Introduction
Breast cancer (BC) is one of the most frequent
tumors affecting women worldwide, with different metastatic
potentials and phenotypes due to inherently different biological
characteristics (1,2). The basal-like BC subtype commonly refers
to any mammary gland that lacks expression of the progesterone
receptor (PR), estrogen receptor (ER), and human epidermal growth
factor receptor (HER) 2, which is widely regarded as
triple-negative BC (TNBC) (3–5). TNBC is characterized as a mesenchymal
phenotype, high proliferation, aggressive clinical behavior and
young age accounts for approximately 8–15% of all BC and (6,7). Recently,
researchers have drawn particular attention to TNBC because of its
poor unfavorable clinical outcome and lower 5-year survival rate
(8). Patients with TNBC are not
expected to benefit from endocrine or anti-HER2 molecularly
targeted therapies, which solely relies on chemotherapy (9,10). A
better investigation of the molecular regulatory mechanisms could
provide effective therapeutic strategies for TNBC treatment.
MicroRNAs (mRNAs) are endogenous 21–24 nucleotide
single-stranded noncoding RNAs, bind to the 3′-untranslated region
(3′UTR) of multiple target mRNAs and have been found to play
important roles in tumorigenesis and progression (11–14).
Numerous miRNAs have been proved to be encoded in cancer-related
gene regions, revealing that alteration of miRNA expression may
have a causal relationship with tumorigenesis (15,16).
Furthermore, the oncogenic properties of miR-182 in various tumors
have been elucidated (17). Previous
studies demonstrated miR-182 clould regulate many suppressor genes
in BC, including BRCA1 (18), RECK
(19), PFN1 (8), FOXO1 (20), ZEB1 and HSF2 (21).
The mesenchymal regulator forkhead-box (FOX)F2
belongs to the FOX transcription factor superfamily, which is
characterized by tissue homeostasis through regulating
epithelial-mesenchymal interaction to maintain epithelium polarity
(22) and regulate various biological
processes (23). Recent studies
indicated that FOXF2 was a tumor suppressor in prostate cancer
(24). Wang et al reported
deficiency of FOXF2 could induce the metastasis of basal-like BC
cells by activating epithelial-mesenchymal transition (EMT) program
(25). However, the pathobiological
effects of miR-182 expression by targeting FOXF2 in TNBC have not
been fully elucidated.
In the present study, we found that miR-182 was
aberrantly upregulated in TNBC cells and tissues and miR-182
promoted TNBC cell proliferation and metastasis. Furthermore, FOXF2
was identified as a functional and direct target of miR-182.
Therefore, this study is expected to present fruitful strategies of
tumor suppressor miR-182 as a potential target for TNBC
therapy.
Materials and methods
Patients and specimens
The TNBC tissues and adjacent relatively normal
tissues were collected in Liaocheng People's Hospital from May 2009
to August 2014. None of the patients had chemotherapy or other
treatment history previously and the TNBC patients were not
afflicted with other inflammatory diseases. Surgically resected
TNBC tissue and adjacent relatively normal breast tissue were
immediately immersed in RNAlater (Ambion; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and stored at −80°C. This study was
approved by the Ethics Committee of Liaocheng People's Hospital, in
accordance with the Helsinki Declaration of 1975 and written
informed consent was given by all participants. The clinical
characteristics are shown in Table
I.
 | Table I.Clinicopathological parameters of
TNBC patients (total cases, n=55). |
Table I.
Clinicopathological parameters of
TNBC patients (total cases, n=55).
|
|
| miR-182
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Cases | Low, n (%) | High, n (%) | P-value |
|---|
| Age (years) |
|
|
| 0.7048 |
|
≤40 | 9 | 2
(22.22) | 7
(77.78) |
|
|
>40 | 46 | 15 (32.61) | 31 (67.39) |
|
| Tumor size
(cm) |
|
|
| 0.7230 |
|
<2 | 10 | 3
(30.00) | 7
(70.00) |
|
| ≥2 | 45 | 19 (42.22) | 26 (57.78) |
|
| Lymph node
metastases |
|
|
| 0.0431 |
| 0 | 3 | 1
(33.33) | 2
(66.67) |
|
|
1–3 | 10 | 5
(50.00) | 5
(50.00) |
|
|
>3 | 42 | 6
(14.29) | 36 (85.71) |
|
|
Differentiation |
|
|
| 0.2788 |
|
Well | 5 | 2
(40.00) | 3
(60.00) |
|
|
Moderate | 10 | 5
(50.00) | 5
(50.00) |
|
|
Poor | 40 | 12 (30.00) | 28 (70.00) |
|
| Tumor stage |
|
|
| 0.0216 |
|
I+II | 16 | 7
(43.75) | 9
(56.25) |
|
|
III+IV | 39 | 5
(12.82) | 34 (87.18) |
|
Cell culture and miR-182
transfection
TNBC cell lines MCF-7, MDA-MB-231 and
non-tumorigenic mammary epithelial cell MCF-10A were obtained from
the American Type Culture Collection (Manassas, VA, USA). Cells
were cultured in cell culture medium consisting of 1:1 Medium199
and MCDB105 medium (both from Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) with 10% heat-inactivated fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and 10 ng/ml
epidermal growth factor (Sigma-Aldrich; Merck KGaA). For stable
infection, MDA-MB-231 cells were evenly seeded in 6-well plates
(Corning Incorporated, Corning, NY, USA) at a concentration of
3×105 cells/ml. After the cells adhered, the
transfection of the miR-182 mimics, miR-182 inhibitor and negative
control were performed with Lipofectamine™ 2000 (Invitrogen Life
Technologies; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol, which clould achieve the
ectopic expression of miRNA. The transfection protocol for siRNA
was the same as that for miR-182 mimics/inhibitor. miR-182
mimics/inhibitor and FOXF2-siRNA were purchased from Guangzhou
RiboBio Co., Ltd. (Guangzhou, China).
RNA isolation and qPCR
Total RNA was extracted using the TRIzol reagents
(Ambion; Thermo Fisher Scientific, Inc.) from culture cells and
cancer tissues according to the manufacturer's instruction. The
cDNA were synthesized from total RNA using the miScript reverse
transcription kit (Qiagen SA, Courtaboeuf Cedex, France). qRT-PCR
was performed using StepOne Plus qPCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). All primers used
were purchased from Guangzhou RiboBio Co., Ltd. The following
cycling conditions were employed for qPCR: 95°C for 10 min,
followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min. The
level of mature miR-182 was normalized relative to U6 endogenous
control and FOXF2 expression was normalized relative to β-actin
(endogenous control) using the 2−∆∆Cq method.
MTT assay
For proliferation rate, cells were uniformly seeded
onto 96-well plates at a density of 5×103 cells per
well. After reaction with 20 µl 5 mg/ml sterile MTT (Sigma-Aldrich;
Merck KGaA) for 4 h at 37°C, culture media was removed and 100 µl
of dimethyl sulphoxide (DMSO; Sigma-Aldrich; Merck KGaA) was added
to each well. The absorbance values were evaluated at 490 nm with
the ELISA reader (BioTek, Winooski, USA). All the reactions were
performed in triplicates.
Migration and invasion assays
After the MCF-7 and MDA-MB-231 cells were
transfected with the miR-182 inhibitor and negative control for 24
h, Transwell assay was performed. Briefly 5–10×104 cells
were seeded into the upper and lower chambers filled with culture
medium containing 20% FBS as a chemoattractant. The cells were then
washed one to two times with PBS and stained with 0.2% crystal
violet. The number of successfully translocated cells was counted
under a light microscope.
In vivo nude mouse tumorigenesis and
metastasis experiments
All animal experiments were approved by the
Liaocheng People's Hospital Animal Care and Use Committee. For
xenograft experiment, cells were injected intravenously into BALB/c
nu/nu female mice aged 4–5 weeks. Upon euthanasia, tumor growth was
measured by calculating tumor volume based on the formula: Volume
(mm3) = L × W2 x ∏/6 where L is tumor length
and W is tumor width. After 28 days (MDA-MB-435s cells), mice were
euthanized and lungs were surgically dissected. Histological
analysis with hematoxylin and eosin was carried out as described
previously (26).
Western blot analysis
Cultured cells were collected and lysed with radio
immune precipitation assay (RIPA) lysis buffer (Beyotime Institute
of Biotechnology, Haimen, China). Protein samples were separated by
SDS-PAGE and then transferred to polyvinyl difluoride membrane (EMD
Millipore, Billerica, MA, USA). The membranes were incubated in
primary antibodies followed by incubation with horseradish
peroxidase-coupled secondary antibody (Santa Cruz Biotechnology,
Inc., Dallas, TX, USA). All the antibodies were obtained from Abcam
(Cambridge, UK). β-actin was used as internal control. The signals
were detected with the ECL western blot analysis system (Amersham;
GE Healthcare, Chicago, IL, USA). The protein bands were visualized
by autoradiography and quantified by ImageJ software (National
Institutes of Health, Bethesda, MD, USA).
Luciferase reporter
The FOXF2 wild- and mutant-type 3′UTR were created
and cloned from human genomic DNA. For luciferase assay, cells were
seeded on 24-well plates the day before transfection. miR-182 mimic
plus WT or MT 3′UTR of FOXF2 were transfected into TNBC cells.
After 48 h, the cells were collected and assayed using Dual
Luciferase Reporter Assay (Promega Corporation, Madison, WI, USA)
according to the manufacture's protocols. Firefly luciferase signal
was normalized to Renilla luciferase signal.
Statistical analysis
Data are all presented as means ± SD in triplicate.
Student's t-test was used for comparisons between two groups of
experiments. The software SPSS 16.0 (IBM Corp., Armonk, NY, USA)
was applied for one-way ANOVA in comparisons among three or more
groups. A P<0.05 was considered to indicate a statistically
significant difference.
Results
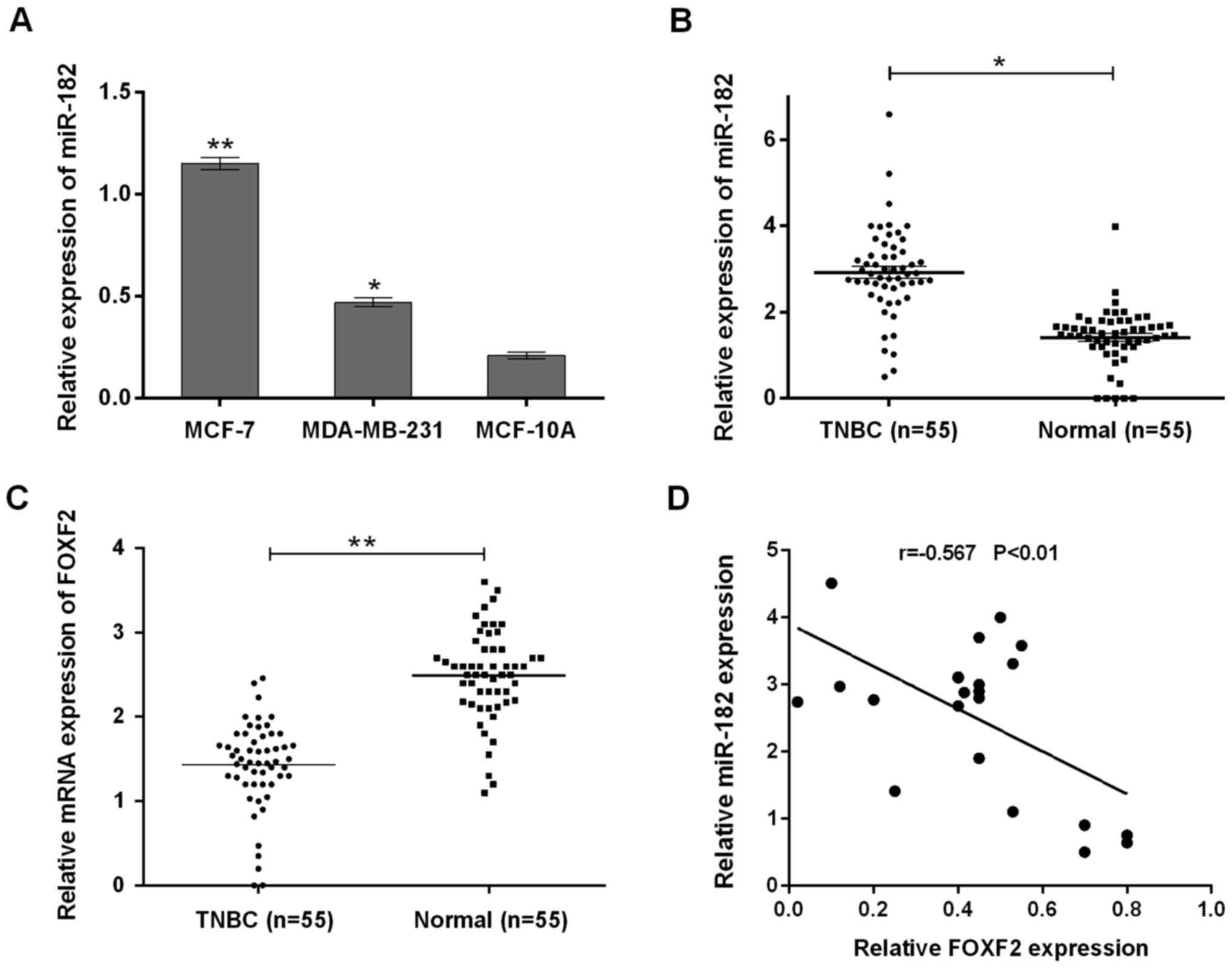
miR-182 expression is upregulated and
inversely correlates with FOXF2 in TNBC tissues and cells
We measured miR-182 expression in two TNBC cell
lines and one immortalized mammary epithelial cell line MCF-10A. As
shown in Fig. 1A, miR-182 expression
levels in MCF-7 and MDA-MB-231 cells were significantly higher than
those in MCF-10A cells (P<0.05). To confirm miR-182 upregulation
in most TNBC tissues, we examined miR-182 expression in larger
clinical samples by qRT-PCR (Fig.
1B). Similar results were obtained that miR-182 was
significantly higher in TNBC tissues (n=65) compared with normal
samples (n=65) (P<0.05). These findings suggested that miR-182
was upregulated in both TNBC cell lines and tissues. Furthermore,
FOXF2 expression levels were measured in TNBC specimens and
adjacent normal tissues. qRT-PCR analysis showed significantly
lower mRNA levels of FOXF2 in TNBC, compared with normal tissue
(Fig. 1C). Spearman's correlation
analysis disclosed an inverse correlation between miR-182
expression and that of FOXF2 (Fig.
1D).
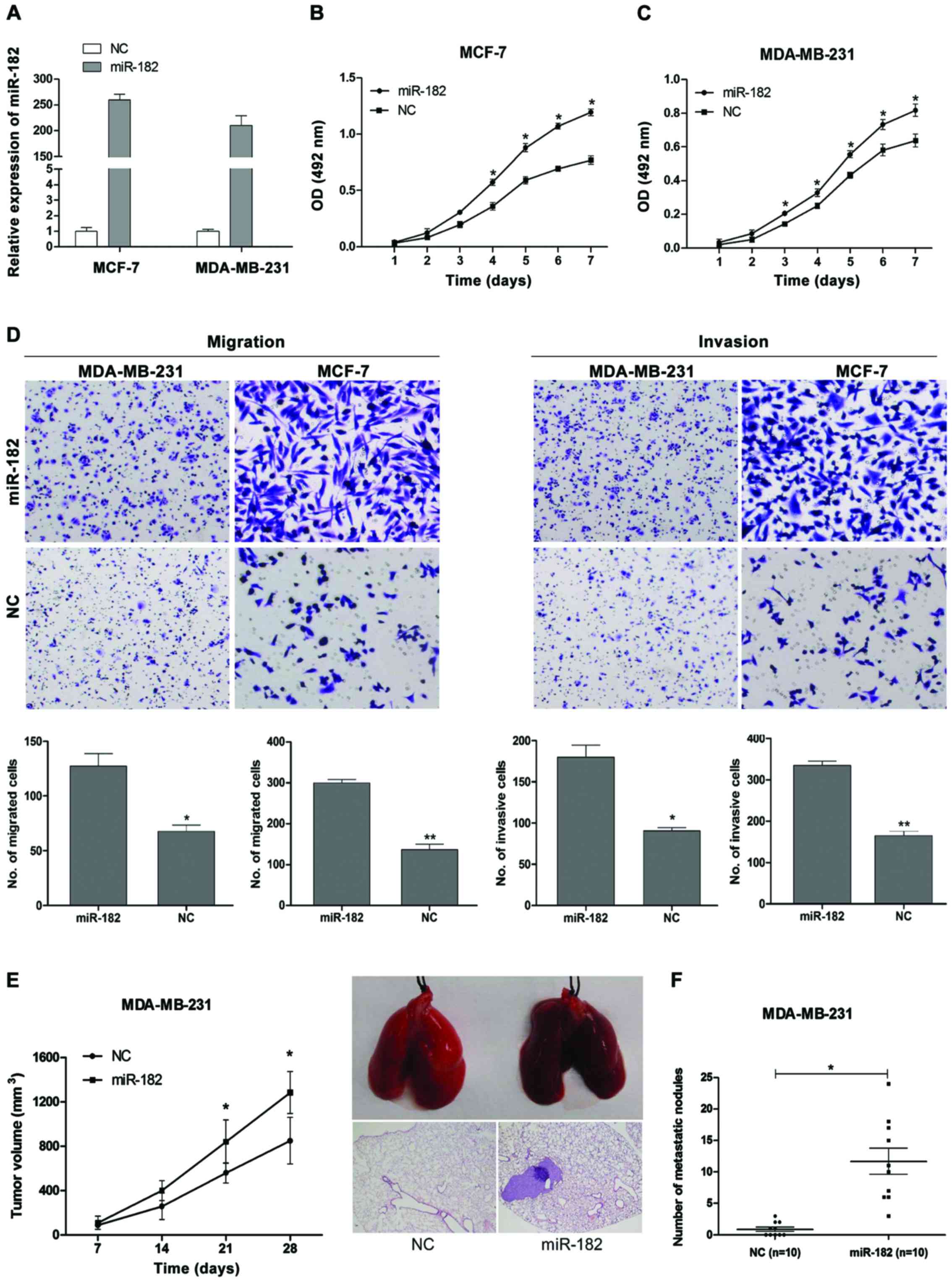
miR-182 promotes proliferation,
migration and invasion of TNBC cells in vitro
Given that the miR-182 was upregulated in TNBC cell
lines, we next assessed the oncogenic potential of miR-182 in
proliferation, migration and invasion by transfection of synthetic
miRNA mimics or negative control in vitro. The successful
re-expression of miR-182 in TNBC cells was confirmed by qRT-PCR
(Fig. 2A). The effects of miR-182 on
the cell proliferation are shown in Fig.
2B and C as determined by MTT assay. The cell growth curves
showed that ectopic expression of miR-182 significantly promoted
the proliferation of MCF-7 and MDA-MB-231 cells, indicating miR-182
promoted cell proliferation of TNBC. The results of migration and
invasion from Transwell assay are shown in Fig. 2D. The overexpression of miR-182
significantly enhanced migration and invasion in both malignant
(MCF-7, MDA-MB-231) cell lines. Therefore, the results suggested
that miR-182 could promote proliferation, migration and invasion of
TNBC cells in vitro.
miR-182 promotes TNBC metastasis in
vivo
To monitor the effect of miR-182 overexpression on
the growth and metastasis of in vivo, we injected the
MDA-MB-231 cells transfected with miR-182 or miR-control into the
tail vein of nude mice (n=10 for each group). We found that tumor
volumes in MDA-MB-231/miR-182 mimic group were larger than NC
groups within 4 weeks (Fig. 2E). In
Fig. 2F, the mice with miR-182
overexpression tumors demonstrated an approximately six-fold
increase in the number of metastatic lung nodules compared with NC
group. These observations demonstrated that miR-182 expression
drives tumor growth and metastasis in TNBC, thereby functioning as
a potential metastatic miRNA.
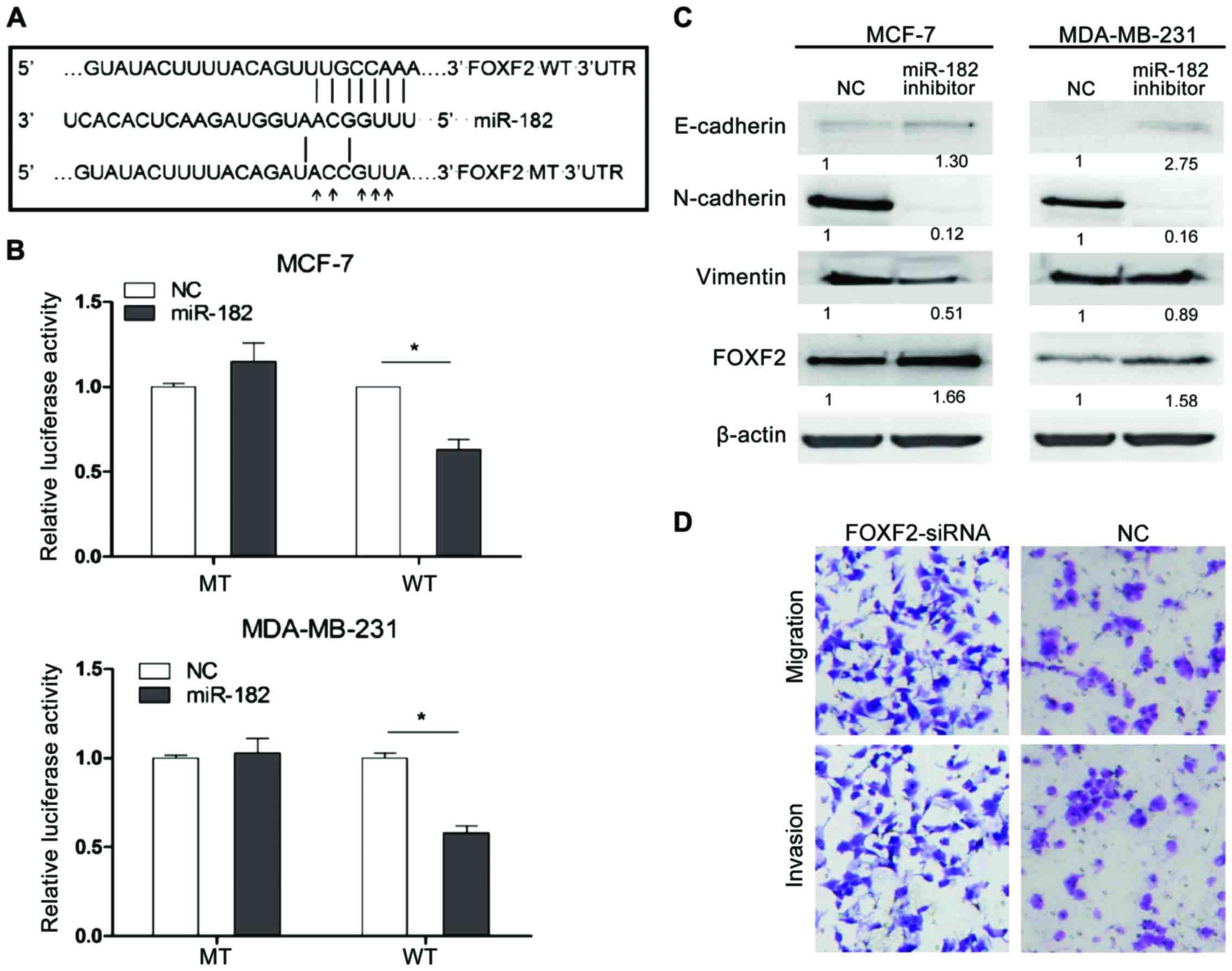
FOXF2 is a direct target of
miR-182
We identified FOXF2 as predicted target gene of
miR-182 from TargetScan and miRanda database. The putative binding
site for miR-182 was found in 688–695 bp of FOXF2 3′UTR (Fig. 3A). To confirm the prediction, a
luciferase reporter assay was performed in MCF-7 and MAD-MB-231
cells. FOXF2 3′UTR WT or MT was cloned into pmiRGLO vector based on
luciferase reporter assay, which were then transfected into cells
with miR-182 mimic or miR-control. The relative luciferase activity
was significantly decreased in cells cotransfected with WT 3′UTR of
FOXF2 and miR-182 mimic compared with negative control cells.
However, the effect was abolished when MT 3′UTR was cotransfected
with miR-182 mimic (Fig. 3B).
Furthermore, we examined the expression of FOXF2
affected by miR-182. The silence of miR-182 significantly increased
endogenous FOXF2 expression at the protein levels in both MCF-7 and
MAD-MB-231 cells (P<0.05) as confirmed by western blot analysis
(Fig. 3C). Furthermore, we detected
the specific effects of siRNA specific to FOXF2 on the cell
proliferation and metastasis of TNBC cells by Transwell assay. The
result showed that FOXF2 inhibits MDA-MB-231 cells invasion and
migration (Fig. 3D). Besides, we
detected the influence of miR-182 on EMT progress in both cell
types. We found that miR-182 inhibitor increased the expression
level of epithelial marker E-cadherin and decreased the levels of
mesenchymal markers (Fig. 3C). It is
probably that the miR-182 promoting migration and invasion was
performed by activation of EMT program in TNBC cells.
Taken together, these results indicated that FOXF2
was a direct downstream target of miR-182 and EMT progression was
inactivated by the miR-182 inhibitor.
Discussion
TNBC refers to a specific subtype of BC that lacks
expression of the ER, PR and HER2 (27). Patients with TNBC have an increased
likelihood of metastasis and distant recurrence compared to
patients with other BCs (28). miRNAs
are critical regulators of gene expression and their dysregulation
has been reported to be closely related to tumorigenesis and cancer
metastasis (29–31). Therefore, there is a major unmet need
to better understand the molecular basis of this type of BC as well
as develop new therapeutic strategies against it.
Although miR-182 is showed to promote apoptosis and
to inhibit the proliferation of lung cancer cells as a tumor
suppressor (32), it has been
revealed to be an oncogene in most malignancies, such as prostate
cancer (24), medulloblastoma
(33), melanoma (34), and glioma (35). Given miR-182 acts as an oncogene in
various cancers and a valuable marker for prognosis of TNBC, we
validated miR-182 expression in TNBC cell lines and tissues by
qRT-PCR. The results showed that the miR-182 expression levels in
the TNBC cells and tissues are significantly higher than those in
the normal cells and tissues, which was consistent with previous
reports (8,36). Furthermore, we observed the depletion
of miR-182 resulted in E-cadherin increase (Fig. 3C). The increase of E-cadherin
expression from adherent junction leads to the inhibitor of
N-catenin into the cytoplasm during EMT progression, which reveal
miR-182 activating EMT program in TNBC cells. The above thus
indicated that miR-182 may act as a cancer promoter in TNBC.
It is well known that FOXF2 may act as a novel
EMT-suppressing factor to induce apoptosis regulation, metastasis,
invasion and poor prognosis in divergent cancer types (25,37–39).
Previous reports have revealed a variety of embryonic and
mesenchymal transcriptional factors function as EMT activators in
BC, including FOXC2 (40) and FOXQ1
(41). Wang et al uncovered
that FOXF2 was a novel EMT-suppressing transcription factor in
basal-like breast cells, whose inhibitor enhanced the metastatic
ability by inducing an EMT phenotype (25). In our study, qRT-PCR analysis showed
significantly lower mRNA levels of FOXF2 in TNBC, compared with
normal tissue (Fig. 1C) and siRNA
specific to FOXF2 revealed FOXF2 inhibits cell invasion and
migration (Fig. 3D), thus indicating
FOXF2 may act as a suppressor in TNBC.
In the present study, we applied both western blot
analysis and luciferase reporter assay to identify FOXF2 as a
direct and functional target of miR-182 in TNBC. Other reports show
FOXF2 was regulated by miR-301 (42)
and miR-183-96-182 cluster (43).
FOXF2 decreased Wnt5a signaling pathway to promote angiogenesis and
cell survival (44). Hirata et
al reported that FOXF2 mRNA has two potential binding sites for
miR-182-5p within its 3′UTR (24). In
this study, we demonstrated that FOXF2 was downregulated by miR-182
by targeting its 3′UTR in TNBC cells.
In conclusion, we revealed that miR-182 can
significantly promote TNBC cell proliferation, invasion and
metastasis. FOXF2 was identified as a direct and functional target
of miR-182. The newly identified miR-182/FOXF2 axis provides new
insight into the pathogenesis of TNBC and represents a potential
therapeutic target for TNBC.
References
|
1
|
Honkoop AH, van Diest PJ, de Jong JS, Linn
SC, Giaccone G, Hoekman K, Wagstaff J and Pinedo HM: Prognostic
role of clinical, pathological and biological characteristics in
patients with locally advanced breast cancer. Br J Cancer.
77:621–626. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Medimegh I, Omrane I, Privat M, Uhrhummer
N, Ayari H, Belaiba F, Benayed F, Benromdhan K, Mader S, Bignon IJ,
et al: MicroRNAs expression in triple negative vs non triple
negative breast cancer in Tunisia: Interaction with clinical
outcome. PLoS One. 9:e1118772014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rakha EA, Elsheikh SE, Aleskandarany MA,
Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet
JS, Akslen LA, et al: Triple-negative breast cancer: Distinguishing
between basal and nonbasal subtypes. Clin Cancer Res. 15:2302–2310.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ishikawa T, Shimizu D, Kito A, Ota I,
Sasaki T, Tanabe M, Yamada A, Arioka H, Shimizu S, Wakasugi J, et
al: Breast cancer manifested by hematologic disorders. J Thorac
Dis. 4:650–654. 2012.PubMed/NCBI
|
|
5
|
Fausto P and Barni S: Benefit of tamoxifen
in estrogen receptor positive DCIS of the breast. Gland Surg.
1:3–4. 2012.PubMed/NCBI
|
|
6
|
Bagaria SP, Ray PS, Sim MS, Ye X, Shamonki
JM, Cui X and Giuliano AE: Personalizing breast cancer staging by
the inclusion of ER, PR, and HER2. JAMA Surg. 149:125–129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Andrés R, Pajares I, Balmaña J, Llort G,
Ramón Y, Cajal T, Chirivella I, Aguirre E, Robles L, Lastra E,
Pérez-Segura P, et al: Association of BRCA1 germline mutations in
young onset triple-negative breast cancer (TNBC). Clin Transl
Oncol. 16:280–284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu H, Wang Y, Li X, Zhang YJ, Li J, Zheng
YQ, Liu M, Song X and Li XR: Expression and regulatory function of
miRNA-182 in triple-negative breast cancer cells through its
targeting of profilin 1. Tumour Biol. 34:1713–1722. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheang MC, Voduc D, Bajdik C, Leung S,
McKinney S, Chia SK, Perou CM and Nielsen TO: Basal-like breast
cancer defined by five biomarkers has superior prognostic value
than triple-negative phenotype. Clin Cancer Res. 14:1368–1376.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gelmon K, Dent R, Mackey JR, Laing K,
McLeod D and Verma S: Targeting triple-negative breast cancer:
Optimising therapeutic outcomes. Ann Oncol. 23:2223–2234. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Di Leva G and Croce CM: The role of
microRNAs in the tumorigenesis of ovarian cancer. Front Oncol.
3:1532013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Korpal M, Ell BJ, Buffa FM, Ibrahim T,
Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua
Y, et al: Direct targeting of Sec23a by miR-200s influences cancer
cell secretome and promotes metastatic colonization. Nat Med.
17:1101–1108. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei
Y, Li Y, Yang Q, Liu J, Wei JJ, et al: miR-145 inhibits tumor
growth and metastasis by targeting metadherin in high-grade serous
ovarian carcinoma. Oncotarget. 5:10816–10829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Link A, Kupcinskas J, Wex T and
Malfertheiner P: Macro-role of microRNA in gastric cancer. Dig Dis.
30:255–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du L and Pertsemlidis A: microRNA
regulation of cell viability and drug sensitivity in lung cancer.
Expert Opin Biol Ther. 12:1221–1239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wall NR: Colorectal cancer screening using
protected microRNAs. J Gastrointest Oncol. 2:206–207.
2011.PubMed/NCBI
|
|
17
|
Saus E, Soria V, Escaramís G, Vivarelli F,
Crespo JM, Kagerbauer B, Menchón JM, Urretavizcaya M, Gratacòs M
and Estivill X: Genetic variants and abnormal processing of
pre-miR-182, a circadian clock modulator, in major depression
patients with late insomnia. Hum Mol Genet. 19:4017–4025. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moskwa P, Buffa FM, Pan Y, Panchakshari R,
Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K,
Weinstock DM, et al: miR-182-mediated down-regulation of BRCA1
impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell.
41:210–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiang CH, Hou MF and Hung WC:
Up-regulation of miR-182 by β-catenin in breast cancer increases
tumorigenicity and invasiveness by targeting the matrix
metalloproteinase inhibitor RECK. Biochim Biophys Acta.
1830:3067–3076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96 and miR-182 in breast cancer
cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li P, Sheng C, Huang L, Zhang H, Huang L,
Cheng Z and Zhu Q: miR-183/−96/−182 cluster is upregulated in most
breast cancers and increases cell proliferation and migration.
Breast Cancer Res. 16:4732014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aitola M, Carlsson P, Mahlapuu M, Enerbäck
S and Pelto-Huikko M: Forkhead transcription factor FoxF2 is
expressed in mesodermal tissues involved in epithelio-mesenchymal
interactions. Dev Dyn. 218:136–149. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hirata H, Ueno K, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: microRNA-182-5p
promotes cell invasion and proliferation by down regulating FOXF2,
RECK and MTSS1 genes in human prostate cancer. PLoS One.
8:e555022013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang QS, Kong PZ, Li XQ, Yang F and Feng
YM: FOXF2 deficiency promotes epithelial-mesenchymal transition and
metastasis of basal-like breast cancer. Breast Cancer Res.
17:302015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pandey V, Perry JK, Mohankumar KM, Kong
XJ, Liu SM, Wu ZS, Mitchell MD, Zhu T and Lobie PE: Autocrine human
growth hormone stimulates oncogenicity of endometrial carcinoma
cells. Endocrinology. 149:3909–3919. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim YJ, Choi JS, Seo J, Song JY, Lee SE,
Kwon MJ, Kwon MJ, Kundu J, Jung K, Oh E, et al: MET is a potential
target for use in combination therapy with EGFR inhibition in
triple-negative/basal-like breast cancer. Int J Cancer.
134:2424–2436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clin Cancer Res. 13:4429–4434. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kasinski AL and Slack FJ: Epigenetics and
genetics. MicroRNAs en route to the clinic: Progress in validating
and targeting microRNAs for cancer therapy. Nat Rev Cancer.
11:849–864. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pencheva N and Tavazoie SF: Control of
metastatic progression by microRNA regulatory networks. Nat Cell
Biol. 15:546–554. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Liu T, Huang Y and Liu J:
microRNA-182 inhibits the proliferation and invasion of human lung
adenocarcinoma cells through its effect on human cortical
actin-associated protein. Int J Mol Med. 28:381–388.
2011.PubMed/NCBI
|
|
33
|
Weeraratne SD, Amani V, Teider N,
Pierre-Francois J, Winter D, Kye MJ, Sengupta S, Archer T, Remke M,
Bai AH, et al: Pleiotropic effects of miR-183~96~182 converge to
regulate cell survival, proliferation and migration in
medulloblastoma. Acta Neuropathol. 123:539–552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Segura MF, Hanniford D, Menendez S, Reavie
L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A,
Bogunovic D, et al: Aberrant miR-182 expression promotes melanoma
metastasis by repressing FOXO3 and microphthalmia-associated
transcription factor. Proc Natl Acad Sci USA. 106:1814–1819. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song L, Liu L, Wu Z, Li Y, Ying Z, Lin C,
Wu J, Hu B, Cheng SY, Li M, et al: TGF-β induces miR-182 to sustain
NF-κB activation in glioma subsets. J Clin Invest. 122:3563–3578.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang W, Qian P, Zhang X, Zhang M, Wang H,
Wu M, Kong X, Tan S, Ding K, Perry JK, et al: Autocrine/paracrine
human growth hormone-stimulated MicroRNA 96–182-183. Cluster
promotes epithelial-mesenchymal transition and invasion in breast
cancer. J Biol Chem. 290:13812–13829. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kong PZ, Yang F, Li L, Li XQ and Feng YM:
Decreased FOXF2 mRNA expression indicates early-onset metastasis
and poor prognosis for breast cancer patients with histological
grade II tumor. PLoS One. 8:e615912013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Z, Liu P, Inuzuka H and Wei W: Roles
of F-box proteins in cancer. Nat Rev Cancer. 14:233–247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mani SA, Yang J, Brooks M, Schwaninger G,
Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL and Weinberg
RA: Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis
and is associated with aggressive basal-like breast cancers. Proc
Natl Acad Sci USA. 104:10069–10074. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi
SC and Yu Q: FOXQ1 regulates epithelial-mesenchymal transition in
human cancers. Cancer Res. 71:3076–3086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shi W, Gerster K, Alajez NM, Tsang J,
Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, et al:
MicroRNA-301 mediates proliferation and invasion in human breast
cancer. Cancer Res. 71:2926–2937. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kundu ST, Byers LA, Peng DH, Roybal JD,
Diao L, Wang J, Tong P, Creighton CJ and Gibbons DL: The miR-200
family and the miR-183~96~182 cluster target Foxf2 to inhibit
invasion and metastasis in lung cancers. Oncogene. 35:173–186.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yamamoto H, Oue N, Sato A, Hasegawa Y,
Yamamoto H, Matsubara A, Yasui W and Kikuchi A: Wnt5a signaling is
involved in the aggressiveness of prostate cancer and expression of
metalloproteinase. Oncogene. 29:2036–2046. 2010. View Article : Google Scholar : PubMed/NCBI
|