Introduction
Sex steroid hormones and sex steroid hormone
receptors play a critical role in breast cancer development and
progression. In particular, androgen and androgen receptor (AR)
have special importance, especially for estrogen receptor-negative,
progesterone receptor-negative, AR-positive (ER−,
PR−, AR+) breast cancers. As patients with
ER−, PR−, AR+ breast cancer obtain
little benefit from anti-estrogen therapy (1), more and more studies are focusing on AR
as a useful therapeutic target, with some encouraging results
(2).
AR is an androgen-dependent transcription factor
activated by binding androgens. Activated AR recognizes androgen
response elements (AREs) located in or near the promoter and
enhancer regions of androgen-dependent genes, thereby activating
the transcriptional machinery, including microRNA transcription
(3–5).
MicroRNAs are evolutionarily conserved
~22-nucleotide-long short non-coding RNA molecules. These molecules
repress target gene expression by binding to complementary
sequences in the 3′-untranslated regions (UTRs) of target mRNAs.
MicroRNAs participate in diverse biological functions, including
development, cell proliferation, differentiation, and apoptosis
(6–9).
As microRNAs play a central role in the regulation of gene
expression, aberrant expression is found in almost all types of
human cancer (10).
As both AR and microRNA have the ability to regulate
genes, their interaction within a cancer cell can take on many
forms. Some studies have suggested a role for AR in the
transcriptional regulation of microRNA expression (11–14),
whereas other studies have shown a role for the regulation of AR
expression by microRNA (15–17). However, most of these regulatory
interactions remain unknown and almost all of the research has been
focused in the field of prostate cancer.
Here, we explored the interaction between miR-30a
and AR, as well as the function of miR-30a, in ER−,
PR−, AR+ MDA-MB-453 breast cancer cells on
the basis of our previous studies (14,18).
Materials and methods
Cell culture and treatment
We chose to use MDA-MB-453 cells [American Type
Culture Collection (ATCC)] because they express high levels of AR
but not ER or PR (19–21). Cells were maintained in
75-cm2 flasks containing minimal essential medium (MEM)
supplemented with 10% fetal bovine serum, 100 IU/ml penicillin, and
100 mg/ml streptomycin at 37°C in a humidified atmosphere
containing 5% CO2. The cells were passaged every 3–4
days (80% confluence) and harvested using 0.25% trypsin/EDTA.
Before each experiment, the cells were grown for 3 days in phenol
red-free (PRF) DMEM containing 5% charcoal-treated fetal calf serum
(PRF-CT). To achieve synchronization, the cells were serum-starved
in PRF DMEM for 24 h. All experiments were performed in 2.5%
PRF-CT.
Cells were treated with 5α-dihydrotestosterone (DHT;
Sigma-Aldrich, MO, USA), a non-aromatizable androgen with the
highest affinity for AR among natural androgens, or vehicle alone
at 10−8 M (14). DHT was
dissolved in 100% ethanol and added to the media immediately before
use.
Western blot analysis
After 48 h, RIPA lysis buffer was used to lyse
charcoal-stripped MDA-MB-453 cells treated with DHT or vehicle.
Proteins were harvested, resolved on 10% SDS denaturing
polyacrylamide gel, and transferred to a nitrocellulose membrane.
The membranes were incubated at 4°C overnight in Blotto with
anti-AR or anti-GAPDH antibody (Saier Biotech, Tianjin, China) and
then washed and incubated with horseradish peroxidase
(HRP)-conjugated secondary antibody (Saier Biotech). Protein
expression was assessed utilizing enhanced chemiluminescence. Band
intensity was determined by Lab Works™ Image Acquisition and
Analysis Software (UVP).
MicroRNA microarray analysis
KangChen Bio-tech, Inc., (Shanghai, China) performed
the microRNA microarray experiments using total RNA extracted from
MDA-MB-453 cells treated with DHT or vehicle alone. TRIzol reagent
(Invitrogen, CA, USA) was used for the extraction according to the
manufacturer's instructions. The quantity and quality of RNA were
assessed using a NanoDrop ND-1000. Denaturing agarose gel
electrophoresis was used to assess RNA integrity and DNA
contamination.
The miRCURY™ Array Power Labeling kit (cat no.
208032-A; Exiqon, Vedbaek, Denmark) was used to label total RNA
with Cy3 or Cy5 fluorescent dye according to the manufacturer's
instructions. Each miRCURY LNA microRNA array (v.11.0; Exiqon),
which consists of 847 capture probes for mature human microRNAs,
was hybridized with a single Cy3- or Cy5-labeled sample. Each group
was hybridized with three miRCURY LNA arrays in triplicate using
independent samples of DHT- or vehicle-treated cells. Images were
scanned by a Gene Pix 4000B scanner using GenePix Pro 6.0 software
(Axon Instruments, Union City, CA, USA). The background was
subtracted and normalization performed. We selected microRNAs with
expression intensities (ForeGround-BackGround) >50 and
expression levels that differed by at least 1.5-fold between DHT-
and vehicle-treated cells.
Predicted AR-targeting microRNA
Computer-aided algorithms to predict microRNAs
targeting AR were obtained from miRanda (August 2010 release,
http://www.microrna.org) and TargetScan (release
5.2, http://www.targetscan.org).
Real-time reverse transcription
PCR
Stem-loop RT-PCR was performed to determine the
level of mature miR-30a as described previously (22). Briefly, 2 µg of small RNA was
reverse-transcribed to cDNA using M-MLV reverse transcriptase
(Promega Corp., Madison, WI, USA) and the following primers
synthesized by BGI, Inc.:
miR-30a-RT,5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACCTTCCAG-3′;
miR-30b-RT, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAGCTGAG-3′;
miR-30c-RT, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACGCTGAGAG−3′;
and U6-RT,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3′,
which can fold into a stem-loop structure. The specific miR-30a,b,c
cDNA fragment and an endogenous control (U6 snRNA) were amplified
using the following primers synthesized by BGI, Inc.: miR-30a-Fwd,
5′-TGCGGTGTAAACATCCTCGACTG-3′; miR-30b-Fwd,
5′-TGCGGTGTAAACATCCTACACTCA-3′; miR-30c-Fwd,
5′-TGCGGTGTAAACATCCTACACTCTC-3′; U6-Fwd,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′; and Reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′, a universal downstream primer. The PCR
reactions were heated to 94°C for 4 min, followed by 40 cycles of
94°C for 30 sec, 50°C for 30 sec, 72°C for 40 sec. The SYBR Premix
Ex Taq™ kit (Takara Bio, Inc., Otsu, Japan) was used as instructed
by the manufacturer and real-time PCR carried out on a 7300
Real-Time PCR system (Applied Biosystems Life Technologies, Foster
City, CA, USA).
For real-time RT-PCR to detect the relative level of
AR transcription, cDNA was generated using M-MLV reverse
transcriptase (Promega) with 5 µg of the large RNA. PCR to amplify
the AR gene, and β-actin gene as an endogenous control, used the
following primers: AR sense, 5′-AAGACGCTTCTACCAGCTCACCAA-3′; AR
antisense, 5′-TCCCAGAAAGGATCTTGGGCACTT-3′; β-actin sense,
5′-CGTGACATTAAGGAGAAGCTG-3′; and β-actin antisense,
5′-CTAGAAGCATTTGCGGTGGAC-3′. The PCR reaction was heated to 94°C
for 5 min, followed by 40 cycles of 94°C for 1 min, 56°C for 1 min,
and 72°C for 1 min. After real-time PCR, the expression of the AR
gene was normalized to the expression of the β-actin gene in the
same sample.
Vector construction
The pcDNA3.1/pri-miR-30a expression vector was
constructed by amplifying a 487 bp DNA fragment carrying
pri-miR-30a from genomic DNA and inserting it into the pcDNA3.1 (+)
vector at the BamHI and EcoRI sites. The PCR primers were
miR-30a-sense, 5′-CGCGGATCCGGAAACACTTGCTGGGATTAC-3′, and
miR-30a-antisense, 5′-CCGGAATTCAACTGCAGAAAGGGCAGGACA-3′.
The enhanced green fluorescent protein (EGFP)
reporter vector (pcDNA3/EGFP/AR) was constructed by amplifying the
EGFP coding region from pEGFP-N2 (Clontech) and inserting it into
pcDNA3.1 (+) at the HindIII and BamHI sites. The AR 3′UTR fragment
containing the predicted miR-30a binding site was then inserted
into pcDNA3.1 (+)/EGFP at the BamHI and XhoI sites. The PCR primers
were EGFP sense, 5′-GCAGCCAAGCTTGCCACCATGTGTAGCAAGGGC-3′; EGFP
antisense, 5′-CGCGGATCCTTTACTTGTACAGCTCGTCC-3′; AR sense,
5′-CGCGGATCCTTTAAATCTGTGATGATCCTC−3′; and AR antisense,
5′-GAGGCCTCGAGTTTGTGTGGCTGGCACAGAG-3′. The mutant enhanced green
fluorescent protein (EGFP) reporter vector (pcDNA3/EGFP/AR-mut) was
constructed by inserting the mutant AR 3′UTR fragment into pcDNA3.1
(+)/EGFP at the same sites. The fragment of mutant AR 3′UTR was
annealed by the following primers. Top-primer:
5-GATCCTTTAAATCTGTGATGATCCTCATATGGCCCAGTGTCAAGTTGTGCTTCTATTCTGCACTACTCTGTGCCAGCCACACAAAC-3′
Bot-primer:
5′-TCGAGTTTGTGTGGCTGGCACAGAGTAGTGCAGAATAGAAGCACAACTTGACACTGGGCCATATGAGGATCATCACAGATTTAAAG-3′.
Transfection
Lipofectamine 2000 (Invitrogen) was used to
transfect MDA-MB-453 cells with vectors (0.5 mg/l) or antisense
oligonucleotide (ASO; final concentration 10−9 M) 24 h
after plating and transfection complexes prepared according to the
manufacturer's instructions. The transfection medium was replaced 4
h after transfection. Oligonucleotides complementary to miR-30a
were synthesized by IDT (Coralville, IA, USA): miR-30a ASO,
5′-CTTCCAGTCGAGGATGTTTACA-3′, and control ASO,
5′-TGACTGTACTGAGACTCGACTG-3′.
EGFP reporter assay
To confirm the direct action of miR-30a on AR,
wild-type and mutant AR experimental groups were further divided
into three subgroups: group 1, cells transiently transfected with
pcDNA3/EGFP/AR; group 2, cells transiently co-transfected with
pcDNA3/EGFP/AR and pcDNA3.1 (+) (control vector); and group 3,
cells transiently co-transfected with pcDNA3/EGFP/AR and
pri-miR-30a. The loading control was RFP expression vector
pDsRed2-N1. Forty-eight h after transfection, the cells were lysed
using radio-immunoprecipitation assay (RIPA) lysis buffer (150 mM
NaCl, 50 mM Tris-HCl, pH Q2 7.2, 1% Triton X-100, and 0.1% SDS).
The intensities of EGFP and RFP were measured using a fluorescence
spectrophotometer (F4500; Hitachi, Tokyo, Japan).
Chromatin immunoprecipitation
assay
Chromatin immunoprecipitation (ChIP) to determine
whether activated AR directly up-regulates miR-30a expression in
MDA-MB-453 cells was performed as described previously (23). Briefly, chromatin DNA was extracted
from harvested cells by sonication and incubated with normal rabbit
serum (IgG) and protein A/G-Sepharose beads. Rabbit anti-AR
antibody (PG-21, Upstate) was used for immunoprecipitation. The
−2843/−2623 and −1513/−1286 fragments were amplified using the
following primers: 5′-CCTAGATTTCTTGCCTTTAG (forward) and
5′-TCCACATAACCTTCTCGCTC (reverse) for −2843/−2623, and
5′-GTAGGGGACCTGTCACTTTG (forward) and 5′-CTTGAGTCAATCCACCAAAC
(reverse) for −1513/−1286.
Cell proliferation assay
MDA-MB-453 cells were plated in 96-well microtiter
plates (7000 cells/well) in replicates of six. Twenty-four h after
seeding, the cells were transfected with vectors or ASO. The 3-(4,5
dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
colorimetric assay was performed to measure cell viability and
proliferation. Over seven consecutive days, MTT was added to the
wells and dimethyl sulfoxide (DMSO) added 4 h later to dissolve the
resulting formazan crystals. The plates were shaken for 20 min and
the absorbance detected at 570 nm using a µQuant Universal
Microplate Spectrophotometer (Bio-Tek Instruments, Winooski,
USA).
Flow cytometry
Forty-eight h after transfection with vectors or
ASO, the MDA-MB-453 cells were detached from the plates using
trypsin, rinsed with PBS, and fixed in 70% (v/v) ethanol. The cells
were rehydrated in PBS and incubated with RNase (100 µg/ml) and
propidium iodide (60 µg/ml; Sigma-Aldrich, MO, USA) prior to
analysis by the FACSCalibur System (BD Biosciences, San Jose, CA,
USA). The cell cycle phase was determined by Cell Quest analysis.
The proliferation index (PI) was calculated as (S+G2/M)/G1, where
S, G2/M, and G1 refer to the percentage of cells in S phase, G2/M
phase, and G1 phase, respectively (24).
Statistical analysis
The data are reported as the mean ± standard
deviation of three independent determinations. The Student's t test
was used for comparisons with corresponding controls. P<0.05 was
considered to indicate a statistically significant difference.
SPSS17.0 was used for all statistical analyses.
Results
Effect of DHT on AR and miR-30a,b,c
expression
AR mRNA and protein were overexpressed when
MDA-MB-453 cells were exposed to 10−8 M DHT for 48 h
compared to vehicle-treated cells (Fig.
1A and B). To identify critical microRNAs potentially related
to androgen-induced AR activating signal in breast cancer, we
examined global microRNA expression in MDA-MB-453 cells. The
microRNA array identified 43 up-regulated microRNAs and 51
down-regulated microRNAs, including miR-30a,b,c (Table I), in DHT-treated cells compared to
vehicle-treated cells.
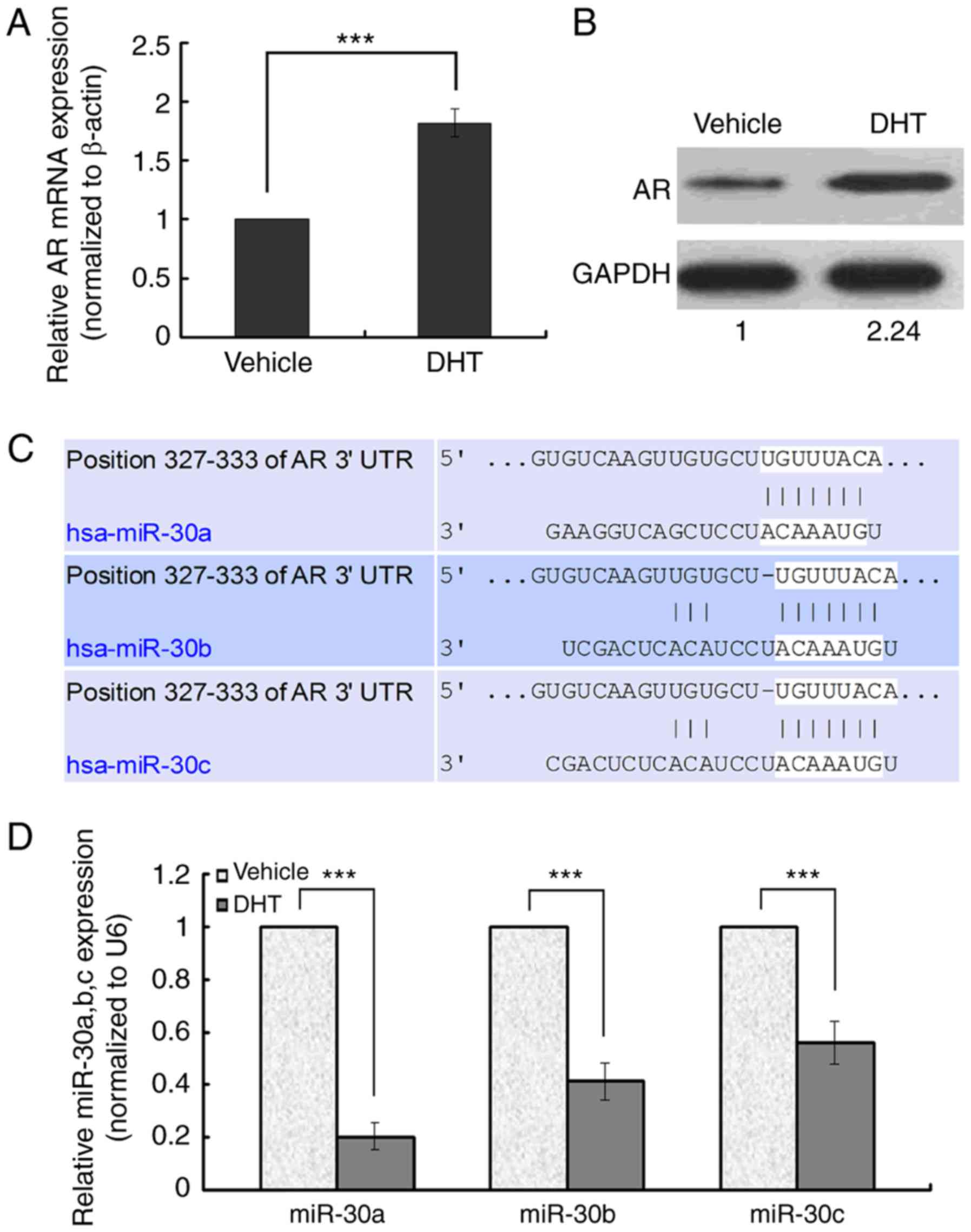 | Figure 1.Effect of activated AR signaling on AR
and miR-30a,b,c expression and TargetScan prediction outcomes. (A
and B) Real-time RT-PCR and Western blot of AR expression. (C)
TargetScan programs predicted that miR-30a,b,c are upstream
microRNAs of AR. (D) The relative miR-30a,b,c mRNA expression level
in DHT- and vehicle-treated MDA-MB-453 cells. In real-time RT-PCR,
the expression level of miR-30a,b,c in the control group was set to
1 and U6 was regarded as an endogenous normalizer. In Western
blots, the expression level of AR protein in the control group was
set to 1 and GAPDH protein regarded as an endogenous normalizer.
Error bars indicate standard deviation. ***P<0.001 compared to
control. |
 | Table I.Differentially expressed miR-30a,b,c
in MDA-MB-453 cells. |
Table I.
Differentially expressed miR-30a,b,c
in MDA-MB-453 cells.
|
|
| Expression
intensity (ForeGround-BackGround) | Normalized |
|---|
|
|
|
|
|
|---|
| MiRNA | Fold change | DHT | Vehicle | DHT | Vehicle |
|---|
| Hsa-miR-30a | 0.591493493 | 1646.5 | 923.5 |
7.925391095 | 4.687817259 |
| Hsa-miR-30b | 0.457648129 | 1919.5 | 833 |
9.239470517 | 4.228426396 |
| Hsa-miR-30c | 0.488154896 | 822 | 380.5 | 3.9566787 | 1.931472081 |
AR may be a target of miR-30a,b,c
Among the microRNAs predicted to target AR,
miR-30a,b,c were identified because the 3′UTR of AR mRNA contained
a putative binding site for miR-30a,b,c (Fig. 1C). In ER−, PR−,
AR+ MDA-MB-453 cells, the validity of ectopic
miR-30a,b,c expression was confirmed by real-time RT-PCR, which
revealed a 5-fold, 2.5-fold, and 2-fold decrease in DHT-treated
MDA-MB-453 cells compared to the vehicle-treated groups (Fig. 1D). We focused on miR-30a and
investigated the relationship between miR-30a and AR.
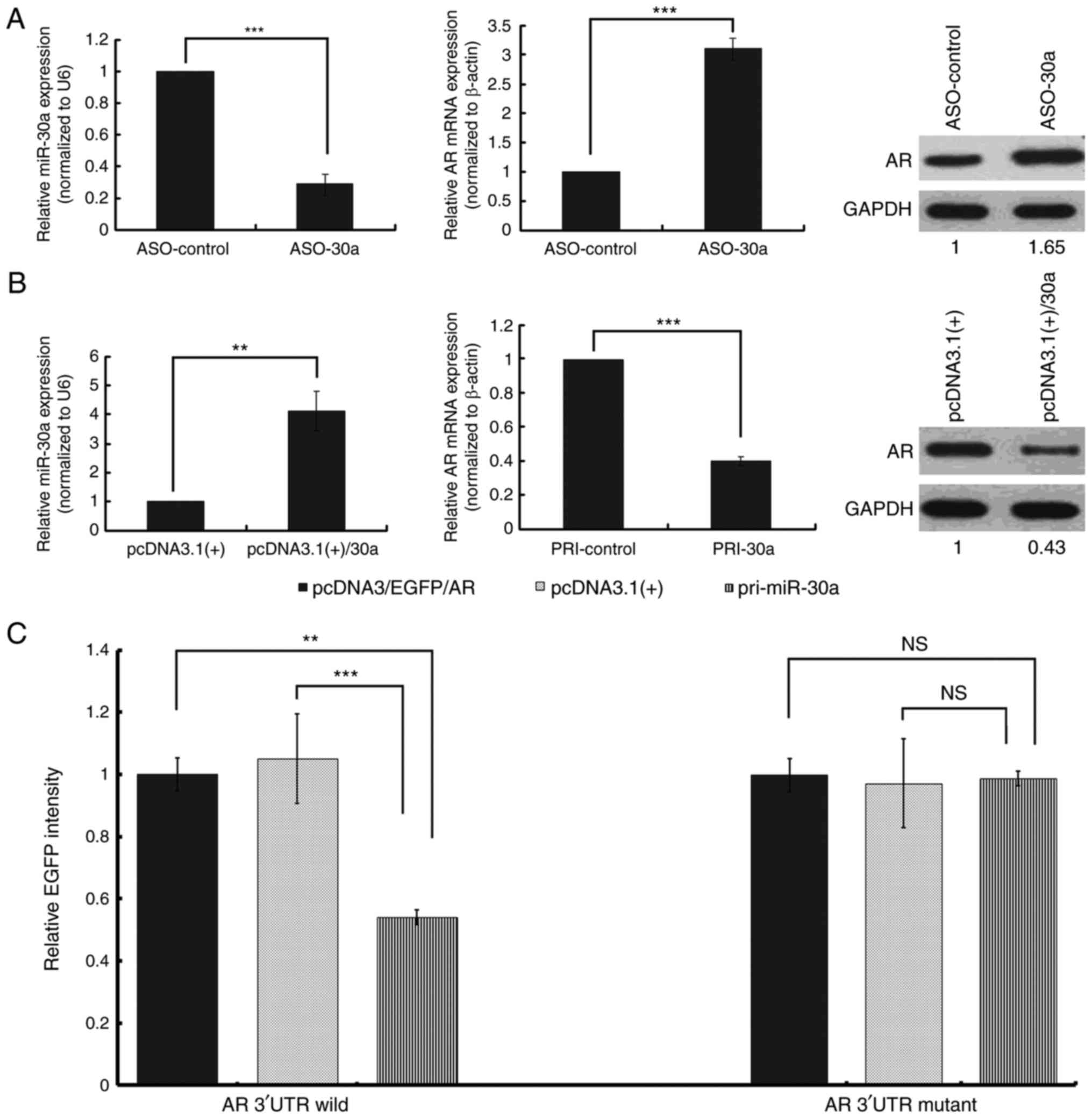
MiR-30a regulates AR at both the mRNA
and protein level
AR expression was elevated at both the mRNA and
protein level in MDA-MB-453 cells transfected with miR-30a ASO
compared to control ASO (Fig. 2A). AR
mRNA and protein levels were also assessed by real-time RT-PCR and
Western blot 48 h after transfecting MDA-MB-453 cells with pcDNA3.1
(+)/30a vector expressing miR-30a or control vector (pcDNA3.1 (+)).
Overexpression of miR-30a reduced the expression of AR at both the
mRNA and protein level (Fig. 2B).
Direct effect of miR-30a on the AR
3′UTR
The alignment of miR-30a with the wild-type or
mutant AR 3′UTR is illustrated in Fig.
1C. Using a vector containing the EGFP gene upstream of the
wild-type AR 3′UTR, EGFP fluorescence was strongly reduced in the
presence of miR-30a overexpression. However, the overexpression of
miR-30a did not affect the fluorescence intensity of the mutant AR
3′UTR (Fig. 2C). Thus, miR-30a
regulates wild-type AR expression directly through the UTR.
Activated AR cannot load the promoter
of miR-30a
We hypothesized that androgen-induced AR activating
signal loaded onto the 5′DNA region of the miR-30a locus may serve
as a transcription factor. A typical TATA box was found in the 5′
region of the miR-30a gene when the length up to 4.0 kb was
analyzed. Using the PROMO 3.0 program, six potential AREs were
identified in the 5′ region of miR-30a, suggesting the presence of
a promoter in the 5′ region. Among these AREs, ARE2 and ARE6 are
the most likely to be loaded by activated AR. To determine AR
loading, ChIP was performed with two primer pairs to amplify the
−2843/−2623 fragment corresponding to ARE2 and the −1513/−1286
fragment corresponding to ARE6. Treatment of MDA-MB-453 cells with
10−8 M DHT did not induce an increase in AR loading at
ARE2 or ARE6. Taken together, the results suggest that androgen-AR
signaling does not directly mediate the regulation of miR-30a in
MDA-MB-453 cells.
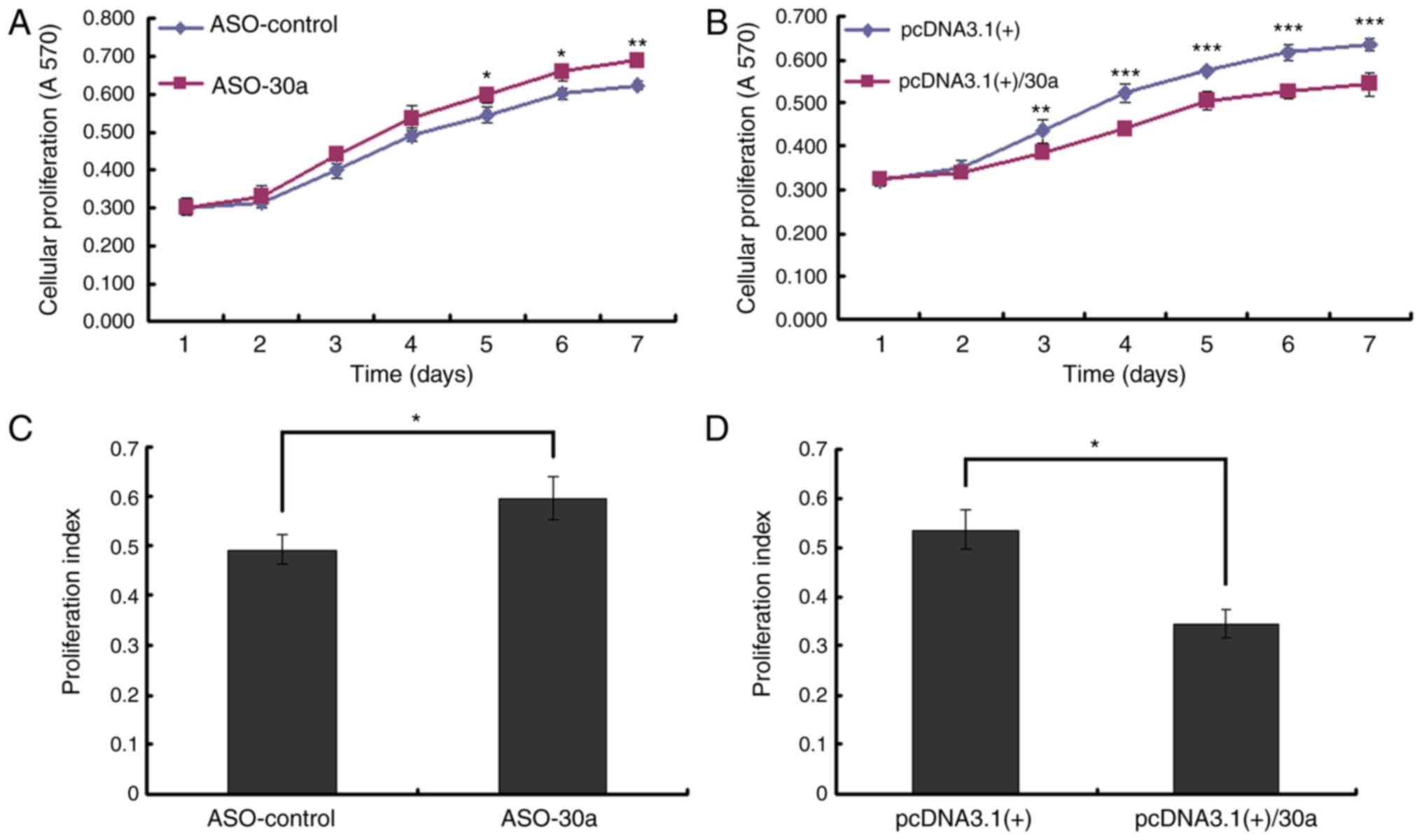
MiR-30a inhibits the growth of
MDA-MB-453 cells
The MTT assay was used to measure viable
proliferating cells 1–7 days after MDA-MB-453 cells were
transfected. Blocking miR-30a with miR-30a ASO increased cell
growth (Fig. 3A). Overexpressing
miR-30a inhibited cell growth, an opposite effect as that observed
with low miR-30a expression (Fig.
3B). Forty-eight h after transfection, the number of cells in S
phase significantly increased and the PI increased when miR-30a was
blocked (Fig. 3C). When miR-30a was
overexpressed, the number of cells in G1 phase significantly
increased, G1-S arrest was obvious, and the PI decreased. An
opposite effect was observed in cells with low levels of miR-30a
overexpression (Fig. 3D).
Discussion
In this study, we found that miR-30a,b,c are
down-regulated by androgen-induced AR activating signal and AR is
up-regulated in MDA-MB-453 cells affected by DHT. Two
bioinformatics programs, miRanda and TargetScan, predicted
miR-30a,b,c as upstream microRNAs of AR because the AR mRNA 3′UTR
contained a putative binding site for miR-30a,b,c. We then chose
miR-30a for further experiments because it is the most
down-regulated among miR-30a,b,c. EGFP reporter assay confirmed
that AR is the downstream target of miR-30a, but ChIP assay showed
that androgen-AR signaling does not directly mediate the regulation
of miR-30a. Therefore, other mechanisms potentially lead to the
down-regulation of miR-30a by AR activating signal. Some studies
have shown that AR and miR-30a are involved in the
epithelial-mesenchymal transition in breast cancer (25–28), and
vimentin is a specific bridge between AR and miR-30a (26,27). Feng
et al reported that AR activated by DHT potentiates the
promoter activity of vimentin and upregulates vimentin expression
(27), whereas Cheng et al
showed that vimentin is the target of miR-30a (26). Activated AR may upregulate vimentin,
resulting in downregulated miR-30a because of the binding between
miR-30a and vimentin. In addition, Guo et al reported that
miR-30a is a novel downstream target of p53 R273H mutant, which
binds to the promoter region to repress miR-30a expression
(29). Therefore, we can assume that
p53 is regulated by AR, and AR indirectly downregulates miR-30a via
p53. These assumptions are worthy of study in the future.
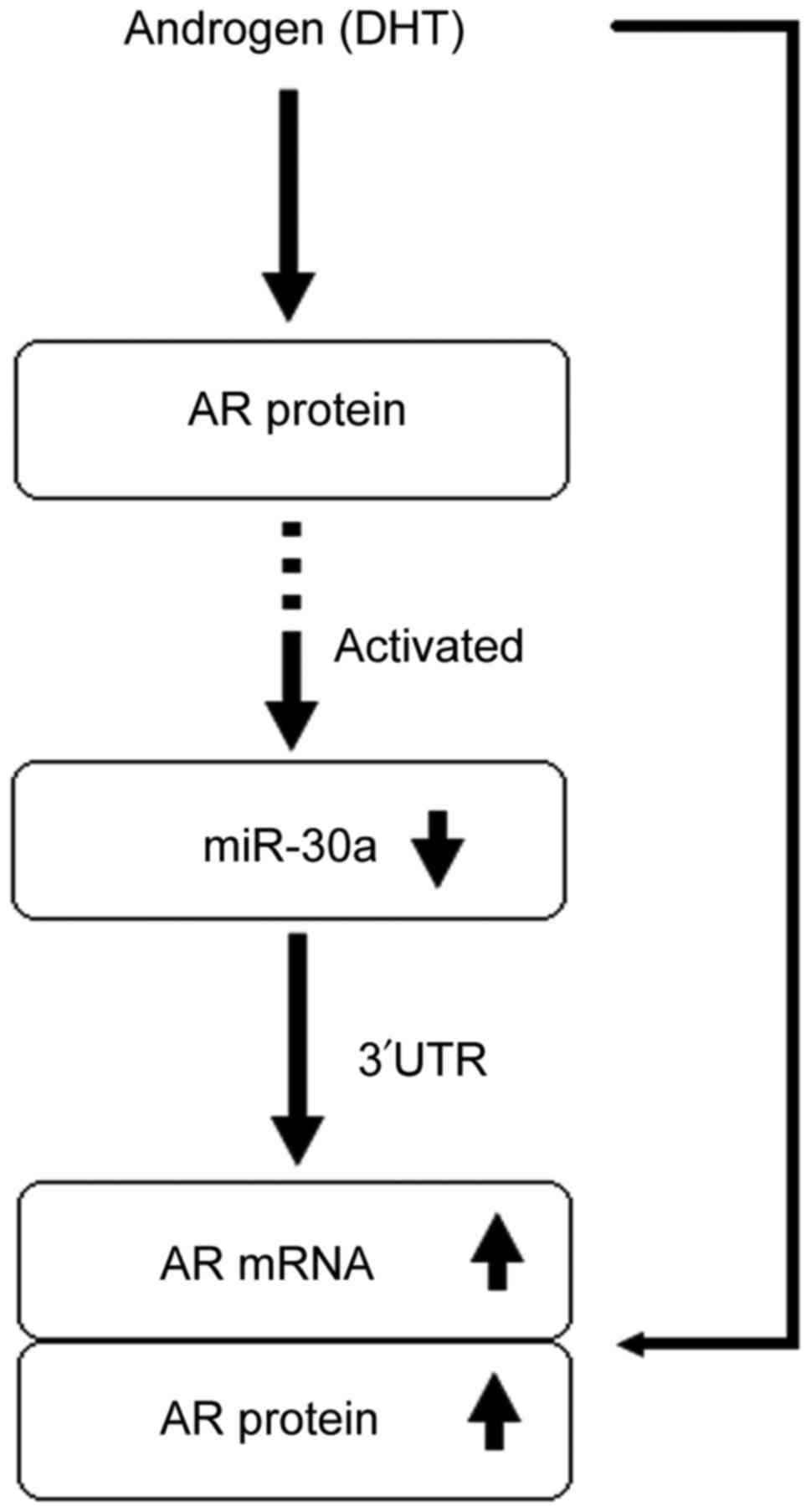
The positive feedback signaling pathway that finally
activates AR activating signal, up-regulating AR, follows a
specific process. First, DHT activates the AR activating signal,
down-regulating miR-30a. Second, the down-regulation of miR-30a
relieves the down-regulating effect of miR-30a on AR. Our previous
study revealed that DHT decreases MDA-MB-453 cell proliferation
(14). In the present study, MTT
assay and flow cytometry revealed that miR-30a inhibits the growth
of MDA-MB-453 cells. The results are similar to other studies in
which miR-30a functioned as a cancer suppressor gene in breast
cancer (26,30,31).
However, in this study, because AR activating signal down-regulates
miR-30a expression, the growth of MDA-MB-453 cells should be
promoted because miR-30a inhibits their growth.
We conclude that miR-30a has two different effects
on MDA-MB-453 cell proliferation via AR activating signal. One
effect is through increased AR expression, as AR activating signal
down-regulates miR-30a expression and then relieves the inhibition
of AR expression by miR-30a. The other effect is through activating
AR signal down-regulating miR-30a expression and relieving the
inhibition of cell growth by miR-30a. The first effect is more
likely than the latter when treating cells with DHT. We also found
that AR activating signal inhibits the proliferation of MDA-MB-453
cells.
In conclusion, we found an interrelation between AR
and miR-30a in human ER−, PR−, AR+
MDA-MB-453 breast cancer cells (Fig.
4). MiR-30a and AR create a positive feedback mechanism in that
leads to AR activating signal inhibiting cell proliferation. In
addition, as a cancer suppressor gene, miR-30a inhibits the growth
of MDA-MB-453 cells and is down-regulated to promote their growth.
The opposite effects of miR-30a in the function of AR activating
signal in MDA-MB-453 cells are very interesting. The results show
that the mechanism underlying AR activating signal regulation of
related microRNAs and AR is complex and waiting to be explored.
Even though the results are for only one cell line, they contribute
to subsequent studies of ER−, PR−,
AR+ breast cancer.
Acknowledgements
This research was supported by the Scientific and
Technological Development Fund of Tianjin Scientific Technological
Committee (09JCYBJC10100), National Natural Science Foundation of
China (81172532), and the Program for Changjiang Scholars and
Innovative Research Team in University (TRT0743).
Glossary
Abbreviations
Abbreviations:
|
ER
|
estrogen receptor
|
|
PR
|
progesterone receptor
|
|
AR
|
androgen receptor
|
References
|
1
|
Gucalp A and Traina TA: Triple-negative
breast cancer: Role of the androgen receptor. Cancer J. 16:62–65.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gucalp A, Tolaney S, Isakoff SJ, Ingle JN,
Liu MC, Carey LA, Blackwell K, Rugo H, Nabell L, Forero A, et al:
Phase II trial of bicalutamide in patients with androgen
receptor-positive, estrogen receptor-negative metastatic Breast
Cancer. Clin Cancer Res. 19:5505–5512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baniahmad A: Nuclear hormone receptor
co-repressors. J Steroid Biochem Mol Biol. 93:89–97. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dehm SM and Tindall DJ: Molecular
regulation of androgen action in prostate cancer. J Cell Biochem.
99:333–344. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Díaz-Chico Nicolás B, Rodríguez Germán F,
González A, Ramírez R, Bilbao C, de León Cabrera A, Jaime Aguirre
A, Chirino R, Navarro D and Díaz-Chico JC: Androgens and androgen
receptors in breast cancer. J Steroid Biochem Mol Biol. 105:1–15.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bussing I, Slack FJ and Grosshans H: let-7
microRNAs in development, stem cells and cancer. Trends Mol Med.
14:400–409. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: Key players in the immune system, differentiation,
tumorigenesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stefani G and Slack FJ: Small non-coding
RNAs in animal development. Nat Rev Mol Cell Biol. 9:219–230. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi XB, Tepper CG and deVere White RW:
Cancerous miRNAs and their regulation. Cell Cycle. 7:1529–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu
M, Tepper CG, Evans CP, Kung HJ and deVere White RW: An
androgen-regulated miRNA suppresses Bak1 expression and induces
androgen-independent growth of prostate cancer cells. Proc Natl
Acad Sci USA. 104:19983–19988. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takayama K, Tsutsumi S, Katayama S,
Okayama T, Horie-Inoue K, Ikeda K, Urano T, Kawazu C, Hasegawa A,
Ikeo K, et al: Integration of cap analysis of gene expression and
chromatin immunoprecipitation analysis on array reveals genome-wide
androgen receptor signaling in prostate cancer cells. Oncogene.
30:619–630. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lyu S, Yu Q, Ying G, Wang S, Wang Y, Zhang
J and Niu Y: Androgen receptor decreases CMYC and KRAS expression
by upregulating let-7a expression in ER-, PR-, AR+ breast cancer.
Int J Oncol. 44:229–237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Epis MR, Giles KM, Barker A, Kendrick TS
and Leedman PJ: miR-331-3p regulates ERBB-2 expression and androgen
receptor signaling in prostate cancer. J Biol Chem.
284:24696–24704. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hagman Z, Haflidadóttir BS, Ceder JA,
Larne O, Bjartell A, Lilja H, Edsjö A and Ceder Y: miR-205
negatively regulates the androgen receptor and is associated with
adverse outcome of prostate cancer patients. Br J Cancer.
108:1668–1676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi XB, Xue L, Ma AH, Tepper CG,
Gandour-Edwards R, Kung HJ and deVere White RW: Tumor suppressive
miR-124 targets androgen receptor and inhibits proliferation of
prostate cancer cells. Oncogene. 32:4130–4138. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Q, Niu Y, Liu N, Zhang JZ, Liu TJ,
Zhang RJ, Wang SL, Ding XM and Xiao XQ: Expression of androgen
receptor in breast cancer and its significance as a prognostic
factor. Ann Oncol. 22:1288–1294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hall RE, Birrell SN, Tilley WD and
Sutherland RL: MDA-MB-453, an androgen-responsive human breast
carcinoma cell line with high level androgen receptor expression.
Eur J Cancer. 30A:484–490. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yeap BB, Krueger RG and Leedman PJ:
Differential posttranscriptional regulation of androgen receptor
gene expression by androgen in prostate and breast cancer cells.
Endocrinology. 140:3282–3291. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vranic S, Gatalica Z and Wang ZY: Update
on the molecular profile of the MDA-MB-453 cell line as a model for
apocrine breast carcinoma studies. Oncol Lett. 2:1131–1137.
2011.PubMed/NCBI
|
|
22
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Louie MC, Yang HQ, Ma AH, Xu W, Zou JX,
Kung HJ and Chen HW: Androgen-induced recruitment of RNA polymerase
II to a nuclear receptor-p160 coactivator complex. Proc Natl Acad
Sci USA. 100:2226–2230. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seifer DB, MacLaughlin DT, Penzias AS,
Behrman HR, Asmundson L, Donahoe PK, Haning RV Jr and Flynn SD:
Gonadotropin-releasing hormone agonist-induced differences in
granulosa cell cycle kinetics are associated with alterations in
follicular fluid mullerian-inhibiting substance and androgen
content. J Clin Endocrinol Metab. 76:711–714. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu YN, Liu Y, Lee HJ, Hsu YH and Chen JH:
Activated androgen receptor downregulates E-cadherin gene
expression and promotes tumor metastasis. Mol Cell Biol.
28:7096–7108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng CW, Wang HW, Chang CW, Chu HW, Chen
CY, Yu JC, Chao JI, Liu HF, Ding SL and Shen CY: MicroRNA-30a
inhibits cell migration and invasion by downregulating vimentin
expression and is a potential prognostic marker in breast cancer.
Breast Cancer Res Treat. 134:1081–1093. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng J, Li L, Zhang N, Liu J, Zhang L, Gao
H, Wang G, Li Y, Zhang Y, Li X, et al: Androgen and AR contribute
to breast cancer development and metastasis: An insight of
mechanisms. Oncogene. 36:2775–2790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chang CW, Yu JC, Hsieh YH, Yao CC, Chao
JI, Chen PM, Hsieh HY, Hsiung CN, Chu HW, Shen CY and Cheng CW:
MicroRNA-30a increases tight junction protein expression to
suppress the epithelial-mesenchymal transition and metastasis by
targeting Slug in breast cancer. Oncotarget. 7:16462–16478. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo F, Chen H, Chang J and Zhang L:
Mutation R273H confers p53 a stimulating effect on the IGF-1R-AKT
pathway via miR-30a suppression in breast cancer. Biomed
Pharmacother. 78:335–341. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J,
Cheng L, Fan Z, Yuan B, Tian P, et al: miR-30a suppresses breast
cancer cell proliferation and migration by targeting Eya2. Biochem
Biophys Res Commun. 445:314–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xiong J, Wei B, Ye Q and Liu W:
MiR-30a-5p/UBE3C axis regulates breast cancer cell proliferation
and migration. Biochem Biophys Res Commun. 2016.(Epub ahead of
print). View Article : Google Scholar
|