Introduction
Lung cancer is one of the most serious cancers
without effective treatment (1). The
incidence rate of lung cancer is the highest among all types of
cancers worldwide. Lung cancer also causes unacceptable high
mortality rate, and accounts for ~25% of the deaths caused by
cancer (2). With the increased number
of smokers and intensified environmental pollution, especially in
industrialized cities, the incidence of lung cancer is rapidly
increasing (3). At present, gene
detection and targeted therapy have attracted more and more
attention. Some targeted drugs have been proved to be able to
prolong the life of patients with lung cancer (4). In recent years, STK33 has been shown to
play an important role in the development of a variety of cancers
and to participate in the regulation of DNA replication, signal
transduction, cell proliferation, cell differentiation, apoptosis
and tumor development. Due to the correlate STK33 and the
well-known oncogene Ras, STK33 has become an active research area
(5). However, the studies on the role
of STK33 in the development of lung cancer are relatively rare, and
the role of STK33 in carcinogenesis and development of lung cancer
is still not clear (6,7). This study investigated the correlation
between STK33 gene expression and pathology and prognosis of lung
cancer to provide reference for clinical diagnosis and
treatment.
Materials and methods
Research subjects
In total 102 lung cancer patients diagnosed by
pathological examinations were randomly selected in Shanghai Jiao
Tong University Affiliated Sixth People's Hospital from February,
2012 to February, 2017. Tumor tissues were collected to serve as
observation group. The patients included 63 males and 39 females,
the age ranged from 33 to 77 years with and average age of 54.3.
There were 42 cases of adenocarcinoma, 29 cases of squamous cell
carcinoma, 16 cases of small cell lung cancer and 15 cases of
pulmonary large cell carcinoma. At the same time, 19 patients with
lung benign lesions were selected and lung tissues were also
collected to serve as control group. Control group contained 8
cases of pneumonia, 5 cases of pulmonary tuberculosis, 4 cases of
benign tumors and 2 cases of lymphoid tissue atypical hyperplasia.
Exclusion criteria: i) Patients with major cardiovascular and
cerebrovascular diseases, and digestive disease; ii) Patients with
mental disorders, or unable to communicate with researchers in a
normal way; iii) Pregnant women; iii) Patients with incomplete
clinical data. All patients were followed up by telephone to
collect data for the analysis of 5-year survival rate. No
significant differences in gender, age and other basic information
were found between the two groups (p>0.05). The study was
approved by the Ethics Committee of Shanghai Jiao Tong University
Affiliated Sixth People's Hospital. All patients or their family
members signed written informed consent.
Reagents
TRIzol (Life Technologies, New York, NY, USA);
chloroform and isopropyl alcohol (Beijing Chemical Co., Ltd.,
Beijing, China); M-MLV reverse transcriptase; DNase I (both from
Life Technologies); SYBR® Premix Ex Taq™ II (Takara Bio
Inc., Liaoning, China); DNA Marker (TransGen Biotech, Beijing,
China); primer synthesis (Beijing Genomics Institute, Guangdong,
China); RIPA protein lysate (Solarbio, Beijing, China); actin and
STK33 rabbit anti-human primary antibodies (Cell Signaling
Technology, Boston, MA, USA); BCA kit (Life Technologies); skim
milk powder (BD Biosciences, New Jersey, NY, USA); horseradish
peroxidase labeled goat anti-rabbit IgG secondary antibody (Cell
Signaling Technology); NC membrane (Millipore, Billerica, MA, USA);
luminescent substrate kit (TransGen Biotech); SP kit (Cell
Signaling Technology).
Real-time reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Lung cancer tissues and benign lesion tissues were
collected by fiberoptic bronchoscopy and stored in liquid nitrogen
before use. Tissues were ground in liquid nitrogen and TRIzοl was
used to extract total RNA according the instructions of the kit.
The concentration of RNA samples was measured and 1 µg RNA and
reverse transcriptase kit were used for reverse transcription to
obtain cDNA. SYBR® Premix Ex Taq™ II and cDNA were used
to prepare PCR reaction system and PCR reaction was performed on
Bio-Rad CFX96 qPCR Instrument to detect the expression level of
STK33 in each sample. Reaction conditions are listed in Table I and primers in Table II.
 | Table I.PCR reaction conditions. |
Table I.
PCR reaction conditions.
| Steps | Temperature | Time | Circle |
|---|
| 1 | 94°C | 15 min | 1 |
| 2 | 94°C | 10 sec | 40 |
|
| 50°C | 30 sec |
|
|
| 72°C | 15 sec |
|
|
| Fluorescence
recording |
|
| 3 | 72°C | 10 min | 1 |
 | Table II.Primers for β-actin and STK33. |
Table II.
Primers for β-actin and STK33.
| Genes | Primer sequences |
|---|
| β-actin | 5′-3′
GTGGACATCCGCAAAGAC |
|
| 3′-5′
GAAAGGGTGTAACGCAACTA |
| STK33 | 5′-3′
GGGAGCCAGATAAACG |
|
| 3′-5′
GCTTCACCCGTTAATT |
Western blot analysis to
quantitatively analyze the expression levels of proteins
Tissues were ground in liquid nitrogen, followed by
lysis with RIPA protein lysate on ice for 30 min. Then the samples
were centrifuged (12,000 × g at 4°C) for 10 min to collect the
supernatant. The supernatant was stored in 1.5 ml EP tube. Protein
concentration was measured using BCA method, and 100 µg protein
from each sample was mixed with loading buffer and denatured in
boiling water for 10 min, followed by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel
electrophoresis under 40 V for 0.5 h. Protein was then transferred
to NC membrane. After blocking with 5% skim milk at 4°C overnight,
membranes were washed 3 times (10 min each time) and incubated with
primary rabbit monoclonal STK33 antibody (dilution, 1:500; cat. no.
ab206296; Abcam, Cambridge, MA, USA) at room temperature for 2 h.
After washing, membranes were incubated withsecondary goat
anti-rabbit (HRP) IgG antibody (dilution, 1:2,000; cat. no. ab6721;
Abcam) at room temperature for 1 h. Fluorescent substrate was then
added for color development in the dark. Imaging results were
quantified using ImageJ software (version X; Media Cybernetics,
Silver Springs, MD, USA).
Immunohistochemistry staining to
analyze the expression of proteins
SP immunohistochemistry staining was performed in
strict accordance with the instructions of the kit. Determination
of positive cells in stained samples: i) Brown particles; ii) color
intensity is higher than the color intensity of background.
Positions of observation: iii) cell cytoplasm, mainly in
macrophages and tumor cells; iv) endothelial cells of interstitial
tubes. Observation method: Five visual fields were randomly
selected (×400). Definition of relative expression: proportion of
positive cells >50%, 3 points; proportion of positive cells
25–49%, 2 points; proportion of positive cells 1–24%, 1 point;
proportion of positive cells <1%, 0 point. Scoring according to
the intensity of staining, dark color was recorded as 3 points,
moderate color as 2 points, and light color as 1 point. The
combination of those two scores was used as the basis for
determining the expression level. Scores ≤3 were recorded as low
expression level, and scores >3 points as high expression level
and positive expression.
Analysis of prognosis
A 5-year follow-up was carried out, and the
recurrence-free survival rate and overall survival rate of patients
with different pathologic types were recorded. Kaplan-Meier
survival curve was also used to show the survival rate of
patients.
Statistical analysis
All data were processed using SPSS 19.0 software
(IBM, Armonk, NY, USA). Measurement data were expressed as mean ±
SD. Comparisons of mean values were performed by t-test and single
factor analysis of variance. Count data were processed using
χ2 test. p<0.05 was considered to be statistically
significant.
Results
Expression of STK33 mRNA in patients
with different pathological types
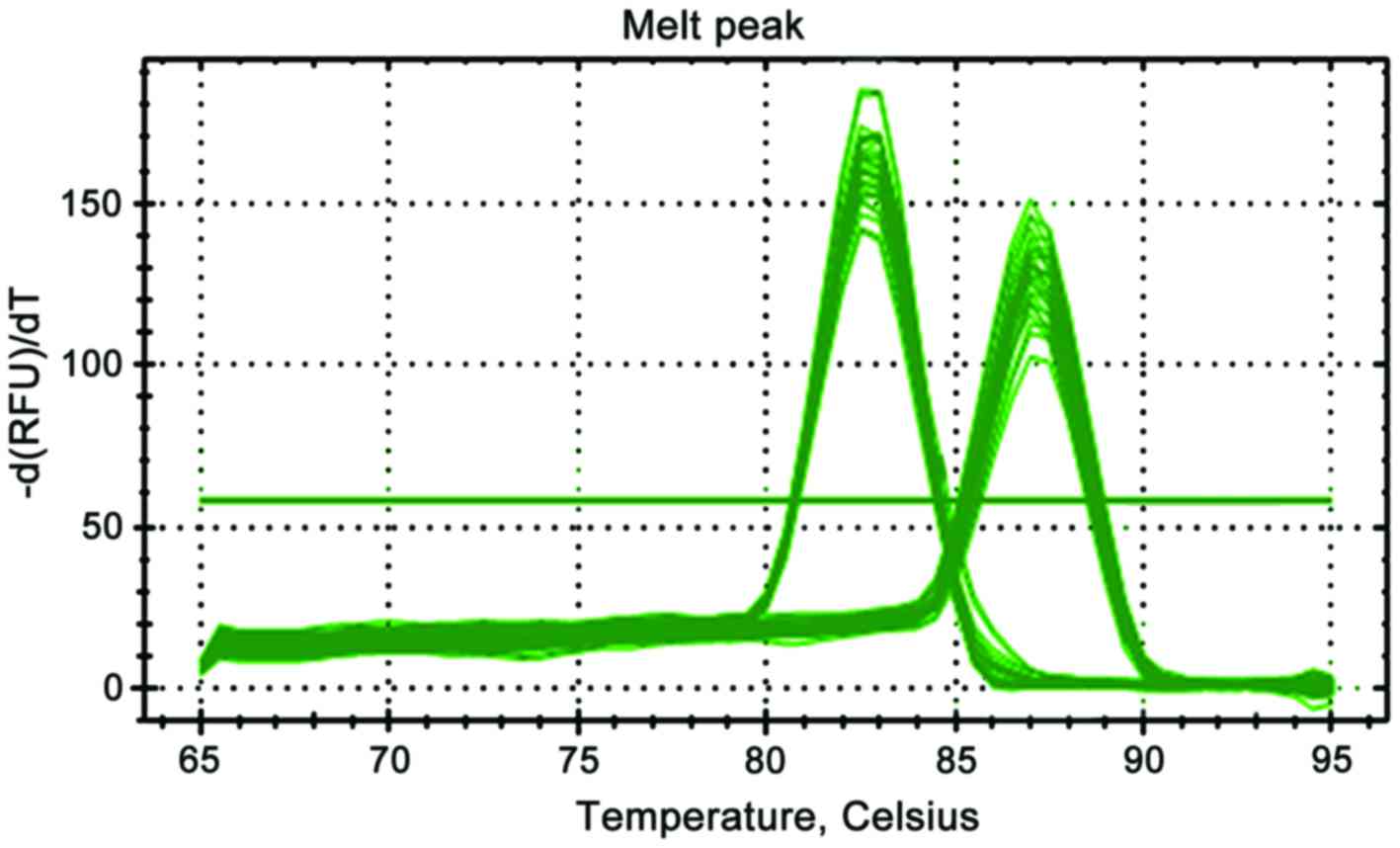
As shown in Fig. 1,
the melting curves of β-actin and STK33 showed single peaks,
indicating the high specificity of the primer and accuracy of
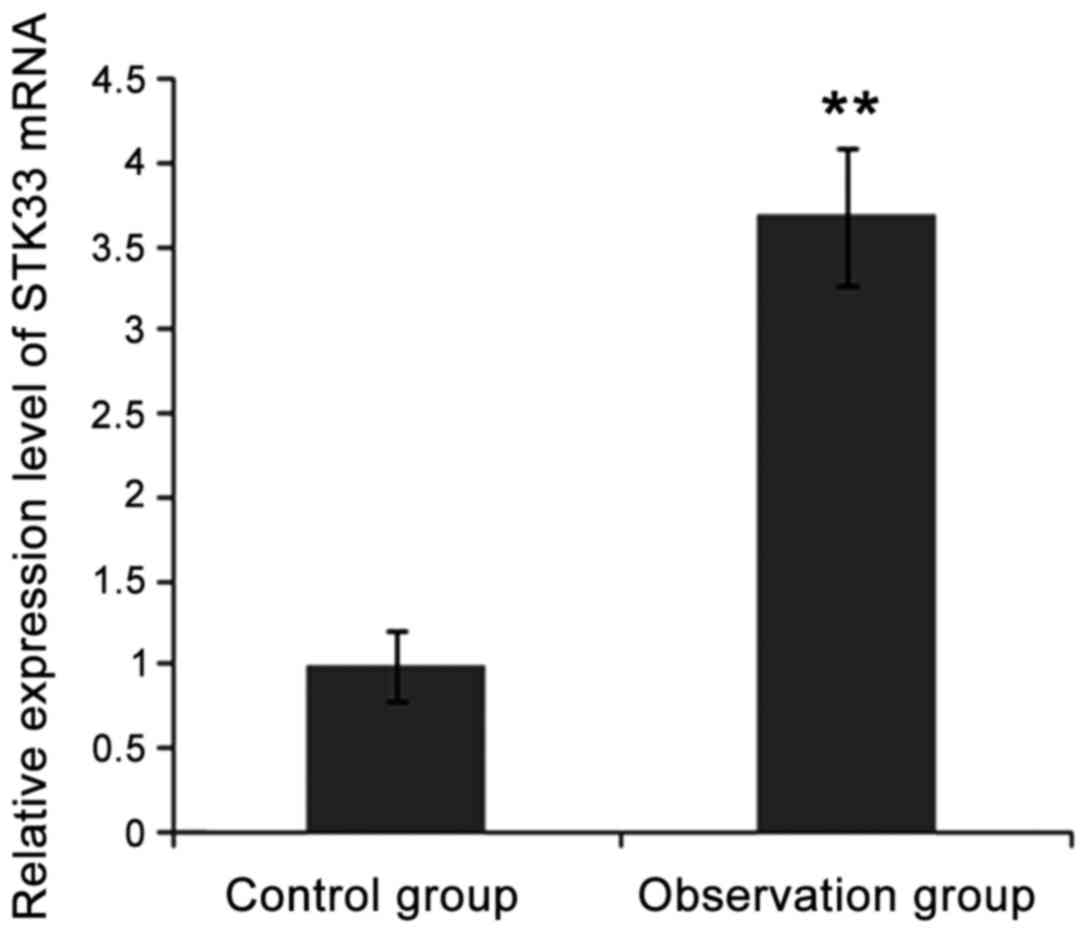
results. As shown in Fig. 2, the
expression level of STK33 mRNA in observation group was
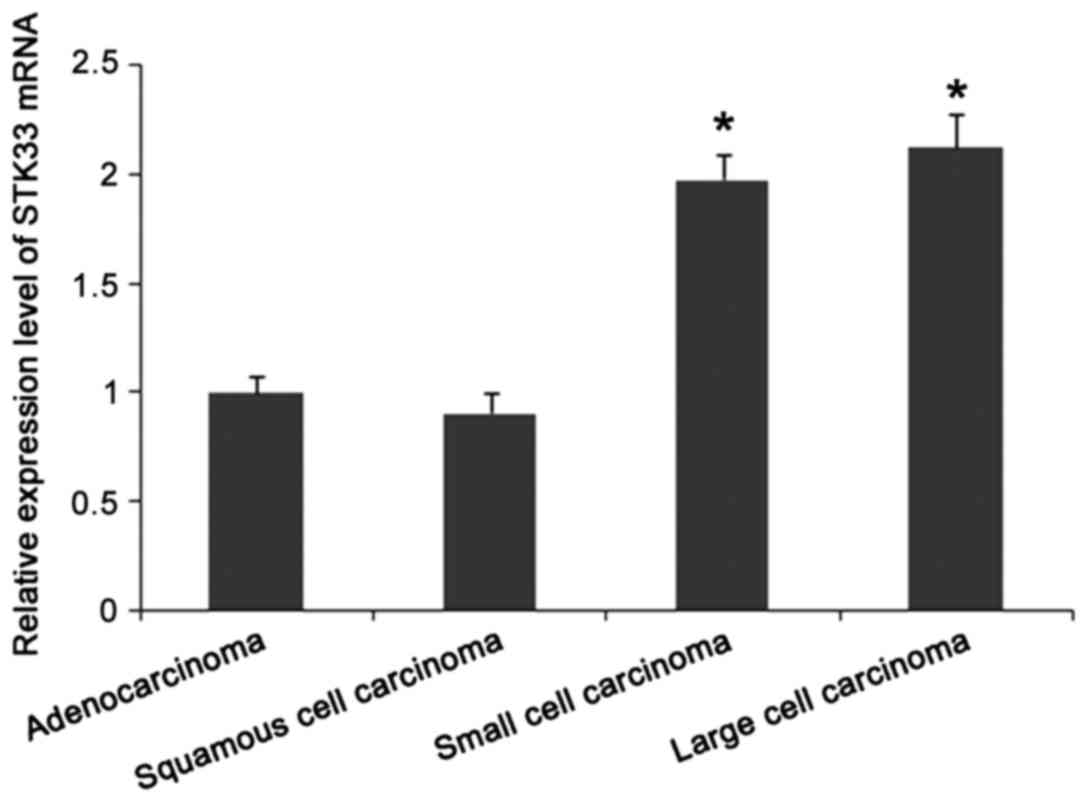
significantly higher than that in control group (p<0.05). As
shown in Fig. 3, expression levels of
STK33 mRNA in lung adenocarcinoma and squamous cell carcinoma were
significantly lower than those in lung small cell carcinoma and
large cell carcinoma (p<0.05).
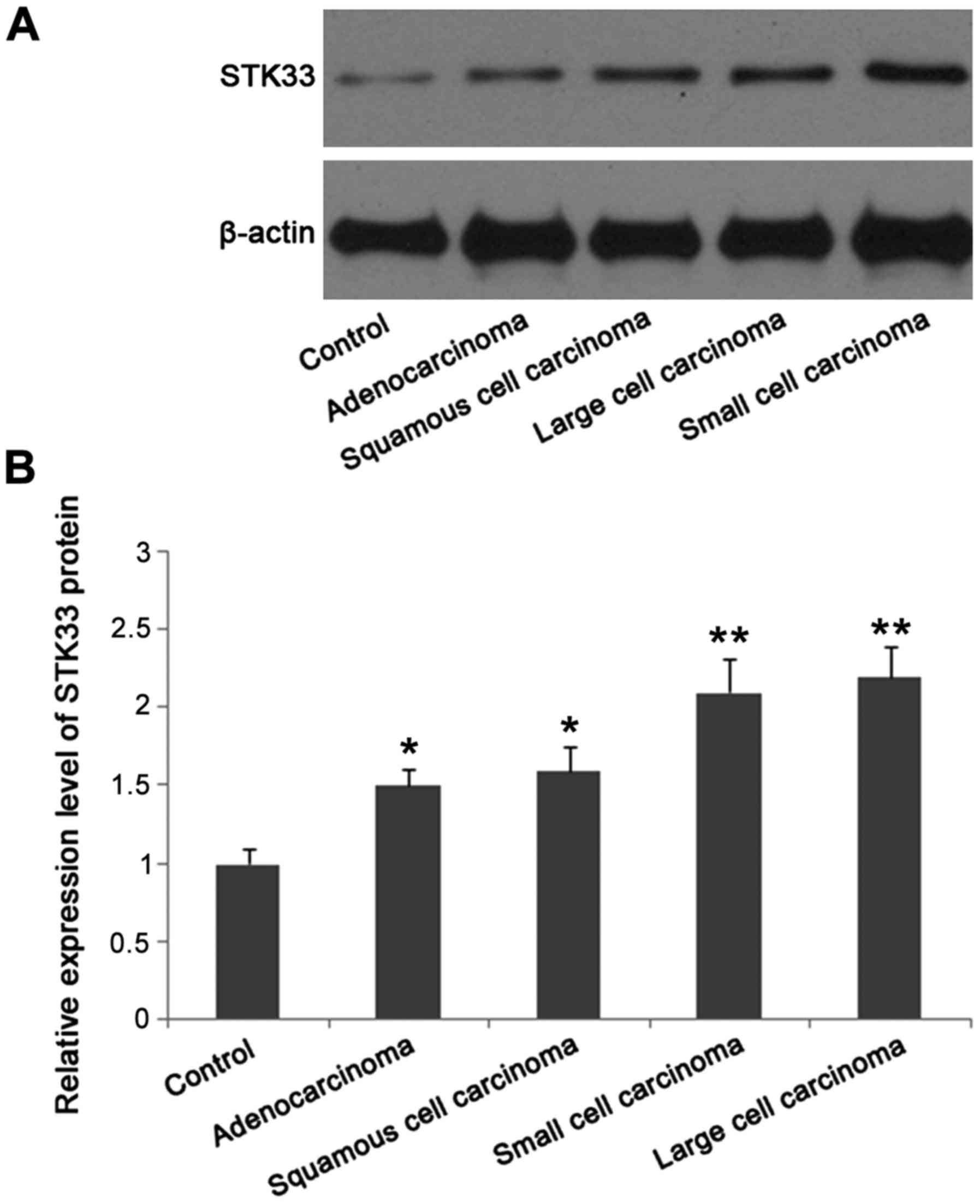
Expression of STK33 protein in
patients with different pathological types detected by western blot
analysis
Expression level of STK33 protein in patients with
four pathological types of lung cancer was significantly higher
than that in patients with benign lesions (p<0.05). In addition,
expression level of STK33 protein in lung small cell carcinoma and
large cell carcinoma was significantly higher than that in lung
adenocarcinoma and squamous cell carcinoma (p<0.05) (Fig. 4).
Expression of STK33 protein detected
by immunohistochemistry staining
Immunohistochemistry staining showed that the
positive rate of STK33 in lung large cell carcinoma (100%) and
small cell carcinoma (100%) was significantly higher than that in
lung adenocarcinoma (88.1%) and squamous cell carcinoma (86.2%),
(p<0.05) (Table III).
 | Table III.Results of immunohistochemistry
staining of lung cancer tissue [n (%)]. |
Table III.
Results of immunohistochemistry
staining of lung cancer tissue [n (%)].
| Types | Cases | Negative | Positive | χ2 | P-value |
|---|
| Adenocarcinoma | 42 | 5 (11.9) | 37 (88.1) | 12.653 | <0.001 |
| Squamous cell
carcinoma | 29 | 4 (13.8) | 25 (86.2) | 14.823 | <0.001 |
| Large cell
carcinoma | 15 | 0 (0) | 15 (100) |
|
|
| Small cell
carcinoma | 16 | 0 (0) | 16 (100) |
|
|
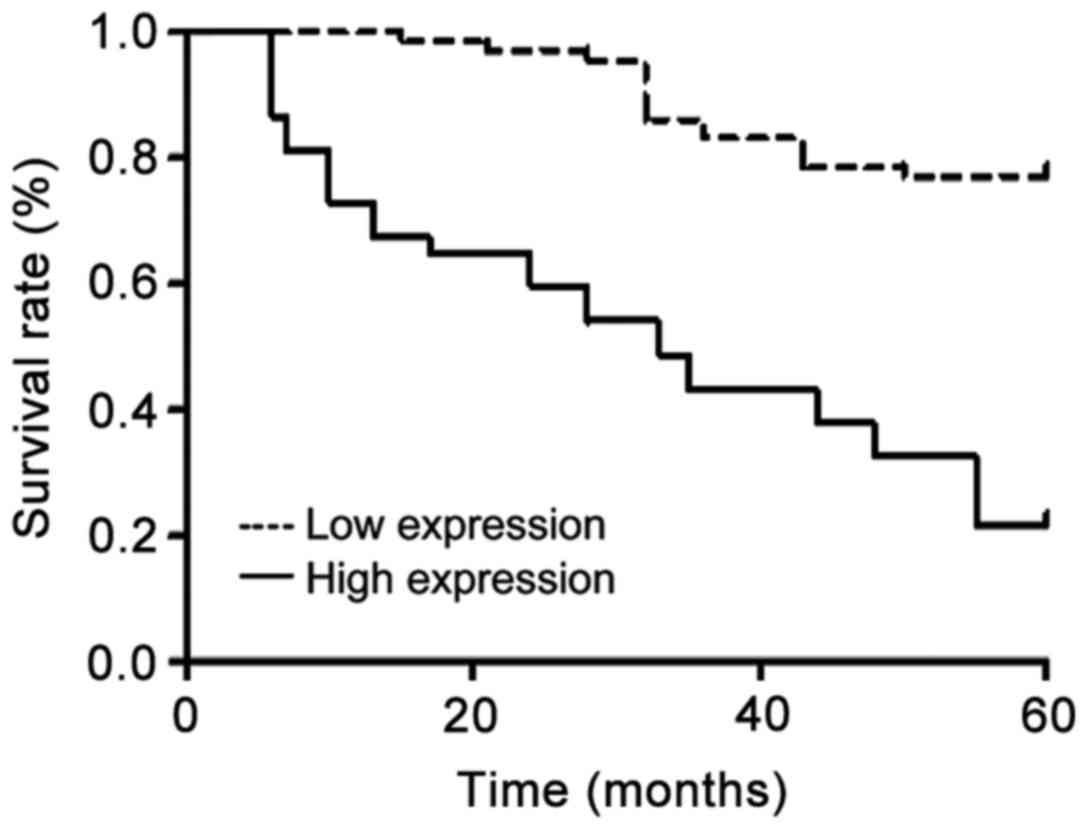
Correlation between STK33 expression and prognosis
of lung cancer patients. According to the results of
immunohistochemistry staining, negative expression group is the low
expression group, and positive expression group is the high
expression group. The 5-year survival rate analysis showed that the
recurrence-free survival rate and overall survival rate of STK33
gene high expression group were significantly lower than those of
low expression group (p<0.05) (Table
IV). Kaplan-Meier survival curves of the two groups are shown
in Fig. 5.
 | Table IV.Comparison of survival rate between
STK33 gene high expression group and low expression group [n
(%)]. |
Table IV.
Comparison of survival rate between
STK33 gene high expression group and low expression group [n
(%)].
| Groups | Cases | Recurrence-free
survival rate | Overall survival
rate |
|---|
| Low expression
group | 71 | 34 (47.9) | 50 (70.4) |
| High expression
group | 31 | 7 (22.6) | 8 (25.8) |
| χ2 |
| 13.92 | 39.841 |
| P-value |
| <0.001 | <0.001 |
Discussion
Lung, as the hub for the gas exchange between the
human body and outside world, can easily be affected by
environment. Lesions or even cancer can develop more easily in lung
than other organs. Lung cancer cells can easily migrate, and the
cure rate is very low. With the development of molecular biology,
the diagnosis and treatment of cancer has moved from chemotherapy
and radiotherapy to individualized therapies at molecular level,
among which the emergence of targeted small molecule drugs for
different genes of different cancers is a typical representative
(8,9).
Lung cancer-related genes of interest include EGFR, KRAS, and BRAF,
and targeted drugs have also been developed to target the genes
(10–12). However, with the deepening of research
and the gradual mining of related molecular signaling pathways,
more and more lung cancer-related genes, such as STK33, have been
identified (13). STK33 is located in
the human chromosome 11p15.3 region. Studies have shown that
multiple genes in this region were associated with the occurrence
and development of a variety of clinical diseases, and the
functional abnormalities of the genes can led to the occurrence of
tumors (14). STK33 gene encodes a
novel serine/threonine protein kinase. Due to the specific
structure, STK33 cannot only activate the protease but also
participate in a variety of life activities by affecting absorption
of calcium (15). STK33, as a newly
discovered gene with unclear mechanism, has attracted increasing
attention.
STK33 can indirectly interact with Ras gene to cause
synthetic lethality of various cancer cells (16). Using RNAi, Scholl et al have
demonstrated that STK33 gene and KRAS gene are two independent and
indispensable genes that affect the occurrence, growth and
metastasis of cancer cells (17).
Some researchers believe that STK33 can participate in
agglutination of mesenchymal cells through specific phosphorylated
proteins to cause changes in cell structure, affect normal
physiological function, and promote the activation and
proliferation of tumor cells (18).
Related studies also showed that the presence of STK33 protein in
colon cancer, lung cancer, breast cancer, pancreatic cancer and
other tumor cells is closely related to occurrence of mutations in
KRAS gene (19,20). However, the mechanism of the role of
STK33 in the pathogenesis of cancer, especially lung cancer is
still unclear.
In this study, the correlation between the
expression of STK33 gene and the clinicopathological features of
lung cancer was investigated, and the effects of the expression of
STK33 on the prognosis of lung cancer were also explored. Results
showed that the expression of STK33 gene was closely correlated
with the pathologic types of lung cancer, and significant
differences were found in expression level of STK33 protein among
patients with different pathologic types. Expression level of STK33
gene in lung cancer group was significantly higher than that in
benign lesion group (p<0.05). Expression level of STK33 mRNA in
lung adenocarcinoma and squamous cell carcinoma was significantly
lower than that in lung small cell carcinoma and large cell
carcinoma (p<0.05). Western blot analysis showed that the
expression level of STK33 protein in lung small cell carcinoma and
large cell carcinoma was significantly higher than that in lung
adenocarcinoma and squamous cell carcinoma (p<0.05).
Immunohistochemistry staining showed that the positive rate of
STK33 in lung large cell carcinoma (100%) and small cell carcinoma
(100%) was significantly higher than that in lung adenocarcinoma
(88.1%) and squamous cell carcinoma (86.2%) (p<0.05). All the
data indicate that STK33 can potentially be used as a biomarker of
non-cancerous lesions. The 5-year survival rate analysis showed
that the recurrence-free survival rate and overall survival rate of
STK33 gene high expression group were significantly lower than
those of low expression group (p<0.05), indicating that this
gene is closely correlated with the degree and clinical staging of
lung cancer.
In conclusion, the differential expression level of
STK33 is correlated with the pathology and prognosis of lung
cancer, which is of great value in clinical diagnosis and prognosis
evaluation.
Acknowledgements
This study was supported by the Natural Science
Foundation of Shanghai Jiao Tong University Affiliated Sixth
People's Hospital (1575).
References
|
1
|
Wakeam E, Acuna SA, Leighl NB, Giuliani
ME, Finlayson SRG, Varghese TK and Darling GE: Surgery versus
chemotherapy and radiotherapy for early and locally advanced small
cell lung cancer: A propensity-matched analysis of survival. Lung
Cancer. 109:78–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lopez-Pastorini A, Riedel R, Koryllos A,
Beckers F, Ludwig C and Stoelben E: The impact of preoperative
elevated serum C-reactive protein on postoperative morbidity and
mortality after anatomic resection for lung cancer. Lung Cancer.
109:68–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu L, Leng D, Cun D, Foged C and Yang M:
Advances in combination therapy of lung cancer: Rationales,
delivery technologies and dosage regimens. J Control Release.
260:78–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen C, Huang L, Zhang G, Li Y, Li L, Bai
X, Liu W, Wang H and Li J: STK33 potentiates the malignancy of
hypopharyngeal squamous carcinoma: Possible relation to calcium.
Cancer Biol Ther. 17:976–984. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Azoitei N, Hoffmann CM, Ellegast JM, Ball
CR, Obermayer K, Gößele U, Koch B, Faber K, Genze F, Schrader M, et
al: Targeting of KRAS mutant tumors by HSP90 inhibitors involves
degradation of STK33. J Exp Med. 209:697–711. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang P, Cheng H, Wu J, Yan A and Zhang L:
STK33 plays an important positive role in the development of human
large cell lung cancers with variable metastatic potential. Acta
Biochim Biophys Sin (Shanghai). 47:214–223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brauksiepe B, Baumgarten L, Reuss S and
Schmidt ER: Co-localization of serine/threonine kinase 33 (Stk33)
and vimentin in the hypothalamus. Cell Tissue Res. 355:189–199.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Milenic DE, Baidoo KE, Kim YS, Barkley R
and Brechbiel MW: Targeted α-particle radiation therapy of
HER1-positive disseminated intraperitoneal disease: An
investigation of the human anti-EGFR monoclonal antibody,
panitumumab. Transl Oncol. 10:535–545. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Groner B and von Manstein V: Jak Stat
signaling and cancer: Opportunities, benefits and side effects of
targeted inhibition. Mol Cell Endocrinol. 451:1–14. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu Y, Bassoff N, Reinshagen C, Bhere D,
Nowicki MO, Lawler SE, Roux J and Shah K: Bi-specific molecule
against EGFR and death receptors simultaneously targets
proliferation and death pathways in tumors. Sci Rep. 7:26022017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen H, Xing C, Cui K and Li Y, Zhang J,
Du R, Zhang X and Li Y: MicroRNA-30a attenuates mutant KRAS-driven
colorectal tumorigenesis via direct suppression of ME1. Cell Death
Differ. 24:1253–1262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lim SY, Menzies AM and Rizos H: Mechanisms
and strategies to overcome resistance to molecularly targeted
therapy for melanoma. Cancer. 123:(S11). 2118–2129. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ye H, Shao M, Shi X, Wu L, Xu B, Qu Q and
Qu J: Predictive assessment in pharmacogenetics of glutathione
S-transferases genes on efficacy of platinum-based chemotherapy in
non-small cell lung cancer patients. Sci Rep. 7:26702017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brace PT, Tezera LB, Bielecka MK, Mellows
T, Garay D, Tian S, Rand L, Green J, Jogai S, Steele AJ, et al:
Mycobacterium tuberculosis subverts negative regulatory pathways in
human macrophages to drive immunopathology. PLoS Pathog.
13:e10063672017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reuss S, Brauksiepe B, Disque-Kaiser U and
Olivier T: Serine/threonine-kinase 33 (Stk33) - Component of the
neuroendocrine network? Brain Res. 1655:152–160. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang L, Chen C, Zhang G, Ju Y, Zhang J,
Wang H and Li J: STK33 overexpression in hypopharyngeal squamous
cell carcinoma: Possible role in tumorigenesis. BMC Cancer.
15:132015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scholl C, Fröhling S, Dunn IF, Schinzel
AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S,
et al: Synthetic lethal interaction between oncogenic KRAS
dependency and STK33 suppression in human cancer cells. Cell.
137:821–834. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mujica AO, Hankeln T and Schmidt ER: A
novel serine/threonine kinase gene, STK33, on human chromosome
11p15.3. Gene. 280:175–181. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang T, Song B, Zhang J, Yang GS, Zhang H,
Yu WF, Wu MC, Lu JH and Shen F: STK33 promotes hepatocellular
carcinoma through binding to c-Myc. Gut. 65:124–133. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Babij C, Zhang Y, Kurzeja RJ, Munzli A,
Shehabeldin A, Fernando M, Quon K, Kassner PD, Ruefli-Brasse AA,
Watson VJ, et al: STK33 kinase activity is nonessential in
KRAS-dependent cancer cells. Cancer Res. 71:5818–5826. 2011.
View Article : Google Scholar : PubMed/NCBI
|