Introduction
Esophageal cancer remains a significant cause of
cancer-related death and its incidence rate has shown a drastic
increase of more than 6-fold worldwide (1,2).
Esophageal squamous cell carcinoma (ESCC) is a dominant
histological type of esophageal malignances (3,4). The poor
prognosis and increasing incidence of ESCC highlight the need for
improved detection, prediction, monitoring, and treatment methods
(2,5).
Existing histopathological terms, such as the pathologic TNM
classification, are insufficient to accurately predict individual
differences in outcome and inform personalized treatment (2,6). Genetic
and epigenetic alterations, such as aberrant gene expression, copy
number alterations, and DNA methylation, are associated with the
development of ESCC, as well as other malignancies, and evidence
for the potential prognostic role of genomic and epigenetic
profiles has been accumulating (7,8). Since
molecular signatures can have clinical application in risk
stratification for prediction of treatment response, metastatic
potential, recurrence, and survival, researchers should continue
their efforts to identify novel ESCC-related molecular events
(9,10).
Non-specific cytotoxic cell receptor protein 1
(NCCRP1) was initially cloned from fish species and
predicted to be a type II/III membrane protein (11). NCCRP1 was believed to be a receptor
expressed in non-specific cytotoxic cells that was responsible for
their cytolytic functions (12).
Later, Kallio et al investigated the human gene and found
that NCCRP1 is expressed intracellularly and is a paralog of the
F-box superfamily of proteins, which are components of the E3
ubiquitin ligase complexes and regulate the cell cycle (13). More importantly, NCCRP1 mRNA
was found to be abundantly expressed in human tissues containing
squamous epithelium and silencing of NCCRP1 caused a
significant decrease in the growth of HeLa cells (13). However, the role of NCCRP1 in
ESCC is unknown.
In the present study, we focused on NCCRP1 as
a candidate ESCC-related gene for the following reasons: (1) NCCRP1 is abundant in the squamous
epithelium; (2) NCCRP1 is involved in
cell proliferation; (3) the
NCCRP1 gene harbors a CpG island in the promoter region
(suggesting the possibility of methylation); (4) NCCRP1 is a paralog of the F-box
superfamily of proteins that regulate the cell cycle (14–16); and,
finally, (5) there are no published
data related to NCCRP1 expression in ESCC. The purpose of
the present study was to evaluate the expression, regulatory
mechanisms, and clinical significance of NCCRP1 in ESCC.
Materials and methods
Ethics
This study conformed to the ethical guidelines of
the World Medical Association Declaration of Helsinki Ethical
Principles for Medical Research Involving Human Subjects. Written
informed consent for the use of clinical samples and data, as
required by the institutional review board at Nagoya University,
Japan, was obtained from all patients.
Sample collection
Nine ESCC cell lines (TE1, TE2, TE3, NUEC1, NUEC2,
NUEC3, TT, TTn, and WSSC) and a control non-tumorigenic epithelial
cell line (FHs74) were obtained from the American Type Culture
Collection (Manassas, VA, USA), Japanese Collection of Research
Bioresources Cell Bank (Osaka, Japan), or were established in our
institute (17). Cells were stored at
−80°C using cell preservative solution (Cell Banker; Mitsubishi
Chemical Medience Corporation, Tokyo, Japan) and cultured in
RPMI-1640 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10%
fetal bovine serum in an atmosphere containing 5% CO2 at
37°C. A total of 213 primary ESCC tissues and adjacent normal
tissues were acquired from patients who underwent radical
esophageal resection at Nagoya University Hospital between October
2001 and January 2016. All tissue samples were diagnosed
histologically as ESCC, frozen immediately after resection, and
stored at −80°C. Specimens were classified histologically using the
seventh edition of the UICC staging system for esophageal cancer.
Patients were questioned to determine their levels of alcohol
consumption, and excessive alcohol consumption was defined as
>210 g/week for ≥3 years (18).
Since 2006, neoadjuvant chemotherapy (fluorouracil combined with
platinum-based drugs) was administered to patients with clinical
stage II/III ESCC unless contraindicated by the patient's condition
or patient refusal (19,20). Postoperative follow-up examinations
included physical examination, measurement of serum tumor markers
every 3 months, and enhanced computed tomography of the chest and
abdominal cavity every 6 months. Adjuvant chemotherapy was
administered to selected patients according to the patient's
condition and the physician's discretion.
Quantitative real-time
reverse-transcription polymerase chain reaction (qRT-PCR)
The levels of NCCRP1 mRNA were determined
using qRT-PCR. Total RNA (10 µg) isolated from ESCC cell lines and
213 primary ESCCs and adjacent normal tissues was used as the
template for cDNA synthesis. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) mRNA levels (TaqMan, GAPDH control
reagents: Applied Biosystems, Foster City, CA, USA) were quantified
to normalize expression levels (9).
qRT-PCR was performed using the SYBR Green PCR Core Reagents Kit
(Applied Biosystems) as follows: One cycle at 95°C for 10 min; 40
cycles at 95°C for 5 sec, and 60°C for 60 sec. All samples were
tested in triplicate, and samples without template were included in
each PCR plate as negative controls. Real-time detection of SYBR
Green fluorescence was conducted using an ABI StepOnePlus Real-Time
PCR System (Applied Biosystems). The expression level of each
sample is shown as the value of the NCCRP1 amplicon divided
by that of GAPDH (21).
Sequences of specific primers are listed in Table I.
 | Table I.Sequences of primers. |
Table I.
Sequences of primers.
| Primer | Experiment | Type | Sequence
(5′-3′) | Product size | Annealing
temperature |
|---|
| NCCRP1 | qRT-PCR | Forward |
AAAGCTCCAGCAGAACCAAA | 104 bp | 60°C |
|
|
| Reverse |
TAATGGCTGGTTGTTCGTCA |
|
|
|
| Bisulfite | Forward |
TTTAGTTAATTTTAGTTTTGTGAAAT | 282 bp | 64°C |
|
| sequencing | Reverse |
CCACTCCTCCAACAACAACTAC |
|
|
| GAPDH | qRT-PCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 bp | 60°C |
|
|
| Probe |
CAAGCTTCCCGTTCTCAGCC |
|
|
|
|
| Reverse |
GAAGATGGTGATGGGATTTC |
|
|
Methylation analysis of NCCRP1
gene
Nucleotide sequence analysis was conducted to
determine the presence of CpG islands around the promoter region of
NCCRP1. CpG islands were defined as follows: ≥200-bp region
with GC content >50% and CpG: Expected CpG ≥0.6 identified using
Methyl Primer Express Software (Applied Biosystems). Genomic DNA
isolated from the cell lines was treated with bisulfite for
bisulfite sequence analysis. Bisulfite DNA from nine ESCC cell
lines and control cell (FHs 74) was amplified with specific primers
(Table I) and sequenced using Big Dye
Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific,
Waltham, MA, USA) and a 3730x l DNA Analyzer (Applied Biosystems)
at Eurofins Genomics Co Ltd, Tokyo, Japan. To assess the
relationship between promoter hypermethylation and NCCRP1
transcription, GC cells (1.5×106 cells) were treated
with 5-aza-2′-deoxycytidine (5-aza-dC; Sigma-Aldrich) to inhibit
DNA methylation and then cultured for 6 days with medium changes on
days 1, 3, and 5 (22). RNA was
extracted and qRT-PCR was performed as described above.
Copy number analysis
NCCRP1 copy number of nine ESCC cell lines
was determined using TaqMan Copy Number Assays (Applied
Biosystems). Twenty nanograms of genomic DNA was amplified using
specific primer pairs according to the manufacturer's instructions
(assay ID: Hs02638838_cn, within exon 6). Data were analyzed using
CopyCaller Software (Life Technologies, Carlsbad, CA) (23). Copy number loss was defined as copy
number value equal to 1 determined in the analyzed region of the
NCCRP1 locus.
Statistical analysis
Numeric variables between the two groups were
compared using the Mann-Whitney U test. The χ2 test was
used to analyze the association between the expression status of
NCCRP1 and clinicopathological parameters. Overall and
disease-free survival rates were calculated using the Kaplan-Meier
method, and differences in survival curves were analyzed using the
log-rank test. We performed multivariable regression analysis to
detect prognostic factors using the Cox proportional hazards model,
and variables with a P-value <0.05 were entered into the final
model. All statistical analysis was performed using JMP 10 software
(SAS Institute Inc., Cary, NC, USA). A P-value <0.05 was
considered statistically significant.
Results
Expression, methylation, and copy
number analysis of cell lines
NCCRP1 harbors a CpG island flanking the
transcription initiation site (length 920 bp, GC 64.9%, CpG 5.7%;
Fig. 1A). NCCRP1 mRNA
expression levels differed among nine ESCC cell lines (Fig. 1B). Bisulfite sequence analysis
revealed that CpG sites in NCCRP1 DNA in all ESCC cells were
CG (complete methylation) and that the corresponding positions in a
control cell line FHs74=were TG (absence of methylation) (Fig. 1C). When we compared the levels of
NCCRP1 mRNA in ESCC cell lines before and after
demethylation, reactivation of NCCRP1 transcription was
detected in all ESCC cells (Fig. 1B).
Moreover, there was no detectable loss of copy number in ESCC cell
lines (Fig. 1B).
Clinical implications of NCCRP1 mRNA
levels in surgically resected esophageal tissues
The median age of the 213 patients was 66 years
(range, 44–84 years). The male:female ratio was 167:16. According
to the UICC staging system (seventh edition), 42, 54, 107, and 10
patients were in pathological stages I, II, III, and IV,
respectively. The median duration of patient follow-up was 35.2
months (range, 4.8–173 months) or until death. In 204 (95.8%)
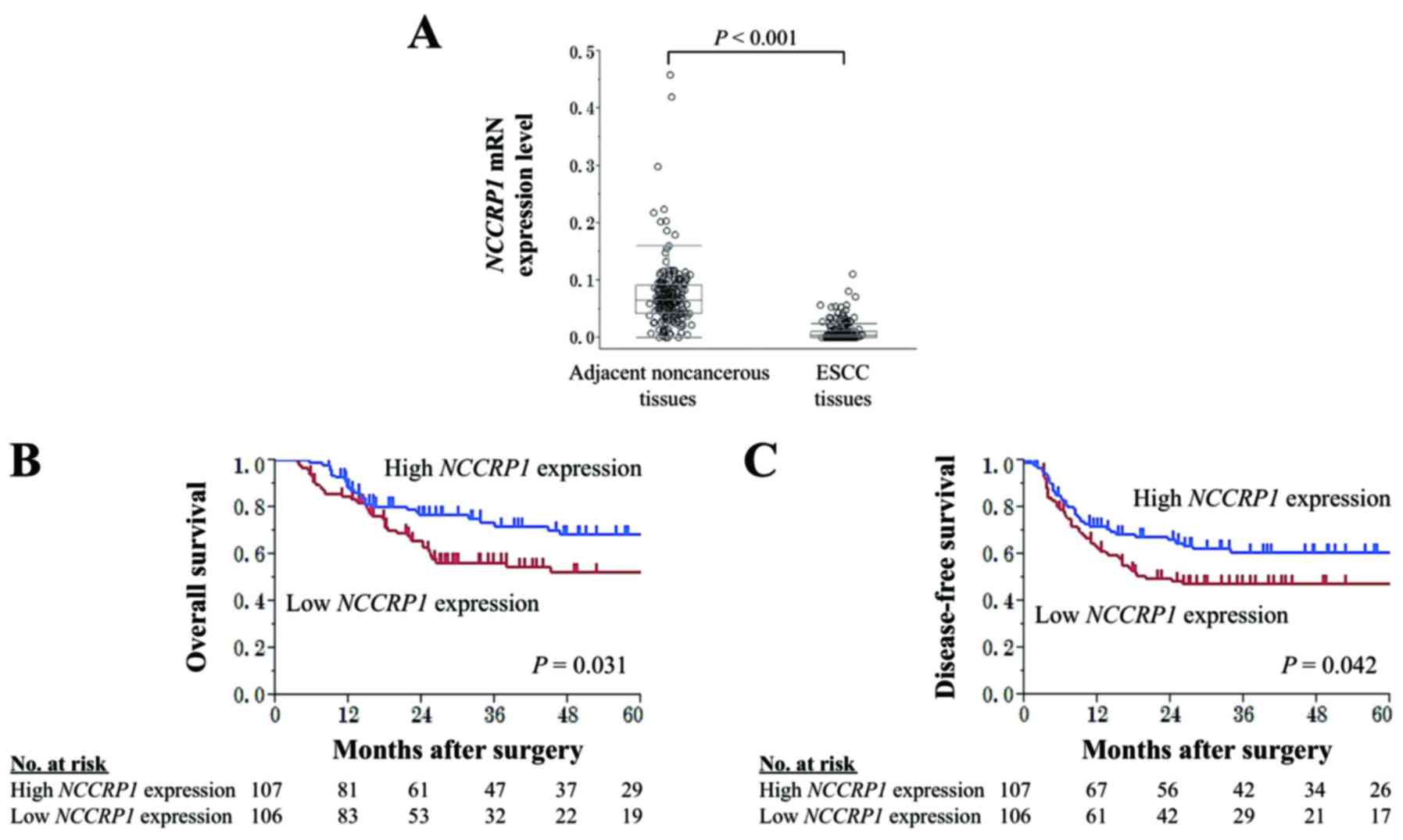
patients, NCCRP1 mRNA expression levels were lower in ESCC
tissues compared with the corresponding non-cancerous adjacent
tissues. The mean expression level of NCCRP1 mRNA was
significantly reduced in ESCC tissues compared with that in
adjacent normal tissues (Fig.
2A).
Patients were assigned to one of two groups
according to their median NCCRP1 mRNA expression level in
ESCC tissues (high NCCRP1 expression group, n=107; low
NCCRP1 expression group, n=106). No significant association
was found between NCCRP1 expression groups and
clinicopathological parameters including patient sex and tumor
size, location, and depth (Table
II). Patients in the low NCCRP1 expression group tended
to have a shorter overall survival (OS) time than those in the high
NCCRP1 expression group (5-year OS rates were 52 and 69% for
the low and high expression groups, respectively; P=0.031; Fig. 2B). In multivariate analysis for
overall survival, low NCCRP1 expression was identified as an
independent prognostic factor (hazard ratio, 1.75; 95% confidence
interval, 1.08–2.87; P=0.022; Table
III). Disease-free survival (DFS) was also significantly poorer
in the low NCCRP1 expression group than in the high
NCCRP1 expression group (3-year DFS rates were 47 and 61%
for the low and high NCCRP1 expression groups, respectively;
P=0.042; Fig. 2C). The frequency of
overall recurrence after radical esophagectomy in the low
NCCRP1 expression group was higher than that of the high
NCCRP1 expression group (49 and 35%, respectively, P=0.032;
Fig. 3A). No appreciable trends were
found in metastasis site as the initial recurrence in comparisons
between low and high NCCRP1 expression groups (Fig. 3A).
 | Table II.Association between the expression of
NCCRP1 mRNA and clinicopathological parameters of 213
patients with squamous cell carcinoma of the esophagus. |
Table II.
Association between the expression of
NCCRP1 mRNA and clinicopathological parameters of 213
patients with squamous cell carcinoma of the esophagus.
| Parameters | Low NCCRP1
expression (n) | High NCCRP1
expression (n) | P-value |
|---|
| Age (years) |
|
|
|
|
<65 | 47 | 46 |
|
|
≥65 | 59 | 61 | 0.843 |
| Sex |
|
|
|
|
Male | 87 | 80 |
|
|
Female | 19 | 27 | 0.194 |
| Preoperative
symptoms |
|
|
|
|
Absent | 24 | 29 |
|
|
Present | 82 | 78 | 0.451 |
| Brinkman index |
|
|
|
|
<1,000 | 66 | 75 |
|
|
≥1,000 | 40 | 32 | 0.227 |
| Excessive alcohol
consumption |
|
|
|
|
Absent | 30 | 29 |
|
|
Present | 76 | 78 | 0.845 |
| Carcinoembryonic
antigen (ng/ml) |
|
|
|
| ≤5 | 94 | 96 |
|
|
>5 | 12 | 11 | 0.807 |
| SCC (ng/ml) |
|
|
|
|
≤1.5 | 65 | 68 |
|
|
>1.5 | 41 | 39 | 0.737 |
| Tumor size
(cm) |
|
|
|
|
<5.0 | 63 | 62 |
|
|
≥5.0 | 43 | 45 | 0.825 |
| Tumor location |
|
|
|
| Ce,
Mt | 65 | 68 |
|
| Lt,
Ae | 41 | 39 | 0.737 |
| UICC pT factor |
|
|
|
|
pT1-2 | 39 | 42 |
|
|
T3-4 | 67 | 65 | 0.712 |
|
Differentiation |
|
|
|
|
Moderate to well | 90 | 94 |
|
|
Poor | 16 | 13 | 0.531 |
| Lymphatic
involvement |
|
|
|
|
Absent | 30 | 28 |
|
|
Present | 76 | 79 | 0.727 |
| Vessel
invasion |
|
|
|
|
Absent | 62 | 68 |
|
|
Present | 44 | 39 | 0.449 |
| Intraepithelial
spread |
|
|
|
|
Absent | 79 | 82 |
|
|
Present | 27 | 25 | 0.720 |
| Intramural
metastasis |
|
|
|
|
Absent | 100 | 97 |
|
|
Present | 6 | 10 | 0.305 |
| Lymph node
metastasis |
|
|
|
|
Absent | 37 | 44 |
|
|
Present | 69 | 63 | 0.350 |
 | Table III.Prognostic factors for overall
survival of 213 patients with squamous cell carcinoma of the
esophagus. |
Table III.
Prognostic factors for overall
survival of 213 patients with squamous cell carcinoma of the
esophagus.
|
|
| Univariate | Multivariable |
|---|
|
|
|
|
|
|---|
| Variable | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65) | 120 | 1.28 | 0.80–2.09 | 0.308 |
|
|
|
| Sex (male) | 167 | 1.30 | 0.74–2.43 | 0.370 |
|
|
|
| Preoperative
symptoms | 160 | 2.05 | 1.12–4.13 | 0.018 | 1.87 | 0.93–4.04 | 0.081 |
| Brinkman index
(≥1,000) | 72 | 1.17 | 0.71–2.02 | 0.540 |
|
|
|
| Excessive alcohol
consumption | 154 | 0.89 | 0.54–1.54 | 0.678 |
|
|
|
| CEA (>5
ng/ml) | 23 | 1.58 | 0.79–2.89 | 0.187 |
|
|
|
| SCC (>1.5
ng/ml) | 80 | 1.36 | 0.84–2.19 | 0.206 |
|
|
|
| Tumor size (≥5.0
cm) | 88 | 1.30 | 0.81–2.07 | 0.281 |
|
|
|
| Tumor location
(Lt/Ae) | 80 | 0.83 | 0.50–1.35 | 0.460 |
|
|
|
| UICC T factor
(T3-4) | 132 | 1.92 | 1.15–3.34 | 0.011 | 1.09 | 0.59–2.08 | 0.794 |
| Tumor
differentiation (poor) | 29 | 1.47 | 0.77–2.61 | 0.226 |
|
|
|
| Lymphatic
involvement | 155 | 3.52 | 1.79–7.98 | <0.001 | 2.87 | 1.37–6.80 | 0.004a |
| Vessel
invasion | 83 | 1.47 | 0.91–2.35 | 0.111 |
|
|
|
| Intraepithelial
spread | 52 | 1.37 | 0.82–2.24 | 0.228 |
|
|
|
| Intramural
metastasis | 16 | 2.00 | 0.92–3.83 | 0.076 |
|
|
|
| Lymph node
metastasis | 132 | 2.51 | 1.47–4.53 | <0.001 | 1.68 | 0.95–3.15 | 0.077 |
| Low NCCRP1
expression | 106 | 1.69 | 1.05–2.75 | 0.031 | 1.75 | 1.08–2.87 | 0.022a |
We conducted a subgroup analysis according to
administration of neoadjuvant chemotherapy (fluorouracil combined
with platinum-based drugs) to further explore the significance of
NCCRP1 expression in ESCC. The prognostic impact of
NCCRP1 expression was similar between patients with and
without neoadjuvant chemotherapy (Fig.
3B).
Discussion
Previous molecular studies have provided evidence
that ESCC arises not only from the combined effects of
environmental factors such as cigarette smoking or excessive
alcohol consumption and susceptible genetic variants, but also from
the accumulation of genetic and epigenetic alterations that play
crucial roles in the process of cellular immortalization and
tumorigenesis (6,7,24).
Understanding of the molecular mechanisms and alterations behind
the initiation and progression of esophageal tumorigenesis is
essential for disease monitoring and identification of novel
therapeutic and clinical targets for ESCC (2,25). To
date, multiple genetic and epigenetic changes in oncogenes and
tumor suppressor genes (TSGs), cell cycle regulators, cell adhesion
molecules, and DNA repair genes have been implicated in esophageal
carcinogenesis (7,26,27).
Nevertheless, the molecular pathogenesis of ESCC is still
incompletely understood and it is vitally important to decipher the
underlying mechanisms of carcinogenesis. We hypothesized that
NCCRP1 is a candidate ESCC-related gene.
The NCCRP1 gene is located on chromosome
19q13.2 and encodes a 31-kDa protein composed of 275 amino acid
residues (11,13). There are no previous reports on
oncological roles of NCCRP1. In this study, we investigated
the expression, methylation status, DNA copy number, and functions
of NCCRP1 in ESCC. Our results suggest that NCCRP1
functions as a TSG that might be responsible, at least in part, for
ESCC carcinogenesis since most ESCCs examined showed reduced
NCCRP1 mRNA expression compared with matched non-cancerous
tissues. We also evaluated the association of NCCRP1
expression with clinical characteristics of ESCC. Patients with low
NCCRP1 expression were likely to have a poor prognosis,
implying a tumor suppressive role of NCCRP1 in ESCC
progression and suggesting that the expression status of
NCCRP1 in ESCC tissues might be a novel biomarker to predict
postoperative outcomes. Of note, the expression of NCCRP1
had no significant association with typical risk factors for ESCC
prognosis, such as tumor depth and lymph node metastasis. This
finding may highlight the utility of NCCRP1 for stratifying
patients at risk of adverse prognosis independent of the TNM
staging system. Since findings of the JCOG9907 phase III study
comparing the survival benefit of pre- or postoperative cisplatin
plus fluorouracil in clinical stage II/III ESCC demonstrated the
superiority of neoadjuvant chemotherapy, neoadjuvant cisplatin plus
fluorouracil followed by esophagectomy has been the standard
treatment for patients with ESCC in Japan (19,20). In
this study, we found that the prognostic impact of NCCRP1
expression was similar between patients who received neoadjuvant
chemotherapy and those who did not. This result emphasized the
clinical utility of NCCRP1 expression to predict
postoperative prognosis regardless of whether the patient received
neoadjuvant chemotherapy.
Promoter hypermethylation leads to transcriptional
silencing of TSGs in various malignancies (28,29). With
respect to regulatory mechanisms, all examined ESCC cell lines
harbored NCCRP1 promoter hypermethylation. Furthermore,
NCCRP1 transcription increased in cells treated with a DNA
methylation inhibitor. To the best of our knowledge, this is the
first report of hypermethylation of NCCRP1. However, none of
the ESCC cell lines had copy number loss at the NCCRP1
locus. These findings indicate that promoter hypermethylation is a
pivotal mechanism that inhibits NCCRP1 transcription in
ESCC. As tumor-specific aberrant DNA methylation can be detected
more stably than mRNA expression levels (30), it has become recognized as a promising
tool for liquid biopsy and assessment of locoregional recurrence at
surgical margin imprints (31,32).
Detection of NCCRP1 methylation in the circulating blood, in
addition to ESCC tissues, would enhance the diagnostic utility of
NCCRP1.
As future perspectives, our findings can be
translated into several clinical applications as follows: i) the
expression and methylation status of NCCRP1 in preoperative
biopsy tissues obtained during endoscopic surveillance may identify
patients requiring intensive perioperative treatment; ii) the
expression levels of NCCRP1 in surgical specimens may
predict recurrence and subsequent adverse prognosis, which will
likely aid efforts to design appropriate postoperative therapeutic
and surveillance strategies; and iii) demethylating agents
targeting NCCRP1 may serve as therapeutics. However, this
study has some limitations. Further studies including pathway
analysis in esophageal carcinogenesis are needed to clarify the
molecular mechanisms underlying the biological activities of
NCCRP1 in ESCC. Also, this study was limited by the
relatively small sample size and lack of external validation of the
reproducibility of the expression assays and their standardization
across laboratories. Finally, this study is limited by its lack of
direct functional analysis of NCCRP1. Better understanding
of the tumor suppressive functions of NCCRP1 would be
elucidated by forced expression of NCCRP1.
Nevertheless, taken together our findings support
the conclusion that NCCRP1 acts as a putative tumor
suppressor gene that is inactivated by promoter hypermethylation
and might serve as a promising biomarker to predict postoperative
prognosis in ESCC.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang FL and Yu SJ: Esophageal cancer:
Risk factors, genetic association and treatment. Asian J Surg. pii:
S1015-S9584. 30201–30209. 2016.(Epub ahead of print).
|
|
3
|
Tanaka H, Kanda M, Koike M, Iwata N,
Shimizu D, Ezaka K, Sueoka S, Tanaka Y, Takami H, Hashimoto R, et
al: Adherens junctions associated protein 1 serves as a predictor
of recurrence of squamous cell carcinoma of the esophagus. Int J
Oncol. 47:1811–1818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hong L, Han Y, Zhang H and Fan D:
Prognostic markers in esophageal cancer: From basic research to
clinical use. Expert Rev Gastroenterol Hepatol. 9:887–889. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Arnal Domper MJ, Arenas Ferrández Á and
Arbeloa Lanas Á: Esophageal cancer: Risk factors, screening and
endoscopic treatment in Western and Eastern countries. World J
Gastroenterol. 21:7933–7943. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oya H, Kanda M, Takami H, Hibino S,
Shimizu D, Niwa Y, Koike M, Nomoto S, Yamada S, Nishikawa Y, et al:
Overexpression of melanoma-associated antigen D4 is an independent
prognostic factor in squamous cell carcinoma of the esophagus. Dis
Esophagus. 28:188–195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kanda M, Nomoto S, Oya H, Takami H,
Shimizu D, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: The expression of melanoma-associated antigen D2 both in
surgically resected and serum samples serves as clinically relevant
biomarker of gastric cancer progression. Ann Surg Oncol. 23 Suppl
2:S214–S221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanda M, Shimizu D, Tanaka H, Tanaka C,
Kobayashi D, Hayashi M, Iwata N, Niwa Y, Yamada S, Fujii T, et al:
Significance of SYT8 for the detection, prediction, and treatment
of peritoneal metastasis from gastric cancer. Ann Surg. Dec
6–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reimers K, Qarn Abu M, Allmeling C, Bucan
V and Vogt PM: Identification of the non-specific cytotoxic cell
receptor protein 1 (NCCRP1) in regenerating axolotl limbs. J Comp
Physiol B. 176:599–605. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cai J, Wei S, Wang B, Huang Y, Tang J, Lu
Y, Wu Z and Jian J: Cloning and expression analysis of nonspecific
cytotoxic cell receptor 1 (Ls-NCCRP1) from red snapper (Lutjanus
sanguineus). Mar Genomics. 11:39–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kallio H, Tolvanen M, Jänis J, Pan PW,
Laurila E, Kallioniemi A, Kilpinen S, Tuominen VJ, Isola J,
Valjakka J, et al: Characterization of non-specific cytotoxic cell
receptor protein 1: A new member of the lectin-type subfamily of
F-box proteins. PLoS One. 6:e271522011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cepeda D, Ng HF, Sharifi HR, Mahmoudi S,
Cerrato VS, Fredlund E, Magnusson K, Nilsson H, Malyukova A,
Rantala J, et al: CDK-mediated activation of the SCF(FBXO) (28)
ubiquitin ligase promotes MYC-driven transcription and
tumourigenesis and predicts poor survival in breast cancer. EMBO
Mol Med. 5:1067–1086. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chiorazzi M, Rui L, Yang Y, Ceribelli M,
Tishbi N, Maurer CW, Ranuncolo SM, Zhao H, Xu W, Chan WC, et al:
Related F-box proteins control cell death in Caenorhabditis
elegans and human lymphoma. Proc Natl Acad Sci USA.
110:3943–3948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang FF, Zhang XJ, Yan YR, Zhu XH, Yu J,
Ding Y, Hu JL, Zhou WJ, Zeng ZC, Liao WT, et al: FBX8 is a
metastasis suppressor downstream of miR-223 and targeting mTOR for
degradation in colorectal carcinoma. Cancer Lett. 388:85–95. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hibino S, Kanda M, Oya H, Takami H,
Shimizu D, Nomoto S, Hishida M, Niwa Y, Koike M, Yamada S, et al:
Reduced expression of DENND2D through promoter hypermethylation is
an adverse prognostic factor in squamous cell carcinoma of the
esophagus. Oncol Rep. 31:693–700. 2014.PubMed/NCBI
|
|
18
|
Mayne ST and Navarro SA: Diet, obesity and
reflux in the etiology of adenocarcinomas of the esophagus and
gastric cardia in humans. J Nutr. 132 (11 Suppl):3467S–3470S.
2002.PubMed/NCBI
|
|
19
|
Ando N, Kato H, Igaki H, Shinoda M, Ozawa
S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, et al: A
randomized trial comparing postoperative adjuvant chemotherapy with
cisplatin and 5-fluorouracil versus preoperative chemotherapy for
localized advanced squamous cell carcinoma of the thoracic
esophagus (JCOG9907). Ann Surg Oncol. 19:68–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kataoka K, Takeuchi H, Mizusawa J, Igaki
H, Ozawa S, Abe T, Nakamura K, Kato K, Ando N and Kitagawa Y:
Prognostic impact of postoperative morbidity after esophagectomy
for esophageal cancer: Exploratory analysis of JCOG9907. Ann Surg.
265:1152–1157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanda M, Oya H, Nomoto S, Takami H,
Shimizu D, Hashimoto R, Sueoka S, Kobayashi D, Tanaka C, Yamada S,
et al: Diversity of clinical implication of B-cell translocation
gene 1 expression by histopathologic and anatomic subtypes of
gastric cancer. Dig Dis Sci. 60:1256–1264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kanda M, Shimizu D, Tanaka H, Shibata M,
Iwata N, Hayashi M, Kobayashi D, Tanaka C, Yamada S, Fujii T, et
al: Metastatic pathway-specific transcriptome analysis identifies
MFSD4 as a putative tumor suppressor and biomarker for hepatic
metastasis in patients with gastric cancer. Oncotarget.
7:13667–13679. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanda M, Tanaka C, Kobayashi D, Tanaka H,
Shimizu D, Shibata M, Takami H, Hayashi M, Iwata N, Niwa Y, et al:
Epigenetic suppression of the immunoregulator MZB1 is associated
with the malignant phenotype of gastric cancer. Int J Cancer.
139:2290–2298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin EW, Karakasheva TA, Hicks PD, Bass AJ
and Rustgi AK: The tumor microenvironment in esophageal cancer.
Oncogene. 35:5337–5349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xin M, Dong XW and Guo XL: Role of the
interaction between galectin-3 and cell adhesion molecules in
cancer metastasis. Biomed Pharmacother. 69:179–185. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee HW, Kwon J, Kang MC, Noh MK, Koh JS,
Kim JH and Park JH: Overexpression of HSP47 in esophageal squamous
cell carcinoma: Clinical implications and functional analysis. Dis
Esophagus. 29:848–855. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shim HJ, Shin MH, Kim HN, Kim JH, Hwang
JE, Bae WK, Chung IJ and Cho SH: The prognostic significance of
FGFR4 Gly388 polymorphism in esophageal squamous cell carcinoma
after concurrent chemoradiotherapy. Cancer Res Treat. 48:71–79.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bird A: Perceptions of epigenetics.
Nature. 447:396–398. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hayashi M, Bernert H, Kagohara LT,
Maldonado L, Brait M, Schoenberg M, Bivalacqua T, Netto GJ, Koch W,
Sidransky D and Hoque MO: Epigenetic inactivation of VGF associated
with Urothelial cell carcinoma and its potential as a non-invasive
biomarker using urine. Oncotarget. 5:3350–3361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hayashi M, Guerrero-Preston R, Okamura J,
Michailidi C, Kahn Z, Li X, Ahn J, Goldsmith M and Koch W:
Innovative rapid gene methylation analysis of surgical margin
tissues in head and neck cancer. Ann Surg Oncol. 21:3124–3131.
2014. View Article : Google Scholar : PubMed/NCBI
|