Introduction
Leukemia is the most prevalent malignant childhood
tumors, accounting for 35% of all malignant tumors in patients
<15 years old with an incidence rate of 4 in 100,000. There are
~15,000 new cases of leukemia in China every year. Acute
lymphoblastic leukemia (ALL), as the most prominent leukemia
subtype, accounts for 75% of pediatric leukemia cases (1). ALL is characterized by the accumulation
of immature lymphoblastic cells due to genomic abnormalities,
blocking the differentiation of early lymphoid progenitors
(2). The genetic changes that
underlie the pathogenesis of ALL have previously been examined in
order to elucidate their relative contributions (3). However, it has been established that
aberrant epigenetic alterations, in particular the DNA methylation
of promoter-associated cytosine-guanine pair (CpG) islands (CGIs),
are frequent events in the progression of ALL and are associated
with an unfavorable prognosis (4–7). A number
of previous studies have indicated that the epigenetic silencing of
tumor suppressor genes due to the DNA methylation of CGIs is an
important mechanism underlying leukemogenesis in ALL (8–10).
The erythroprotein-producing hepatoma amplified
sequence (Eph) receptors and their ephrin ligands comprise the
largest subfamily of receptor tyrosine kinases and are involved in
various cellular developmental processes, including embryonic,
hematopoietic and vascular development, as well as tumorigenesis
(11,12) Previous studies have suggested that Eph
receptors exhibit a tumor suppressor function in certain types of
cancer, including colorectal and breast cancer (13,14).
Aberrant DNA methylation has been reported to be an important
contributor to the inactivation of Eph receptors and ephrin genes,
a process which subsequently promotes tumor progression (6,15,16). Kuang et al (17) performed a comprehensive analysis of
Eph/ephrin methylation using bisulfite pyrosequencing, revealing
that 15 of the Eph/ephrin family genes are frequently
hypermethylated and are associated with gene inactivation in ALL
bone marrow tissues and leukemia cell lines. These genes include
ephrin type-B receptor 4 (EPHB4), which is hypermethylated
in ALL, resulting in transcriptional silencing (16). In addition, apoptosis induction and
the growth suppression of tumor cells are observed following the
restoration of EPHB4 gene expression levels using lentiviral
transduction (16).
The EPHB4 gene was originally identified to
be abundantly expressed in human bone marrow CD34+ cells, and is
highly expressed by primary T cells (18). Previous studies have reported that the
interaction between EPHB4 and ephrin-B2 is involved in the
inhibition of T-cell proliferation by activating the Src,
phosphoinositide 3-kinase (PI3K), Abelson murine leukemia (Abl) and
N-terminal kinase signaling pathways (19). It has also been suggested that T cells
are important in mediating antitumor immune responses; therefore,
insufficient priming of CD4+ T cells may result in impaired
specific anti-leukemia immunity and subsequently enhance the
invasive ability of leukemic cells (20,21). The
role of EPHB4 in the modulation of T-cell physiology implies
that EPHB4 gene methylation may be associated with ALL
pathogenesis (22). A previous study
demonstrated that the EPHB4 gene was highly methylated and
transcriptionally silenced in CEM cell lines, whereas the
suppression of cell growth and promotion of apoptosis was observed
upon addition of 5-aza-2′-deoxycytidine, a demethylating agent
(23). Therefore, the EPHB4
gene is a promising candidate tumor suppressor in ALL. In the
present study, the DNA methylation status of promoter-associated
CGIs in the EPHB4 gene, and the mRNA and protein expression
levels of EPHB4, were analyzed in newly diagnosed cases of
childhood ALL. Furthermore, the two-year disease-free survival
(DFS) of the patients was examined in order to reveal the clinical
relevance of aberrant EPHB4 methylation and its role in the
pathogenesis and prognosis of childhood ALL.
Materials and methods
Clinical samples
The present study included 40 newly diagnosed
patients with ALL who were treated at the Division of Hematology
and Oncology at Shenzhen Children's Hospital, Department of
Pediatrics (Shenzhen, China), between 1st October 2010 and 30th
September 2012. Bone marrow biopsy samples were obtained from the
patients with ALL at the time of initial diagnosis, prior to
chemotherapy administration. All patients participated in the
ongoing Multicenter Trial of GD-2008 ALL protocol for childhood ALL
(trial no. NCT00846703). All the patients were diagnosed and
classified according to the ALL International
Berlin-Frankfurt-Münster 2002 protocol (24) and the duration of follow-up was 2
years (patient characteristics are summarized in Table I). Treatments administered to the
patients with ALL were carried out according to the GD-2008 ALL
protocol (25) which is based on a
modification of the ALL IC-BFM 2002 protocol. For all patients with
ALL the duration of therapy was 104 weeks. During the first stage
of remission induction, all patients were treated with vincristine
intravenously (1.5 mg/m2/day; Shanghai Hualian Pharmacy
Co., Ltd., Shanghai, China), dexamethasone orally or intravenously
(6 mg/m2/day; SPH Sine Pharmaceutical Co., Ltd.,
Shanghai, China), daunorubicin intravenously guttae (30
mg/m2/day; Zhejiang Hisun Pharmaceutical Co., Ltd.,
Zhejiang, China) and L-asparaginase intravenously guttae (5,000
IU/m2/day; GuangZhou BaiYunShan Pharmaceutical Holdings
Co., Ltd, Guangzhou, China), following prednisone prophase. In the
second stage of remission induction, patients were treated with
cyclophosphamide intravenously guttae (1,000 mg/m2/day;
Shanghai Hualian Pharmacy Co., Ltd.), Ara-c intravenously or
intramuscularly (75 mg/m2/day; Pfizer, Inc., New York,
NY, USA.), 6-mercaptopurine orally (6-MP; 60 mg/m2/day;
Zhejiang Zhebei Pharmaceutical Co., Ltd., Zhejiang, China) and
methotrexate intrathecally (age-adapted dose; SPH Sine
Pharmaceutical Co., Ltd.). Oral 6-MP (25 mg/m2/day) and
intravenous methotrexate (5,000 mg/m2) were used in
consolidation therapy following induction therapy. The subsequent
reintroduction and maintenance therapy was based on the same scheme
as the introduction and consolidation stages. Control bone marrow
samples were obtained from 10 patients with idiopathic
thrombocytopenic purpura (ITP) of a similar age range to the
patients with ALL, who did not exhibit hematological malignancy or
any other tumorous disease. The Ethics Committee of Shenzhen
Children's Hospital approved the study. Written informed consent
for recruitment to the study was provided by patients and/or their
parents, following the Shenzhen Children's Hospital Ethical
Guidelines in accordance with the Declaration of Helsinki. The bone
marrow samples were aspirated and stored in heparinized tubes;
mononuclear cells were separated using Ficoll-Paque density
centrifugation at 300–540 × g for 15 min at 4°C (Shanghai Yuanye
Biotechnology Co., Ltd., Shanghai, China) and stored at −80°C.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Characteristic | ALL | ITP |
|---|
| Mean age, years
(range) | 4.2 (0.5–12.6) | 5.5 (2.0–10.5) |
| Sex |
|
|
|
Male | 18 (40) | 4 (10) |
|
Female | 22 (40) | 6 (10) |
| Median WBC count,
109/l | 15.8 | 9.3 |
|
Immunophenotype |
|
|
|
T-ALL | 8 (40) |
|
|
B-ALL | 32 (40) |
|
|
Complete remission | 38 (40) |
|
|
Relapse | 3 (40) |
|
DNA extraction and bisulfite
sequencing polymerase chain reaction (BSP)
DNA was extracted from the mononuclear cells using
the Wizard® Genomic DNA Purification kit (Promega
Corporation, Madison, WI, USA). The genomic DNA was treated with
sodium bisulfite using the EpiTect Bisulfite kit (Qiagen, Inc.,
Valencia, CA, USA), according to a previous protocol (26). Bisulfite treatment induces deamination
of unmethylated cytosines, converting unmethylated CpG sites to
uracil-guanine pairs (UpG) with no effect on the methylated sites
(27). Previously described primer
sequences for EPHB4 (23) were
used for bisulfite pyrosequencing [reaction mixes (final volume,
150 µl) included: 30 µl RNA-free water, 35 µl DNA Protect buffer
(Qiagen, Inc.), DNA (between 1 ng and 2 µg), 85 µl bisulfite mix]
and primer sequences are provided in Table II. The thermocycling conditions were
as follows: 95°C for 10 min; 40 cycles of 95°C for 30 sec, 60°C
(~5°C below the melting temperature of the primers) for 15 sec and
72°C for 1 min. To evaluate the pyrosequencing results of the
mononuclear cell samples, BSP was performed using the same forward
and reverse primers as applied for pyrosequencing. The PCR products
were cloned into a pMD19-T vector (Takara Biotechnology Co., Ltd.,
Dalian, China), and individual clones were sequenced at BGI Genomic
Organization (Shenzhen, China). Five clones were sequenced in each
ALL and ITP sample. The methylation density was calculated as the
mean value of the five clones.
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| A, Primers used for
bisulfite pyrosequenscing |
|---|
|
|---|
| Primer | Sequence | Amplification
product size | Amplified
region | Covered CpG
sites |
|---|
| EPHB4
forward (5–3) |
GTTTGTTTTGGGGGTTTTTGGGTTTTAG | 329bp | (−305, +24) | 31 |
| EPHB4
reverse (5–3) |
AACAAAAACTAAACTACACTAAAACTAAACC |
|
|
|
|
| B, Primers for
quantitative polymerase chain reaction |
|
| Primer |
| Sequence |
|
| EPHB4
forward (5–3) |
|
GTCCTGGTGGTCATTGTGGT |
| EPHB4
reverse (5–3) |
|
TGTCCGTGTTTGTCCGAATA |
| GADPH
forward (5–3) |
|
AGAAGGCTGGGGCTCATTTG |
| GADPH
reverse (5–3) |
|
AGGGGCCATCCACAGTCTTC |
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA (1 µg) was extracted using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and reverse transcription was performed
using RevertAid Reverse Transcriptase (Thermo Fisher Scientific,
Inc.) and random hexamer primers (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany), according to the manufacturer's protocols.
SYBR®-Green Real-Time PCR Master mix (Toyobo Co., Ltd.,
Osaka, Japan) and the aforementioned EPHB4 primers in
Table II were used for RT-qPCR
analysis of mRNA expression levels, according to the manufacturer's
protocol (Toyobo, Co., Ltd.). Data were analyzed using the
2−ΔΔCq method (28) and
normalized to the reference gene GAPDH with primers as described in
Table II. The thermocycling
conditions were as follows: 95°C for 10 min; 40 cycles of 95°C for
30 sec, 60°C (5°C below the melting temperature of the primers) for
15 sec and 72°C for 32 sec.
Western blot analysis
The protein samples were analyzed by western
blotting using a previously described method (29). Briefly, the mononuclear cell samples
were washed with PBS and lysed using a radioimmunoprecipitation
assay buffer containing a protease inhibitor (1:100; Thermo Fisher
Scientific, Inc.), 50 mM Tris-Cl (pH 7.4), 0.15 M NaCl, 1% Na
deoxycholate, 0.5 M EDTA and 0.1% NP-40. The protein lysates were
rotated using a rotator at 50 rmp at 4°C for 1 h and the insoluble
proteins were subsequently removed by centrifugation at 13,400 × g
for 10 min at 4°C. A bicinchoninic acid protein assay kit was used
to determine the soluble protein concentration. Protein products
(8–10 µg/µl; 50–60 µg total) were separated using 10% SDS-PAGE on a
10% gel and subsequently transferred overnight onto a
polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA,
USA) using SDS-transfer buffer (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The membrane was blocked for 1 h using a
western-blocking reagent (Bio-Rad Laboratories, Inc.) at room
temperature, prior to protein detection using a specific monoclonal
EPHB4 antibody (1:1,000; cat. no. sc-130081; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C, followed by
incubation with an horseradish peroxidase-conjugated anti-mouse
secondary antibody (1:1,000; cat. no. 6120-05; SouthernBiotech,
Birmingham, AL, USA) overnight at 4 °C. The Amersham™
ECL™ Prime Western Blotting Detection Reagent (GE
Healthcare Life Sciences, Shanghai, China) was used to visualize
the blots, according to the manufacturer's protocol, with a 5 min
exposure to SuperRX X-ray film (Fujifilm Investment Co., Ltd.,
Shanghai, China). Protein band density was quantified used ImageJ
(version 2.1.4.7, National Institutes of Health, Bethesda, MD,
USA). Anti-GAPDH (1:1,000; cat. no. ab9485; Abcam, Cambridge, UK)
antibody, incubated at 4°C for 1 h, was used as a loading
control.
Statistical analysis
Statistical analysis was performed using SPSS
version 22.0 (IBM SPSS, Armonk, NY, USA). Results were presented as
the mean ± standard deviation of values from each group. The
χ2 test and Fisher's exact test were used to compare
gene methylation frequencies between the ALL group and the ITP
control group. One-way analysis of variance with the Bonferroni
correction was performed to determine the variation in relative
mRNA expression levels between methylated ALL, unmethylated ALL and
ITP groups. Pearson correlation analysis was applied to determine
the correlation between DNA methylation and EPHB4 mRNA
expression levels. Kaplan-Meier analysis was performed to analyze
the association between the gene methylation status and DFS in
patients with ALL, and the log-rank test was used to examine the
variation between the methylated and unmethylated ALL groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
EPHB4 methylation status in patients
with ALL, compared with the control ITP group
BSP was used to detect the methylation profile of
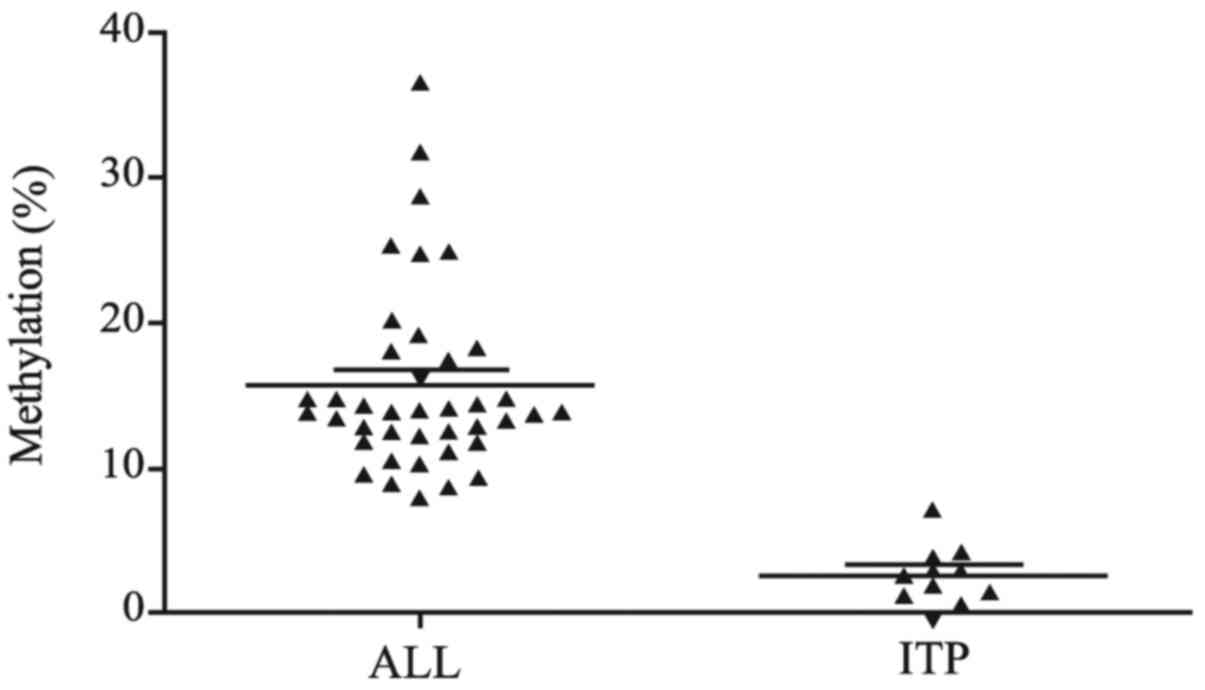
EPHB4 in patients with ALL and ITP. The methylation density
(proportion of methylated CpG sites within a specific promoter
region) of the EPHB4 gene in the ALL bone marrow samples
ranged from 7.94 to 36.50%, and the average methylation density was
15.05%. A 15% methylation density was used as the cut-off value to
classify a bone marrow sample as methylated (16). The number of methylated cases in the
ALL group was 12 (30%), whereas the EPHB4 gene was determine
to be unmethylated in all the ITP bone marrow samples, in which the
methylation density ranged from 0–7.14% and the average methylation
density was 3.47%. The frequency of EPHB4 gene methylation
was significantly higher in the ALL samples compared with the ITP
samples (P=0.046). The status of methylation density and the
bisulfite sequencing results for EPHB4 are presented in
Figs. 1 and 2. The sequence data for the EPHB4
gene have been submitted to the GenBank® database under
the accession number NM_004444.
The expression levels of the EPHB4
gene in the patients with ALL and the control ITP group
In order to investigate the association between
EPHB4 DNA methylation and gene expression, mRNA levels were
detected using RT-qPCR in the ALL and the control ITP bone marrow
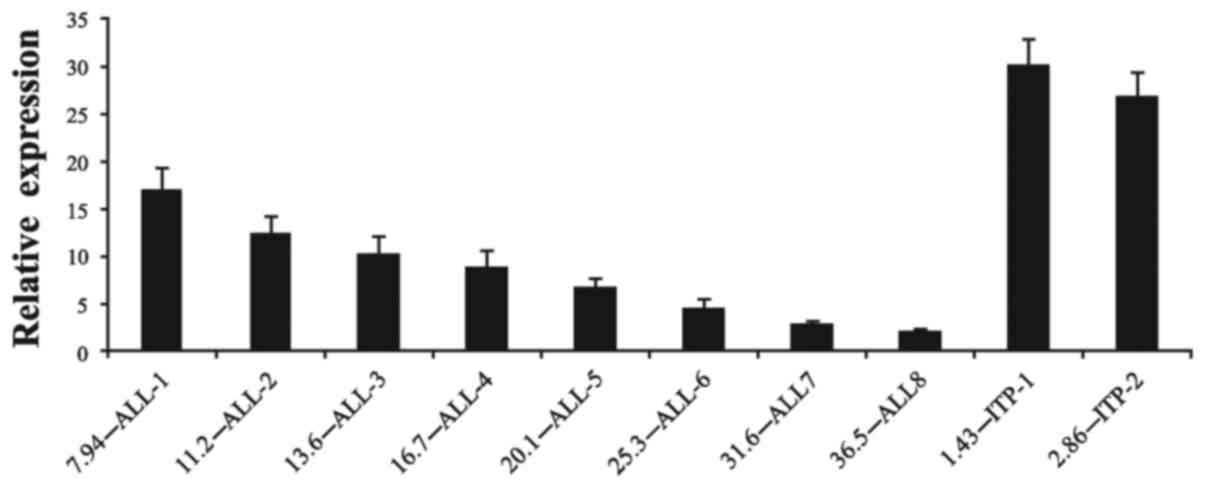
samples. The relative EPHB4 mRNA expression levels in the
ITP patients and the patients with unmethylated ALL (25.08±4.03 and
12.33±2.16, respectively) were significantly higher (P<0.01),
compared with those observed in the patients with methylated ALL
(6.48±2.73). The mRNA expression levels in the ALL samples were
markedly lower (P<0.01), as compared with the ITP samples
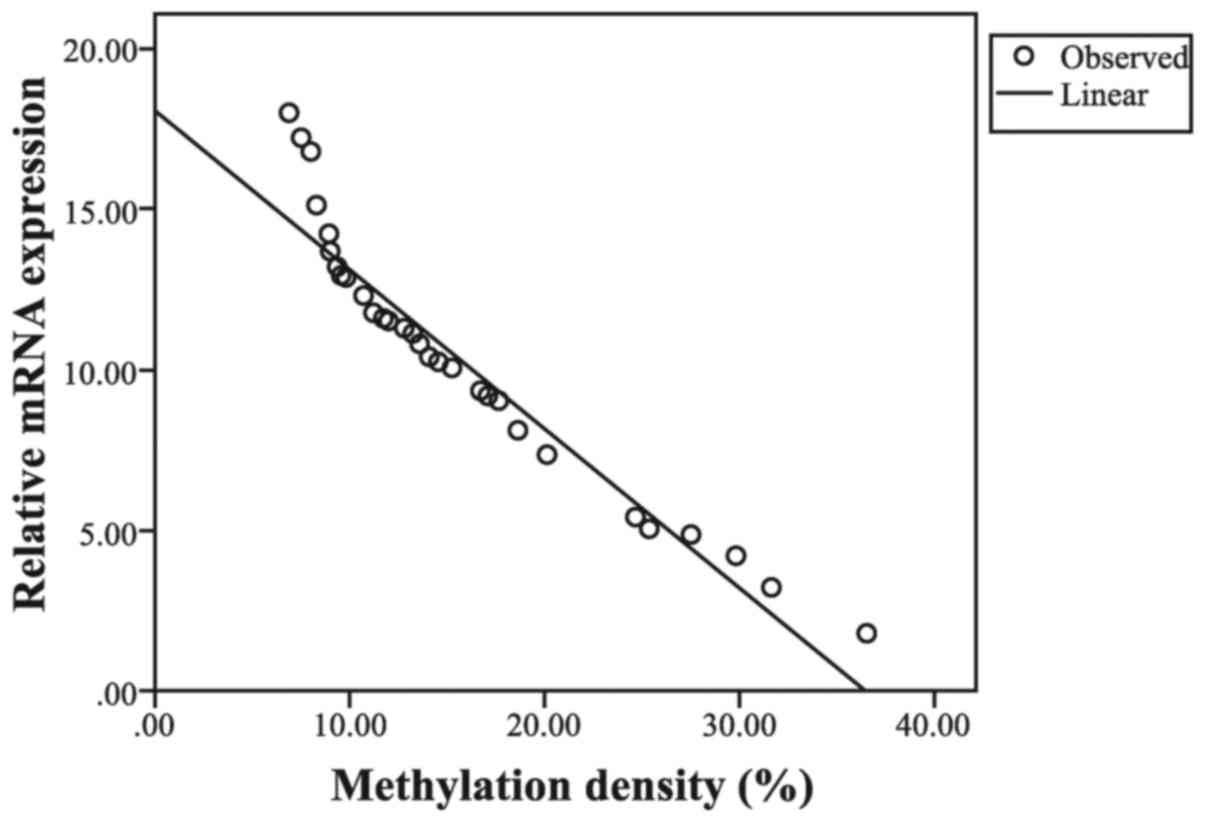
(Fig. 3). Pearson correlation
analysis revealed a negative linear correlation between
EPHB4 gene methylation and its expression (r=−0.957;
P<0.001; Fig. 4). Additionally,
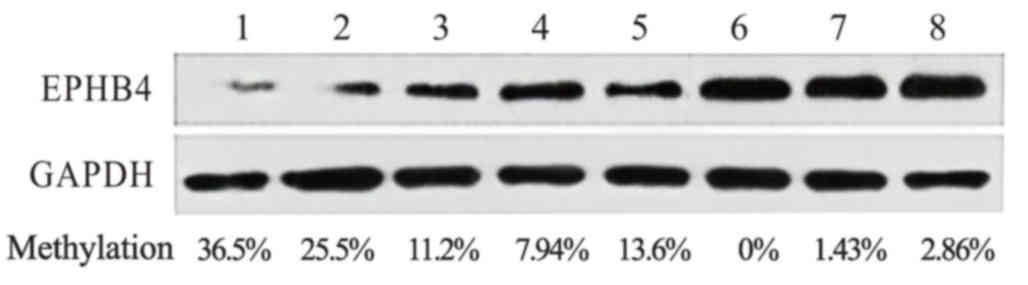
western blot analysis was performed to examine the protein
expression levels of the EPHB4 gene, demonstrating that its
protein production was negatively associated with the methylation
level of the gene (Fig. 5).
Collectively, these results suggested that the DNA methylation of
the EPHB4 gene is associated with the suppression of its
expression.
The impact of EPHB4 gene methylation
on the prognosis of patients with ALL
To evaluate the clinical impact of EPHB4
methylation on patients with ALL, the present study analyzed the
association between EphB4 methylation and the following variables:
Age, sex, white blood cell count, French-American-British
classification, immunophenotype and Berlin-Frankfurt-Münster risk
classification. No significant correlation was observed between
EPHB4 gene methylation and any of these variables (Table III). Kaplan-Meier analysis was
performed to investigate the impact of gene methylation status on
the overall survival of patients with ALL. These patients were
divided into methylated and unmethylated groups by using a
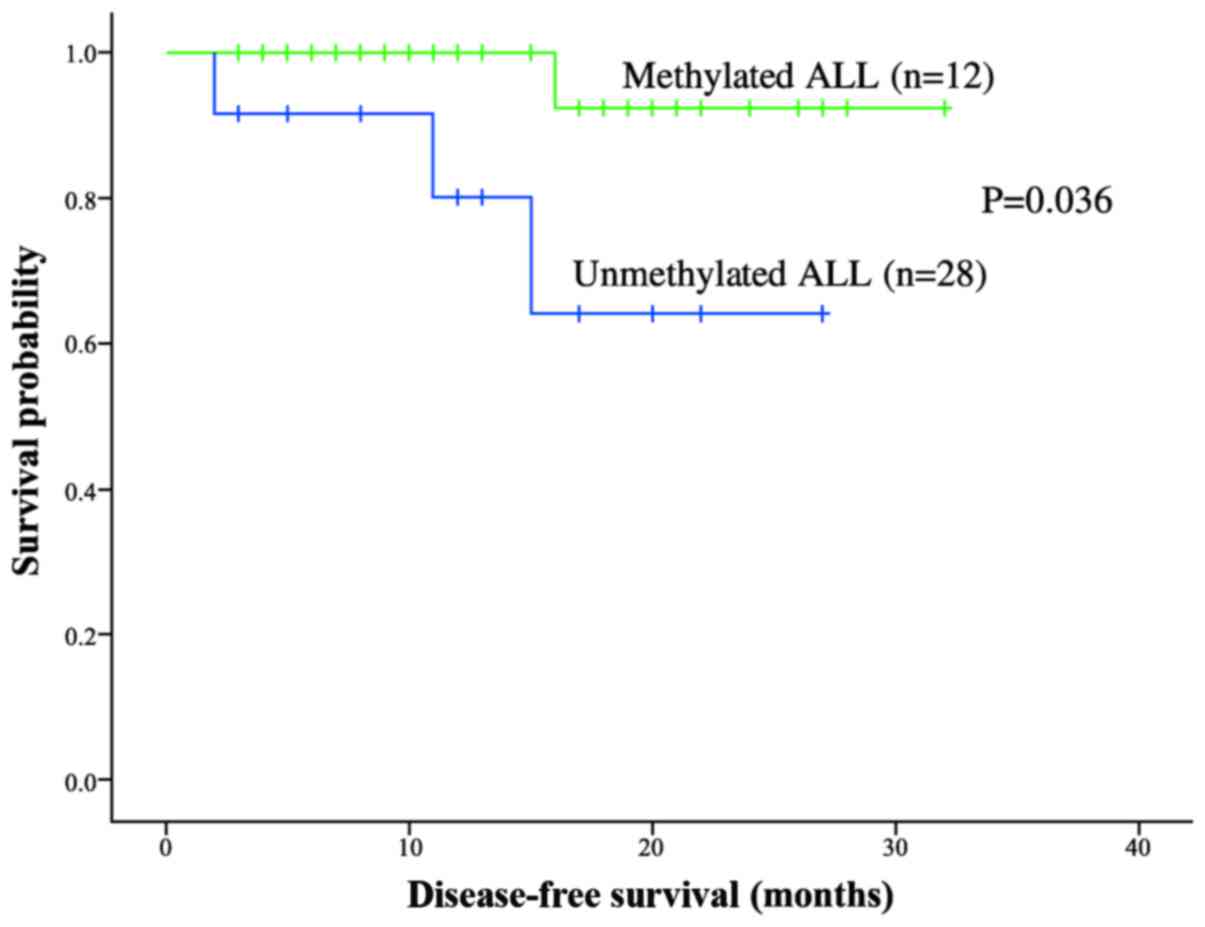
methylation density of 15% as the cut-off value. The two-year DFS
rate of the patients with unmethylated ALL (92.3±7.4%) was
significantly higher, compared with that of the methylated patients
with ALL (68.4±1.58%; P=0.036; Fig.
6). These results suggest a trend toward a poorer prognosis in
patients with EPHB4 gene methylation. However, an increased
observation time and a larger sample size are required in order to
evaluate the predictive role of EPHB4 methylation in ALL
prognosis.
 | Table III.Correlations between EPHB4
methylation status and clinical variables. |
Table III.
Correlations between EPHB4
methylation status and clinical variables.
|
| EPHB4
methylation status |
|
|
|---|
|
|
|
|
|
|---|
| Clinical
variables | + | − | χ2 | P-value |
|---|
| Age, years |
|
|
| 0.833 |
|
<6 | 8 | 16 | 0.045 |
|
|
>6 | 4 | 12 |
|
|
| Sex, n |
|
|
| 0.267 |
|
Male | 7 | 11 | 1.231 |
|
|
Female | 5 | 17 |
|
|
| WBC count |
|
|
| 0.499 |
|
<50×109/l | 9 | 25 | 0.458 |
|
|
≥50×109/l | 3 | 3 |
|
|
| FAB classification,
n |
|
|
| 0.348 |
| L1 | 7 | 18 | 2.111 |
|
| L2 | 3 | 9 |
|
|
| L3 | 2 | 1 |
|
|
| Immunophenotype,
n |
|
|
| 0.204 |
|
T-ALL | 4 | 3 | 1.616 |
|
B-ALL | 8 | 25 |
|
|
| BFM risk, n |
|
|
| 0.956 |
| SR | 4 | 8 | 0.091 |
| IR | 6 | 15 |
|
|
| HR | 2 | 5 |
|
|
Discussion
DNA hypermethylation in promoter CGIs has been
revealed as a predominant mechanism, by which tumor suppressors are
inactivated in various types of cancer (8). The potential mechanisms underlying the
loss of EPHB4 expression include promoter hypermethylation,
chromosomal abnormalities and transcriptional repression (30). Gene inactivation caused by promoter
hypermethylation in the Eph family has been implicated in the
pathogenesis and progression of various tumors (30). In the present study, the methylation
status of the EPHB4 gene was detected in bone marrow samples
from 40 newly diagnosed patients with ALL, and 10 patients with
ITP. Using BSP, it was demonstrated that the EPHB4 gene was
methylated significantly more frequently in the ALL samples,
compared with the ITP controls (P=0.046). The methylation density
in the ALL bone marrow samples ranged from 7.94 to 36.50%, the
average methylation density was 15.05% and methylation was present
in 30% of the samples. By contrast, the EPHB4 gene was
classified as unmethylated in all the ITP controls, with the
methylation density ranging from 0–7.14%. These data suggest that
EPHB4 gene methylation is prevalent in patients with ALL,
concordant with a previous study (17). The present study provided additional
evidence that EPHB4 gene methylation may be important in the
development of childhood ALL; however, the methylation level in the
present study was slightly lower compared with that detected by
Kuang et al (17). This may be
due to variation in patient ethnic background or in the CpG sites
examined. A previous study reported that the expression levels of
EPHB4 varied significantly between Caucasian and
African-American patients (31).
Therefore, a larger scale study, including additional geographic
regions and performed under homogenous conditions, is required to
demonstrate that the hypermethylation of EPHB4 gene is a
frequent event during the pathogenesis of childhood ALL.
The EPHB4 expression profile was subsequently
evaluated using RT-qPCR. The relative mRNA expression levels of
EPHB4 in patients with ITP and ALL without methylation were
significantly higher, compared with those detected in the patients
with methylated ALL (P<0.01). Pearson analysis revealed a
significant negative linear correlation between EPHB4 gene
methylation and its expression levels (r=−0.957; P<0.01;
Fig. 3). As presented in Fig. 4, there were decreased EPHB4
protein expression levels, as examined by western blot analysis, in
the methylated ALL samples, with higher levels of expression in the
ITP samples. A previous study demonstrated that the EPHB4
gene in the CEM T-cell line was able to be restored by the
methyltransferase inhibitor 5-aza-2′-deoxycytidine, resulting in
increased mRNA expression levels and protein production (23). Furthermore, decreased cell
proliferation and increased cell apoptosis were observed upon the
reintroduction of EPHB4 gene expression (21). Taken together, these results support
the hypothesis that promoter hypermethylation and epigenetic gene
silencing may be an important molecular mechanism in the
progression of ALL, and that the EPHB4 gene may be
considered as a candidate tumor suppressor. Regarding the overall
mechanisms underlying Eph family gene inactivation, the role of
transcriptional regulation appears to be more complex, and the
variation in the EPHB4 gene caused by chromosomal
abnormalities has yet to be elucidated (30). Previous studies have reported that
EPHB2 is suppressed by REL but induced by Wnt,
β-catenin and T-cell factor family transcription factors in
colorectal cancer (32,33). However, the effects of β-catenin on
EPHB2 and EPHB4 gene expression vary according to the
stage of colorectal cancer progression (32,33). In
the present study, only the mechanism of methylation-associated
gene silencing was investigated. However, other mechanisms,
including mutations and chromosomal changes in mRNA stability, also
regulate Eph and ephrin expression in cancer, which may affect the
process of EPHB4 gene inactivation, leading to diverse gene
methylation levels between individuals (30).
A previous study, based on >9 years of follow-up
data, revealed that EPHB4 is a valuable prognostic marker in
colorectal cancer as low EPHB4 expression levels are
significantly associated with a shorter survival time (34). Kuang et al (17) investigated the association between the
methylation of Eph receptors and overall survival in patients with
ALL, observing that patients with methylation of numerous Eph genes
exhibited a shorter median survival time. However, to the best of
our knowledge, the impact of EPHB4 gene expression on the
prognosis of childhood ALL has yet to be investigated. The present
study revealed, using Kaplan-Meier survival analysis, that patients
with a methylated EPHB4 gene exhibited a shorter
disease-free survival time, as compared with patients with an
unmethylated EPHB4 gene (P=0.036), establishing a
correlation between the methylation status of the EPHB4 gene
and an unfavorable prognosis. This suggests that EPHB4
methylation is a potential prognostic factor for childhood ALL.
However, longer-term observation is required for a thorough
elucidation of the role of EPHB4 in the prognosis of
childhood ALL.
The current understanding of the mechanisms
underlying the tumor suppressor effect of the EPHB4 receptor
is primarily based on its forward signaling pathway, which is
involved in tumor pathogenesis and progression (22,35).
Previous studies have indicated that the EPHB4 gene exhibits
a bidirectional role in the regulation of tumorigenesis in
epithelial cancer, depending on the presence or absence of the
ephrin-B2 ligand (29,30). The binding of ephrin-B2 to the
EPHB4 receptor leads to the modulation of certain signaling
pathways underlying tumor suppression, including the Harvey rat
sarcoma-extracellular-signal related kinase, phosphoinositide
3-kinase-protein kinase B (Akt) and Abl-Crk signaling pathways,
inhibiting tumor cell proliferation, migration and invasion
(30,36,37).
Previous studies have indicated that this bidirectional signaling
mechanism may serve a particular role in inducing tumor dormancy,
which consequently reduces tumor progression (30,38,39).
According to research by Kuang et al (17), the modulation of the Akt signaling
pathway is attributed to EPHB4-mediated cell growth
inhibition in ALL cell lines following stimulation by ephrin-B2.
However, this result does not incorporate data from bone marrow
samples from patients with ALL. The function of the downstream
signaling pathway of EPHB4 during leukemogenesis, and the
interactions between EPHB4 and its ligand, have not yet been
fully elucidated. Further clinical studies in addition to long-term
survival observations, are required in order to provide a clear
profile of the involvement of the EPHB4 gene in ALL
pathogenesis. The established knowledge of the suppressive effect
of the EPHB4 gene on oncogenesis, and the potential positive
correlation between EPHB4 gene expression levels and a
favorable ALL prognosis (19,29,40),
indicates that the EPHB4 gene may be a candidate tumor
suppressor and prognostic biomarker in childhood ALL. Due to the
reversible status of gene methylation, the present study has
revealed the promising value of EPHB4 as a novel target for
demethylation treatment.
In conclusion, the present study demonstrated that
EPHB4 gene methylation is a frequent event in childhood ALL.
The methylation-associated inactivation of the EPHB4 gene
leads to transcriptional silencing, resulting in reduced or absent
gene expression levels. Further survival analysis performed in
patients with ALL revealed a trend of positive association between
EPHB4 gene inactivation and poor prognosis. Therefore, it is
hypothesized that the EPHB4 gene may function as a tumor
suppressor in ALL pathogenesis, and may be utilized as a novel
prognostic biomarker during ALL therapy. However, larger scale
investigations are required in order to clarify the function of
Eph/ephrins in the development of ALL and hematological
malignancies.
Acknowledgements
This work was supported by a Grant-in-Aid for
Medical Research from The Technology Innovation Project (grant no.
CXZZ20130320172336579) launched by The Science Technology and
Innovation Committee of Shenzhen Municipality.
References
|
1
|
Wu MY and Hu YM: LeukemiaZhu futang
practice of pediatrics. Hu YM and Jiang ZF: People's medical
publishing house Co., Ltd.; Beijing: pp. 2351. 2015
|
|
2
|
Pui CH, Robison LL and Look AT: Acute
lymphoblastic leukaemia. Lancet. 371:1030–1043. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pui CH, Carroll WL, Meshinchi S and Arceci
RJ: Biology, risk stratification, and therapy of pediatric acute
leukemias: An update. J Clin Oncol. 29:551–565. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garcia-Manero G, Yang H, Kuang SQ, O'Brien
S, Thomas D and Kantarjian H: Epigenetics of acute lymphocytic
leukemia. Semin Hematol. 46:24–32. 2009. View Article : Google Scholar
|
|
5
|
Issa JP, Baylin SB and Herman JG: DNA
methylation changes in hematologic malignancies: Biologic and
clinical implications. Leukemia. 11 (Suppl 1):S7–S11. 1997.
|
|
6
|
Kuang SQ, Tong WG, Yang H, Lin W, Lee MK,
Fang ZH, Wei Y, Jelinek J, Issa JP and Garcia-Manero G: Genome-wide
identification of aberrantly methylated promoter associated CpG
islands in acute lymphocytic leukemia. Leukemia. 22:1529–1538.
2008. View Article : Google Scholar
|
|
7
|
Roman-Gomez J, Jimenez-Velasco A,
Castillejo JA, Agirre X, Barrios M, Navarro G, Molina FJ, Calasanz
MJ, Prosper F, Heiniger A and Torres A: Promoter hypermethylation
of cancer-related genes: A strong independent prognostic factor in
acute lymphoblastic leukemia. Blood. 104:2492–2498. 2004.
View Article : Google Scholar
|
|
8
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar
|
|
9
|
Kuang SQ, Ling X, Sanchez-Gonzalez B, Yang
H, Andreeff M and Garcia-Manero G: Differential tumor suppressor
properties and transforming growth factor-beta responsiveness of
p57KIP2 in leukemia cells with aberrant p57KIP2 promoter DNA
methylation. Oncogene. 26:1439–1448. 2007. View Article : Google Scholar
|
|
10
|
Takeuchi S, Matsushita M, Zimmermann M,
Ikezoe T, Komatsu N, Seriu T, Schrappe M, Bartram CR and Koeffler
HP: Clinical significance of aberrant DNA methylation in childhood
acute lymphoblastic leukemia. Leuk Res. 35:1345–1349. 2011.
View Article : Google Scholar
|
|
11
|
Hafner C, Schmitz G, Meyer S, Bataille F,
Hau P, Langmann T, Dietmaier W, Landthaler M and Vogt T:
Differential gene expression of Eph receptors and ephrins in benign
human tissues and cancers. Clin Chem. 50:490–499. 2004. View Article : Google Scholar
|
|
12
|
Himanen JP and Nikolov DB: Eph receptors
and ephrins. Int J Biochem Cell Biol. 35:130–134. 2003. View Article : Google Scholar
|
|
13
|
Batlle E, Bacani J, Begthel H, Jonkheer S,
Gregorieff A, van de Born M, Malats N, Sancho E, Boon E, Pawson T,
et al: EphB receptor activity suppresses colorectal cancer
progression. Nature. 435:1126–1130. 2005. View Article : Google Scholar
|
|
14
|
Vaught D, Brantley-Sieders DM and Chen J:
Eph receptors in breast cancer: Roles in tumor promotion and tumor
suppression. Breast Cancer Res. 10:2172008. View Article : Google Scholar
|
|
15
|
Nosho K, Yamamoto H, Takahashi T, Mikami
M, Taniguchi H, Miyamoto N, Adachi Y, Arimura Y, Itoh F, Imai K and
Shinomura Y: Genetic and epigenetic profiling in early colorectal
tumors and prediction of invasive potential in pT1 (early invasive)
colorectal cancers. Carcinogenesis. 28:1364–1370. 2007. View Article : Google Scholar
|
|
16
|
Wang J, Kataoka H, Suzuki M, Sato N,
Nakamura R, Tao H, Maruyama K, Isogaki J, Kanaoka S, Ihara M, et
al: Downregulation of EphA7 by hypermethylation in colorectal
cancer. Oncogene. 24:5637–5647. 2005. View Article : Google Scholar
|
|
17
|
Kuang SQ, Bai H, Fang ZH, Lopez G, Yang H,
Tong W, Wang ZZ and Garcia-Manero G: Aberrant DNA methylation and
epigenetic inactivation of Eph receptor tyrosine kinases and ephrin
ligands in acute lymphoblastic leukemia. Blood. 115:2412–2419.
2010. View Article : Google Scholar
|
|
18
|
Jin W, Luo H and Wu J: Effect of reduced
EPHB4 expression in thymic epithelial cells on thymocyte
development and peripheral T cell function. Mol Immunol. 58:1–9.
2014. View Article : Google Scholar
|
|
19
|
Nguyen TM, Arthur A, Hayball JD and
Gronthos S: EphB and Ephrin-B interactions mediate human
mesenchymal stem cell suppression of activated T-cells. Stem Cells
Dev. 22:2751–2764. 2013. View Article : Google Scholar
|
|
20
|
Noyan F, Lieke T, Taubert R, Sievers M,
Dywicki J, Hapke M, Falk CS, Manns MP, Jaeckel E and
Hardtke-Wolenski M: Naive tumour-specific CD4+ T cells were
efficiently primed in acute lymphoblastic leukaemia. Scand J
Immunol. 80:161–168. 2014. View Article : Google Scholar
|
|
21
|
Hegazy AN and Klein C: Ex vivo priming of
CD4 T cells converts immunological tolerance into effective
antitumor immunity in a murine model of acute lymphoblastic
leukemia. Leukemia. 22:2070–2079. 2008. View Article : Google Scholar
|
|
22
|
Takahashi Y, Itoh M, Nara N and Tohda S:
Effect of EPH-ephrin signaling on the growth of human leukemia
cells. Anticancer Res. 34:2913–2918. 2014.
|
|
23
|
Li YH, Wen FQ, Chen YX, Li CG, Zhang ZX,
Chen XW and Li B: Effect of methylation inhibitor on EphB4 gene
expression, proliferation and apoptosis in CEM cells. Zhongguo Dang
Dai Er Ke Za Zhi. 14:205–209. 2012.(In Chinese).
|
|
24
|
Stary J, Zimmermann M, Campbell M,
Castillo L, Dibar E, Donska S, Gonzalez A, Izraeli S, Janic D,
Jazbec J, et al: Intensive chemotherapy for childhood acute
lymphoblastic leukemia: Results of the randomized intercontinental
trial ALL IC-BFM 2002. J Clin Oncol. 32:174–184. 2014. View Article : Google Scholar
|
|
25
|
Mai H, Liu X, Chen Y, Li C, Cao L, Chen X,
Chen S, Liu G and Wen F: Hypermethylation of p15 gene associated
with an inferior poor long-term outcome in childhood acute
lymphoblastic leukemia. J Cancer Res Clin Oncol. 142:497–504. 2016.
View Article : Google Scholar
|
|
26
|
Shen L, Toyota M, Kondo Y, Obata T, Daniel
S, Pierce S, Imai K, Kantarjian HM, Issa JP and Garcia-Manero G:
Aberrant DNA methylation of p57KIP2 identifies a cell-cycle
regulatory pathway with prognostic impact in adult acute
lymphocytic leukemia. Blood. 101:4131–4161. 2003. View Article : Google Scholar
|
|
27
|
Adusumalli S, Omar Mohd MF, Soong R and
Benoukraf T: Methodological aspects of whole-genome bisulfite
sequencing analysis. Brief Bioinform. 16:369–379. 2015. View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real- time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
29
|
Rutkowski R, Mertens-Walker I, Lisle JE,
Herington AC and Stephenson SA: Evidence for a dual function of
EphB4 as tumor promoter and suppressor regulated by the absence or
presence of the ephrin-B2 ligand. Int J Cancer. 131:E614–E624.
2012. View Article : Google Scholar
|
|
30
|
Pasquale EB: Eph receptors and ephrins in
cancer: Bidirectional signalling and beyond. Nat Rev Cancer.
10:165–180. 2010. View Article : Google Scholar
|
|
31
|
Ferguson BD, Liu R, Rolle CE, Tan YH,
Krasnoperov V, Kanteti R, Tretiakova MS, Cervantes GM, Hasina R,
Hseu RD, et al: The EphB4 receptor tyrosine kinase promotes lung
cancer growth: A potential novel therapeutic target. PLoS One.
8:e676682013. View Article : Google Scholar
|
|
32
|
Fu T, Li P, Wang H, He Y, Luo D, Zhang A,
Tong W, Zhang L, Liu B and Hu C: c-Rel is a transcriptional
repressor of EPHB2 in colorectal cancer. J Pathol. 219:103–113.
2009. View Article : Google Scholar
|
|
33
|
Kumar SR, Scehnet JS, Ley EJ, Singh J,
Krasnoperov V, Liu R, Manchanda PK, Ladner RD, Hawes D, Weaver FA,
et al: Preferential induction of EphB4 over EphB2 and its
implication in colorectal cancer progression. Cancer Res.
69:3736–3745. 2009. View Article : Google Scholar
|
|
34
|
Davalos V, Dopeso H, Castaño J, Wilson AJ,
Vilardell F, Romero-Gimenez J, Espin E, Armengol M, Capella G,
Mariadason JM, et al: EPHB4 and survival of colorectal cancer
patients. Cancer Res. 66:8943–8948. 2006. View Article : Google Scholar
|
|
35
|
Kullander K and Klein R: Mechanisms and
functions of Eph and ephrin signalling. Nat Rev Mol Cell Biol.
3:475–486. 2002. View
Article : Google Scholar
|
|
36
|
Xiao Z, Carrasco R, Kinneer K, Sabol D,
Jallal B, Coats S and Tice DA: EphB4 promotes or suppresses
Ras/MEK/ERK pathway in a context-dependent manner: Implications for
EphB4 as a cancer target. Cancer Biol Ther. 13:630–637. 2012.
View Article : Google Scholar
|
|
37
|
Noren NK, Foos G, Hauser CA and Pasquale
EB: The EphB4 receptor suppresses breast cancer cell tumorigenicity
through an Abl-Crk pathway. Nat Cell Biol. 8:815–825. 2006.
View Article : Google Scholar
|
|
38
|
Almog N, Ma L, Raychowdhury R, Schwager C,
Erber R, Short S, Hlatky L, Vajkoczy P, Huber PE, Folkman J and
Abdollahi A: Transcriptional switch of dormant tumors to
fast-growing angiogenic phenotype. Cancer Res. 69:836–844. 2009.
View Article : Google Scholar
|
|
39
|
Jørgensen C, Sherman A, Chen GI,
Pasculescu A, Poliakov A, Hsiung M, Larsen B, Wilkinson DG, Linding
R and Pawson T: Cell-specific information processing in segregating
populations of Eph receptor ephrin-expressing cells. Science.
326:1502–1509. 2009. View Article : Google Scholar
|
|
40
|
Dopeso H, Mateo-Lozano S, Mazzolini R,
Rodrigues P, Lagares-Tena L, Ceron J, Romero J, Esteves M, Landolfi
S, Hernández-Losa J, et al: The receptor tyrosine kinase EPHB4 has
tumor suppressor activities in intestinal tumorigenesis. Cancer
Res. 69:7430–7438. 2009. View Article : Google Scholar
|