seachrist D, Tochtrop GP and Noy N: Genetic ablation of the fatty
acid-binding protein FABP5 suppresses HER2-induced mammary
tumorigenesis. Cancer Res. 73:4770–4780. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harris FT, Rahman SM, Hassanein M, Qian J,
Hoeksema MD, Chen H, Eisenberg R, Chaurand P, Caprioli RM, Shiota
M, et al: Acyl-coenzyme A-binding protein regulates Beta-oxidation
required for growth and survival of non-small cell lung cancer.
Cancer Prev Res (Phila). 7:748–757. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Q, Wang S, Xu H and Zhang S:
Expressions and significances of CRABPII and E-FABP in non-small
cell lung cancer. Zhongguo Fei Ai Za Zhi. 16:12–19. 2013.(In
Chinese). PubMed/NCBI
|
|
20
|
Celis JE, Rasmussen HH, Vorum H, Madsen P,
Honoré B, Wolf H and Orntoft TF: Bladder squamous cell carcinomas
express psoriasin and externalize it to the urine. J Urol.
155:2105–2112. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Adamson J, Morgan EA, Beesley C, Mei Y,
Foster CS, Fujii H, Rudland PS, Smith PH and Ke Y: High-level
expression of cutaneous fatty acid-binding protein in prostatic
carcinomas and its effect on tumorigenicity. Oncogene.
22:2739–2749. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morgan EA, Forootan SS, Adamson J, Foster
CS, Fujii H, Igarashi M, Beesley C, Smith PH and Ke Y: Expression
of cutaneous fatty acid-binding protein (C-FABP) in prostate
cancer: Potential prognostic marker and target for
tumourigenicity-suppression. Int J Oncol. 32:767–775.
2008.PubMed/NCBI
|
|
23
|
Jeong CY, Hah YS, Cho BI, Lee SM, Joo YT,
Jung EJ, Jeong SH, Lee YJ, Choi SK, Ha WS, et al: Fatty
acid-binding protein 5 promotes cell proliferation and invasion in
human intrahepatic cholangiocarcinoma. Oncol Rep. 28:1283–1292.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang LY, Wong TY, Chiang WF and Chen YL:
Fatty-acid-binding protein 5 promotes cell proliferation and
invasion in oral squamous cell carcinoma. J Oral Pathol Med.
39:342–348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rauch J, Ahlemann M, Schaffrik M, Mack B,
Ertongur S, Andratschke M, Zeidler R, Lang S and Gires O: Allogenic
antibody-mediated identification of head and neck cancer antigens.
Biochem Biophys Res Commun. 323:156–162. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Huang C, Bai S, Pan X, Zhou R, Wei Y
and Zhao X: Prognostic evaluation of epidermal fatty acid-binding
protein and calcyphosine, two proteins implicated in endometrial
cancer using a proteomic approach. Int J Cancer. 123:2377–2383.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brouard MC, Saurat JH, Ghanem G and
Siegenthaler G: Urinary excretion of epidermal-type fatty
acid-binding protein and S100A7 protein in patients with cutaneous
melanoma. Melanoma Res. 12:627–631. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ogawa R, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Mori Y, Mori R, Tomoda K, Katada T, Harada K, et al:
Identification of candidate genes involved in the radiosensitivity
of esophageal cancer cells by microarray analysis. Dis Esophagus.
21:288–297. 2008. View Article : Google Scholar : PubMed/NCBI
|