Introduction
Brain and central nervous system (CNS) tumors are
the second most common type of cancer in children, comprising ~21%
of cases, and the third most common cancer type in adolescents,
contributing to ~10% of cases (1). It
is estimated that >23,800 (13,450 males and 10,350 females) new
cases of brain and CNS tumors will be diagnosed and 16,700 (9,620
men and 7,080 women) brain and CNS tumor-associated mortalities
will occur in the United States in 2017 (http://www.cancer.org) (2,3). The
incidence of brain tumors in Malaysia was relatively low,
accounting for ~1.95% of all cancer cases prior to 2003 (4). Since then, the incidence rate has been
increasing rapidly, and brain and CNS tumors have become the third
most common type of pediatric cancer in Malaysia, behind leukemia
and lymphoma (5,6). In 2012, the incidence rate was
4.6/100,000 individuals/year (6).
The overall prognosis for brain tumors is based on
tumor pathology or grade (7,8). The majority of patients who are
diagnosed with malignant primary brain tumors have a poor prognosis
(7,8).
Therefore, it is crucial to identify novel potential bio-tumor
markers for brain tumors in order to improve the diagnosis,
prognosis and treatment of the disease.
Mitochondria have long been considered as crucial
organelles, since they contain their own DNA (9). Prior research shows that mitochondria
have a variety of roles in energy metabolism and cellular
homeostasis, including ATP production, reactive oxygen species
(ROS) production, metabolic homeostasis and apoptosis (10,11). Human
mitochondrial DNA (mtDNA) is a closed circular, double-stranded
molecule of ~16.5 kb (12). It
contains genes coding for 13 polypeptide components of respiratory
chain enzyme complexes (complex I, III, IV and V), two ribosomal
RNAs (rRNAs) and 22 transfer RNAs (tRNAs), which are components of
the mitochondrial protein translation system (12,13). mtDNA
is also composed of a non-coding region known as the
displacement-loop (D-loop), located between nucleotides 16,024 and
576, which contain essential elements that are responsible for the
transcription and replication of the mitochondrial genome (14). Furthermore, mtDNA is considered to
have a much higher mutation rate than nuclear DNA and is more
sensitive to oxidative damage (15).
As mtDNA is in close proximity to the respiratory chain, it is
constantly exposed to endogenous ROS produced by the mitochondria
that can damage DNA (16).
The first report of mtDNA mutations was described in
1998 by Polyak et al (17) in
human colorectal cancer. Since then, numerous studies have been
reported with >200 mtDNA mutation and/or alteration cases in
cancer published worldwide (18,19).
Somatic mutations of mtDNA have been identified in multiple cancer
types, including breast cancer, colorectal cancer, hepatocellular
carcinoma, gastric cancer and lung cancer (19–24).
Therefore, mtDNA is considered to serve an important role in tumor
progression and carcinogenesis. The mtDNA D-loop has been
identified as a hot spot of genetic alterations in human cancer
(25–27). Accumulation of mtDNA D-loop
alterations may contribute to altered replication and/or
transcription of mitochondrial genes, which may lead to
mitochondrial dysfunction and excessive cellular ROS
production.
Although there have been several alterations in
mtDNA reported in patients with brain tumors (28–30), the
contribution of mtDNA mutations to brain tumorigenesis remains
unclear and requires additional exploration. As data on
mtDNA alterations was not available for Malaysian patients with
brain tumors, the D-loop mtDNA alterations were examined in
patients with brain tumors of diverse types and grades. The aims of
the present study were to identify mtDNA D-loop alterations, and to
assess their association with clinicopathological features of brain
tumors.
Materials and methods
Tumor specimens
Paired samples of brain tumor tissue and blood
samples from 49 patients were collected during elective
neurosurgical procedures at the Department of Neurosciences,
Hospital Universiti Sains Malaysia, Kubang Kerian, Kelantan,
Malaysia between July 2010 and December 2014. Peripheral blood
specimens were obtained from the same patients as a the control.
The study population consisted of 29 males (59.2%) and 20 females
(40.8%) with age of patients ranging between 2 and 74 years (mean,
44.2 years). The histopathological diagnosis of the brain tumor
samples was determined according to the World Health Organization
(WHO) criteria (31) by a consultant
neuropathologist. These neoplasms comprised 6 pilocytic astrocytoma
WHO grade I (PA), 2 astrocytoma WHO grade II (A II), 5 anaplastic
astrocytoma WHO grade III (AA III), 16 glioblastoma multiforme WHO
grade IV (GBM IV), 3 oligodendroglioma WHO grade II (ODG), 2
ependymoma WHO grade II (EP) and 15 meningioma WHO grade I. The
protocol was approved by the Research Ethics Committee of
Universiti Sains Malaysia, and all patients provided written
informed consent for participation. The tumor tissue biopsies and
peripheral blood specimens (5 ml), obtained from all patients, were
snap-frozen in liquid nitrogen and stored at −80°C until DNA
extraction.
DNA extraction
Genomic DNA was extracted from brain tumor tissues
and blood samples of patients using the QIAamp DNA mini kit (Qiagen
GmbH, Hilden, Germany), according to the manufacturer's protocol.
The concentration and quality of extracted DNA was measured using a
NanoDrop ND1000 spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Inc., Pittsburgh, PA, USA) and 1% agarose gel
electrophoresis stained with 10 mg/ml ethidium bromide (Invitrogen;
Thermo Fisher Scientific, Inc.).
Polymerase chain reaction (PCR)
amplification of the mtDNA D-loop
Three sets of primers were designed to amplify or
target three independent but overlapping fragments that span the
whole D-loop region, as listed in Table
I. Briefly, PCR was performed on the SureCycler 8800 Thermal
Cycler (Agilent Technologies, Inc., Santa Clara, CA, USA), in a 50
µl reaction mixture containing 100 ng DNA template, 200 µM each
dNTP, 20 pMol each primer, 10 µl 5X Phusion HF buffer (Thermo
Fisher Scientific, Inc.) and 2 U Phusion high-fidelity DNA
polymerase (Thermo Fisher Scientific, Inc.).
 | Table I.Polymerase chain reaction primers and
the expected sizes of amplicons. |
Table I.
Polymerase chain reaction primers and
the expected sizes of amplicons.
| Primer | Forward primer,
5′-3′ | Reverse primer,
5′-3′ | Band size, bp |
|---|
| Mito-D-loop 1 |
5′-CCTATGTCGCAGTATCTGTC-3′ |
5′-TGCTTTGAGGAGGTAAGCTA-3′ | 491 bp (np.
113-603)a |
| Mito-D-loop 2 |
5′-GTCTTGTAAACCGGAGATGA-3′ |
5′-GAGCGAGGAGAGTAGCAC-3′ | 539 bp (np.
15,915–16,453)a |
| Mito-D-loop 3 |
5′-TACAGTCAAATCCCTTCTCG-3′ |
5′-AATAGGATGAGGCAGGAATC-3′ | 383 bp (np.
16,342–155)a |
The cycling profile was set at 98°C initial
denaturation for 1 min, followed by 35 cycles of denaturation at
98°C for 20 sec, annealing at 56°C for 20 sec, extension at 72°C
for 20 sec and a final extension at 72°C for 5 min. The amplified
PCR fragments were analyzed via 2% agarose gel electrophoresis
using a GeneRuler 100 bp DNA ladder (Thermo Fisher Scientific,
Inc.) in order to determine the expected size of the amplified
PCR fragments. The PCR products with the expected size were
purified with the QIAquick PCR Purification kit (Qiagen GmbH)
according to the manufacturer's protocol and stored at −20°C until
use for DNA sequencing analysis.
Direct sequencing of the mtDNA
D-loop
Purified PCR products were sequenced in both
directions using the same primers as described for the PCR
reactions (Table I). Sequencing was
performed using a Big Dye Terminator cycle sequencing kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions, on an ABI Prism 3700 DNA Analyzer
automated sequencer (Applied Biosystems; Thermo Fisher Scientific,
Inc.). DNA sequencing and electropherogram results were analyzed
manually and aligned using BLAST software from the NCBI site
(http://www.ncbi.nlm.nih.gov/blast),
and then compared with the published revised Cambridge Reference
Sequence (rCRS) of the human mtDNA (NC_012920) in the MITOMAP
database (http://www.mitomap.org).
Statistical analysis
The data were analyzed using GraphPad Prism software
version 5 (GraphPad Software, Inc., La Jolla, CA, USA) with
Fisher's exact test to calculate the significance of associations
between mtDNA D-loop mutations and the clinicopathological
parameters of brain tumor samples. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
Brain tumor tissue specimens from 49 Malaysian
patients were obtained at the same time as blood samples, which
were used as controls. If the mtDNA sequence in the tumor differed
from the corresponding blood sample, this was described as a
somatic mutation. The mean age of the patients at the time of
initial surgery was 44.2 years (range, 2–74 years). Of these, 29
were males (59.2%) and 20 were females (40.8%). In this
retrospective cohort, GBM IV represents 32.7% (n=16) of all primary
brain tumors, followed by meningioma (n=15, 30.6%), PA I (n=6,
12.2%), AA III (n=5, 10.2%), ODG (n=3, 6.1%), A II (n=2, 4.1%) and
EP (n=2, 4.1%).
Somatic mtDNA D-loop mutations status
in brain tumors
A total of three overlapping fragments that spanned
the whole 1,122 bp D-loop region were amplified in 49 brain tumor
tissue samples and corresponding blood samples. The successful
amplification of three fragments resulted in the amplified
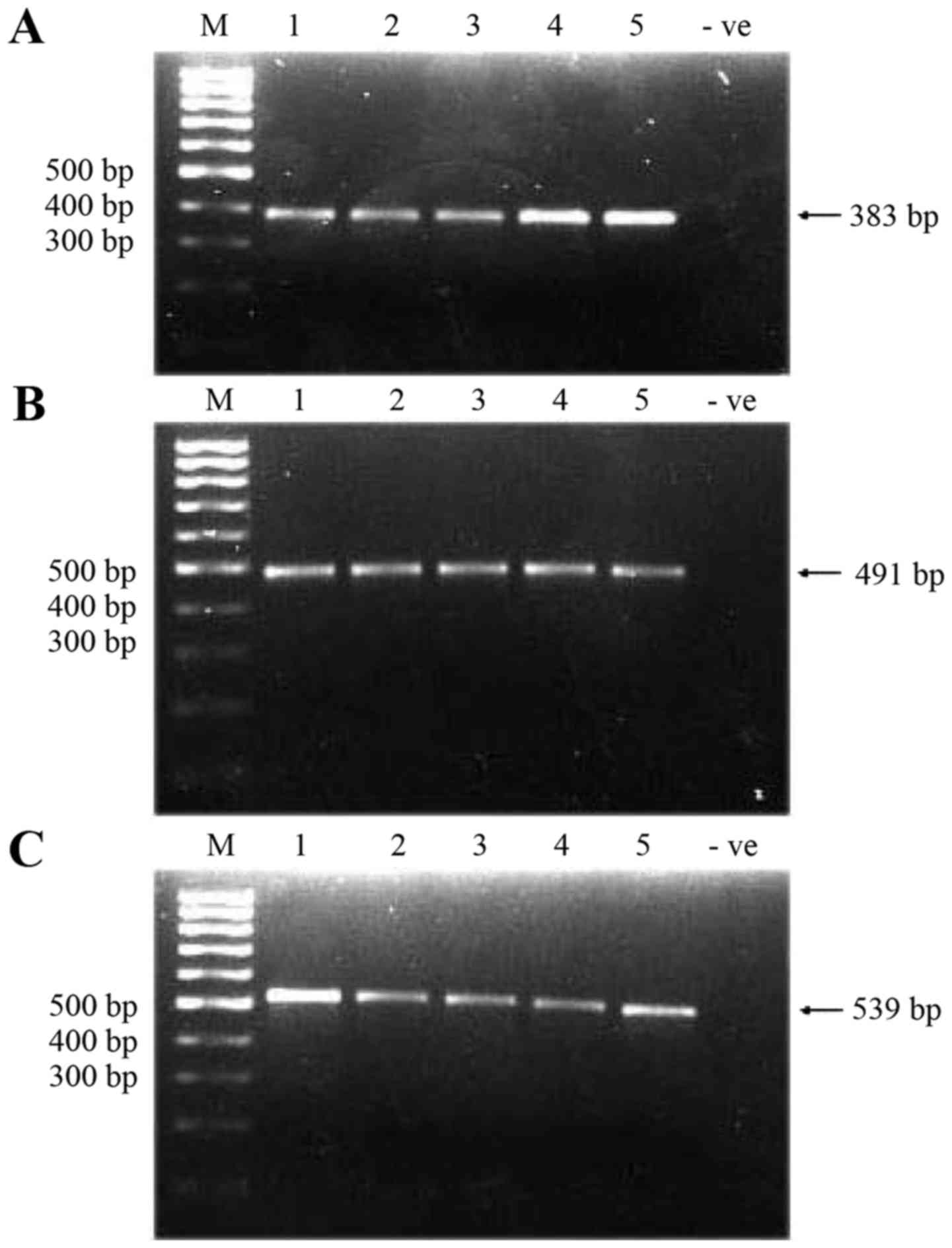
fragments of sizes 383, 491 and 539 bp (Fig. 1) (the total size of amplified
fragments was 1,413 bp, 79% more than the entire D-loop due to the
overlapping regions). Amplified fragments/products were then
sequenced, and the sequenced data obtained were compared with the
rCRS in the MITOMAP database.
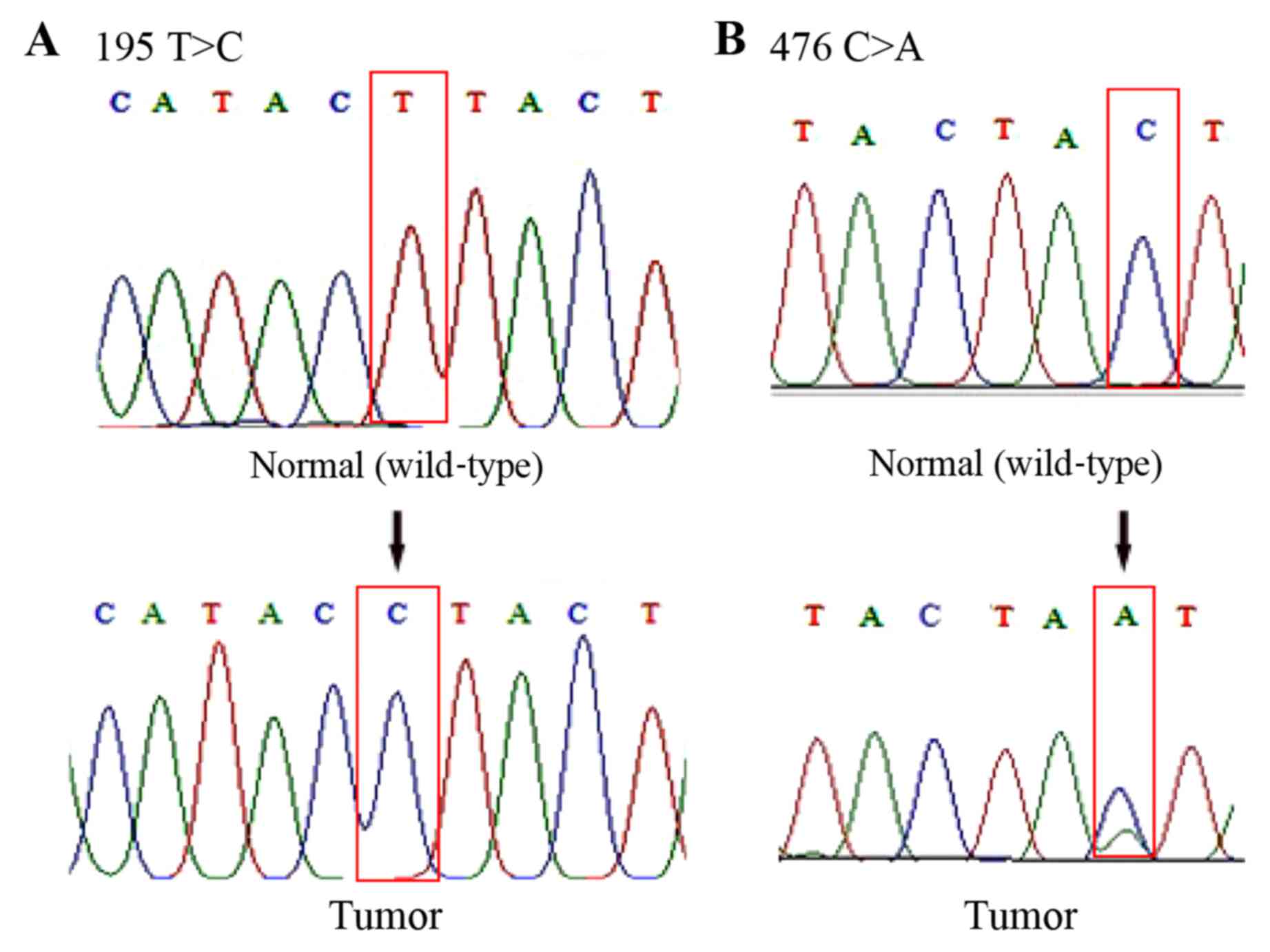
By analyzing the sequencing data, it was observed
that 51% (25/49) of the patients carried a total of 48 somatic
mutations at 27 positions in the D-loop of mtDNA (Table II). Among these mutations, four (176
A-deletion, 476C>A, 566C>A and 16405 A-deletion) had not
previously been recorded in the MITOMAP database (Fig. 2). Therefore, it was assumed that they
are novel mutations. Mutations in nucleotide 195T>C, 146T>C,
152 T>C, 204 T>C, 303 CC-insertion, 311 C-insertion and
16519T>C were also found, which have previously been reported in
brain tumors (28,29,32). In
addition, 94% (45/48) of these mutations were homoplasmic and 19%
(9/48) of them were located in the D310 mononucleotide repeat (np
303–315). Representative DNA sequence chromatograms are presented
in Fig. 2.
 | Table II.Mitochondrial DNA D-loop somatic
mutations in patients with brain tumors. |
Table II.
Mitochondrial DNA D-loop somatic
mutations in patients with brain tumors.
| Patient code | Tumor type | Nucleotide
position | Somatic
mutation |
Homoplasmy/heteroplasmy | Novel/reported | (Refs.) |
|---|
| ID003 | GBM IV | 16265 | A-G | Homoplasmy | Bladder | (33) |
|
|
|
|
|
| Leukemia | (34) |
|
|
|
|
|
| Prostate
cancer | (35) |
| ID004 | GBM IV | 303 | CC insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Gastric cancer | (36) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Oral | (38 |
|
|
| 414 | T-G | Homoplasmy | Colorectal | (39) |
|
|
| 476a | C-A/C | Heteroplasmy | Novela | Present study |
| ID005 | GBM IV | 195 | T-C | Homoplasmy | Brain tumor | (28,29) |
|
|
|
|
|
| Ovarian | (40) |
|
|
|
|
|
| Breast | (41) |
|
|
|
|
|
| Leukemia | (42) |
| ID006 | AA III | 16356 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Breast | (41) |
| ID007 | Meningioma I | 249 | A deletion | Homoplasmy | Nasopharyngeal | (43) |
|
|
|
|
|
| carcinoma | (44) |
|
|
|
|
|
| Breast | (35a) |
|
|
|
|
|
| Prostate
cancer |
|
|
|
| 73 | A-G | Homoplasmy | Prostate
cancer | (45) |
|
|
|
|
|
| Brain tumor | (29) |
|
|
|
|
|
| Breast | (41) |
|
|
|
|
|
| Leukemia | (42) |
| ID008 | GBM IV | 249 | A deletion | Homoplasmy | Nasopharyngeal | (43) |
|
|
|
|
|
| carcinoma | (44) |
|
|
|
|
|
| Breast | (35a) |
|
|
|
|
|
| Prostate |
|
|
|
| 311 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
|
|
| 73 | A-G | Homoplasmy | Prostate
cancer | (45) |
|
|
|
|
|
| Brain tumor | (29) |
|
|
|
|
|
| Breast | (41) |
|
|
|
|
|
| Leukemia | (42) |
|
|
| 16356 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Breast | (41) |
| ID011 | PA | 303 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Gastric cancer | (36) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Oral | (38) |
|
|
| 311 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
| ID013 | PA | 16325 | T-C | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Leukemia | (34) |
|
|
|
|
|
| Breast | (37) |
| ID014 | AA III | 249 | A deletion | Homoplasmy | Nasopharyngeal | (43) |
|
|
|
|
|
| carcinoma | (44) |
|
|
|
|
|
| Breast | (35a) |
|
|
|
|
|
| Prostate |
|
|
|
| 311 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
| ID017 | GBM IV | 311 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
|
|
| 511 | C-T | Homoplasmy | Cervical
cancer | (46) |
| ID019 | Meningioma I | 146 | T-C | Homoplasmy | Brain tumor | (29) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
|
|
|
|
|
| Oral | (38) |
|
|
| 204 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Ovarian | (47) |
|
|
|
|
|
| Leukemia | (34,42) |
|
|
| 16519 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Leukemia | (34) |
|
|
|
|
|
| Gastric cancer | (36) |
|
|
|
|
|
| Breast | (41) |
| ID020 | GBM IV | 195 | T-C | Homoplasmy | Brain tumor | (28,29) |
|
|
|
|
|
| Ovarian | (40) |
|
|
|
|
|
| Breast | (41) |
|
|
|
|
|
| Leukemia | (42) |
|
|
| 303 | CC insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Gastric cancer | (36) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Oral | (38) |
|
|
| 411 | C-G/C | Heteroplasmy | Leukemia | (34) |
|
|
| 16381 | T-C | Homoplasmy | Reported in
MITOMAP | MITOMAP |
| ID021 | Meningioma I | 176a | A deletion | Homoplasmy | Novela | Present study |
|
|
| 16265 | A-G | Homoplasmy | Bladder | (33) |
|
|
|
|
|
| Leukemia | (34) |
|
|
|
|
|
| Prostate
cancer | (35) |
| ID022 | Meningioma I | 411 | C-G | Homoplasmy | Leukemia | (34) |
|
|
| 432 | A-C | Homoplasmy | Reported in
MITOMAP | MITOMAP |
| ID024 | GBM IV | 186 | C-G | Homoplasmy | Oral cancer | (48) |
| ID026 | GBM IV | 71 | G deletion | Homoplasmy | Oral cancer | (49) |
|
|
| 311 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
| ID027 | GBM IV | 61 | C-A | Homoplasmy | Reported in
MITOMAP | MITOMAP |
|
|
| 16381 | T-C | Homoplasmy | Reported in
MITOMAP | MITOMAP |
|
|
| 16405a | A deletion | Homoplasmy | Novela | Present study |
| ID030 | GBM IV | 311 | C insertion | Homoplasmy | Brain tumor | (32) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
|
|
| 523–524 | AC deletion | Homoplasmy | Reported in
MITOMAP | MITOMAP |
| ID036 | Meningioma I | 503 | A-G | Homoplasmy | Reported in
MITOMAP | MITOMAP |
|
|
| 566a | C-A | Homoplasmy | Novela | Present study |
| ID038 | Meningioma I | 146 | T-C | Homoplasmy | Brain tumor | (29) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
|
|
|
|
|
| Oral | (38) |
|
|
| 152 | T-C/T | Heteroplasmy | Brain tumor | (29) |
|
|
|
|
|
| Breast | (20,41) |
|
|
|
|
|
| Laryngeal
carcinoma | (50) |
| ID039 | Meningioma I | 472 | A-G | Homoplasmy | Reported in
MITOMAP | MITOMAP |
| ID042 | GBM IV | 16519 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Leukemia | (34) |
|
|
|
|
|
| Gastric cancer | (36) |
|
|
|
|
|
| Breast | (41) |
| ID044 | GBM IV | 146 | T-C | Homoplasmy | Brain tumor | (29) |
|
|
|
|
|
| Breast | (37) |
|
|
|
|
|
| Leukemia | (42) |
|
|
|
|
|
| Oral | (38) |
|
|
| 204 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Ovarian | (47) |
|
|
|
|
|
| Leukemia | (34,42) |
| ID045 | Meningioma I | 176a | A deletion | Homoplasmy | Novela | Present study |
| ID047 | Ependymoma | 16519 | T-C | Homoplasmy | Brain tumor | (28) |
|
|
|
|
|
| Leukemia | (34) |
|
|
|
|
|
| Gastric cancer | (36) |
|
|
|
|
|
| Breast | (41) |
mtDNA D-loop polymorphism status in
brain tumors
Any DNA sequence variation present in the peripheral
blood and tumor tissue of patients was classified as a
polymorphism. In the present study, 210 polymorphisms were
identified at 26 nucleotide positions (Table III). The majority of polymorphisms
identified in the present study were found in the nucleotides
263A>G, 16189T>C, 16261C>T and 16271T>C.
 | Table III.Mitochondrial DNA D-loop
polymorphisms in patients with brain tumors. |
Table III.
Mitochondrial DNA D-loop
polymorphisms in patients with brain tumors.
| Nucleotide
position | Base variation | Cases, n |
|---|
| 143 | G→A | 2 |
| 189 | A→G | 1 |
| 199 | T→C | 3 |
| 263 | A→G | 20 |
| 318 | T→C | 3 |
| 16093 | T→C | 10 |
| 16129 | G→A | 13 |
| 16162 | A→G | 11 |
| 16172 | T→C | 4 |
| 16189 | T→C | 23 |
| 16223 | C→T | 12 |
| 16256 | C→T | 10 |
| 16259 | T→C | 5 |
| 16261 | C→T | 24 |
| 16271 | T→C | 19 |
| 16278 | C→T | 5 |
| 16288 | T→C | 8 |
| 16290 | C→T | 2 |
| 16298 | T→C | 2 |
| 16304 | T→C | 9 |
| 16309 | A→G | 5 |
| 16311 | T→C | 3 |
| 16319 | G→A | 6 |
| 16325 | T→C | 2 |
| 16362 | T→C | 3 |
| 16390 | G→A | 5 |
Associations between D-loop mutation
status and clinicopathological parameters of brain tumors
Table IV describes
the D-loop mutation status and its association with patient gender,
age, race and histological tumor type. No significant association
was observed between the D-loop mutation status and gender (P=1.0);
however, the number of patients with D-loop mutations tends to be
higher in males (n=15/25, 60%) than females (n=10/25, 40%).
 | Table IV.Clinicopathological characteristics
of patients with brain tumors and D-Loop mutation status. |
Table IV.
Clinicopathological characteristics
of patients with brain tumors and D-Loop mutation status.
|
| D-loop status, n
(%) |
|---|
|
|
|
|---|
| Parameter | Total patients, no.
(%) | Mutation | No mutation | P-value |
|---|
| No. of
patients | 49 (100) | 25 (51.0) | 24 (49.0) |
|
| Sex |
|
|
|
|
|
Male | 29 (59.2) | 15 (51.7) | 14 (48.3) | 1.000 |
|
Female | 20 (40.8) | 10 (50.0) | 10 (50.0) |
|
| Age, years |
|
|
|
|
|
<45 | 20 (40.8) | 7
(35.0) | 13 (65.0) | 0.0845 |
|
≥45 | 29 (59.2) | 18 (62.1) | 11 (37.9) |
|
| Ethnicity |
|
|
|
|
|
Malaysian | 42 (85.7) | 22 (52.4) | 20 (47.6) | 0.8716 |
|
Chinese | 5
(10.2) | 2
(40.0) | 3
(60.0) |
|
|
Indian | 2
(4.1) | 1
(50.0) | 1
(50.0) |
|
| Histological tumor
types (grade) |
|
|
|
|
|
Pilocytic astrocytoma (I) | 6
(12.2) | 2
(33.3) | 4
(66.7) | 0.1282 |
|
Astrocytomas (II) | 2 (4.1) | 0 (0) | 2 (100) |
|
|
Anaplastic astrocytomas
(III) | 5
(10.2) | 2
(40.0) | 3
(60.0) |
|
|
Glioblastomas multiform
(IV) | 16 (32.7) | 12 (75.0) | 4
(25.0) |
|
|
Ependymoma | 2 (4.1) | 1
(50.0) | 1
(50.0) |
|
|
Oligodendroglioma | 3 (6.1) | 0 (0) | 3 (100) |
|
|
Meningioma (I) | 15 (30.6) | 8
(53.3) | 7
(46.7) |
|
In the <45 and ≥45 year groups, the association
was determined to be non-significant, although D-loop mutations
were observed to be more evident in the ≥45 year group (n=18/25,
72%) compared with in the <45 year group (n=7/25, 28%). In
addition, mutations of the D-loop had no significant association
with the ethnicity of patients.
As presented in Table
IV, the D-loop mutations were frequent in GBM IV (n=12/25,
48%), followed by distribution in meningioma (n=8/25, 32%), and
equal in PA I and AA III (n=2/25, 8%). Furthermore, no significant
association between D-loop mutations and any of these histological
tumor types was observed.
Discussion
DNA mutation in mitochondria is one of the most
common genetic alterations in cancer (51,52) and
has been widely investigated (51–53). mtDNA
alterations were reported to be frequently activated via certain
nucleotide changes in the control region that is known as the
D-loop region (17,18,54). This
area has been identified as a hot spot region for somatic mtDNA
mutations in various types of human cancer, including breast,
colorectal and lung cancer (55–58). Thus
far, the majority of research on tumor-driving mutations and
associated mutations in mtDNA has focused on alterations of the
D-loop in the mitochondrial genome (26,28,38). Lin
et al (38) detected somatic
D-loop mutations in 62.5% patients with oral squamous cell
carcinoma. Rahmani et al (41)
reported the presence of mtDNA D-loop mutations in 52% patients
with breast cancer. In cases of brain tumors, Montanini et
al (59) identified sequence
alterations in 36% of patients with malignant gliomas, primarily in
the D-loop region.
Until now, there were no published studies or data
available on mtDNA mutations in the brain tumors of Malaysian
patients. As the majority of the available studies on mtDNA
mutations in brain tumors were conducted on cohorts in Western
countries, it was assumed that this is the first study involving
Malaysian patients. In the current study, the frequency of mtDNA
mutations was identified in 51% of the patients tested, which was
high when compared with three prior studies of German patients with
brain tumors: 41% glioblastoma (28),
43% neurofibromatosis type I (60)
and 40% medulloblastoma (61).
However, this rate was lower than that reported for pilocytic
astrocytoma (84%) by Lueth et al (32). The difference in the frequency of
mtDNA mutations may be due to sample size, or the genetic
differences among the studied populations.
The majority of nucleotide changes reported in the
present study have been previously described; mutations at
nucleotide positions 303, 311, 146, 152, 204, 16,356 and 16,519
have been reported in patients with brain tumors (28,29,32). All
of these reported mutations were also observed in our brain tumor
samples. In addition, changes in these nucleotides were observed in
several types of cancer, including breast cancer, leukemia, gastric
cancer and oral cancer (Table
II).
The mtDNA D-loop contains three hypervariable
segments: HVI (16024–16383), HVII (57–372) and HVIII (438–574),
which are highly polymorphic (18,62,63). In
the present study, the most frequent D-loop mutation was in the
HVII segment, particularly in a polycytosine (poly-C)
mononucleotide repeat tract located between nucleotides 303 and
315, termed D310 (55,64,65). The
present findings confirm the previous observation that base
deletions or insertions in D310, are the most common mutations of
mtDNA in human cancer, including brain cancer (55,59,65). The
D-loop was reported to be more sensitive to oxidative stress than
other mtDNA regions (66). Damage in
poly-C sequences by extensive oxidative stress may lead to slippage
and/or misincorporation during the replication or repair of mtDNA
by mitochondrial DNA polymerase γ, and subsequently give rise to
mtDNA mutations in cancer cells (67).
In the present study, four mutations were identified
in the D-loop, including 176 A-deletion (in 2 cases), 476C>A (in
1 case), 566C>A (in 1 case) and 16,405 A-deletion (in 1 case),
which have not previously been reported in MITOMAP databases, and
they were considered to be novel mutations in the mtDNA D-loop.
In the present study, 210 germline nucleotide
changes were identified in peripheral blood and tumor tissue
samples of patient cohort. All of these changes were considered to
be polymorphisms. Polymorphisms in 263A>G, 16189T>C,
16261C>T and 16271T>C were the most frequently observed in
the present study. Yacoubi Loueslati et al (68) determined that Tunisian female patients
who harbored a 263A>G germline polymorphism exhibited a weak
protective effect against breast cancer risk. Previously, attention
has been paid to the carriers of the 16189T>C polymorphism in
oncological studies, due to their susceptibility to endometrial
cancer progression (69–71). The 16189T>C polymorphism has been
hypothesized to affect mtDNA replication and its cellular copy
number (72). In addition to
endometrial cancer, the 16189T>C polymorphism has been
identified in pilocytic astrocytoma (32), breast cancer (26,73) and
coronary artery disease (74). Thus
far, no strong association between these polymorphisms and the
etiology of brain tumors has been observed.
By analyzing the association between D-loop
mutational status and clinicopathological characteristics, no
significant difference was observed between the D-loop mutation
group and the non-mutation group with regard to age, gender, race
and histological tumor type. However, it was suggested that these
mtDNA D-loop mutations are capable of initiating and promoting
tumorigenesis in the brain.
As only a limited number of studies have analyzed
the role of mtDNA mutations in brain tumors, it remains unclear
whether the mtDNA mutation has prognostic value. In a prior study,
Montanini et al (59) reported
that mtDNA mutations have no prognostic effect in gliomas.
Similarly, Vidone et al (75)
suggested that mtDNA genotyping may not be an efficient molecular
tool to predict prognosis. There is still controversy regarding the
precise prognostic role of mtDNA alterations in brain tumors
(30,59,75).
Additional studies with larger populations are required to clarify
the prognostic impact of the mtDNA alteration status in brain
tumors.
In summary, the present study identified a high
frequency of D-loop region mtDNA mutations in Malaysian patients
with brain tumors. Although no significant association was observed
between mtDNA mutations and clinicopathological parameters, the
present study is able to provided novel data (local data as well as
global data) of an association between mtDNA mutations and the
pathogenesis of brain tumors. This may also provide important
information as to how mtDNA defects lead to cancer. The alterations
of mtDNA in tumorigenesis may be used in the future as novel
potential target biomarkers for the diagnosis, prognosis and
treatment of brain tumors.
Acknowledgements
The present study was financially supported by a
Short Term Grant (grant no. 304/PPSP/61310010) from the Universiti
Sains Malaysia.
References
|
1
|
Ward E, DeSantis C, Robbins A, Kohler B
and Jemal A: Childhood and adolescent cancer statistics, 2014. CA
Cancer J Clin. 64:83–103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Facts & Figures, . 2017 from
the American Cancer Society. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2017/cancer-facts-and-figures-2017.pdf
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim CC, Yahya H and Lim TO: Second report
of the national cancer registry, cancer incidence in Malaysia.
Kuala Lumpur: National Cancer Registry; 2003
|
|
5
|
Lim CC, Rampal S, Halimah Y and Har YC:
Cancer incidence in Peninsular Malaysia. Kuala Lumpur: National
Cancer Registry: 2003–2005; 2008
|
|
6
|
Goh CH, Lu YY, Lau BL, Oy J, Lee HK, Liew
D and Wong A: Brain and spinal tumour. Med J Malaysia. 69:261–267.
2014.PubMed/NCBI
|
|
7
|
McConigley R, Halkett G, Lobb E and Nowak
A: Caring for someone with high-grade glioma: A time of rapid
change for caregivers. Palliat Med. 24:473–479. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Visser O, Ardanaz E, Botta L, Sant M,
Tavilla A and Minicozzi P: EUROCARE-5 Working Group: Survival of
adults with primary malignant brain tumours in Europe; Results of
the EUROCARE-5 study. Eur J Cancer. Sep 5–2015. View Article : Google Scholar
|
|
9
|
Krishnan KJ and Turnbull DM: Mitochondrial
DNA and genetic disease. Essays Biochem. 47:139–151. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang X: The expanding role of mitochondria
in apoptosis. Genes Dev. 15:2922–2933. 2001.PubMed/NCBI
|
|
11
|
Wallace DC, Fan W and Procaccio V:
Mitochondrial energetic and therapeutics. Annu Rev Pathol.
5:297–348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Anderson S, Bankier AT, Barrell BG, de
Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA,
Sanger F, et al: Sequence and organization of the human
mitochondrial genome. Nature. 290:457–465. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Liu D, Lu J and Bai Y: Physiology
and pathophysiology of mitochondrial DNA. Adv Exp Med Biol.
942:39–51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Andrews RM, Kubacka I, Chinnery PF,
Lightowlers RN, Turnbull DM and Howell N: Reanalysis and revision
of the Cambridge reference sequence for human mitochondrial DNA.
Nat Genet. 23:1471999. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Larsen NB, Rasmussen M and Rasmussen LJ:
Nuclear and mitochondrial DNA repair: Similar pathways?
Mitochondrion. 5:89–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alexeyev M, Shokolenko I, Wilson G and
LeDoux S: The maintenance of mitochondrial DNA integrity-critical
analysis and update. Cold Spring Harb Perspect Biol. 5:a0126412013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Polyak K, Li Y, Zhu H, Lengauer C, Willson
JK, Markowitz SD, Trush MA, Kinzler KW and Vogelstein B: Somatic
mutations of the mitochondrial genome in human colorectal tumours.
Nat Genet. 20:291–293. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Penta JS, Johnson FM, Wachsman JT and
Copeland WC: Mitochondrial DNA in human malignancy. Mutat Res.
488:119–133. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chatterjee A, Mambo E and Sidransky D:
Mitochondrial DNA mutations in human cancer. Oncogene.
25:4663–4674. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tseng LM, Yin PH, Yang CW, Tsai YF, Hsu
CY, Chi CW and Lee HC: Somatic mutations of the mitochondrial
genome in human breast cancers. Genes Chromosomes Cancer.
50:800–811. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li LH, Kang T, Chen L, Zhang W, Liao Y,
Chen J and Shi Y: Detection of mitochondrial DNA mutations by
high-throughput sequencing in the blood of breast cancer patients.
Int J Mol Med. 33:77–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu CC, Lee HC and Wei YH: Mitochondrial
DNA alterations and mitochondrial dysfunction in the progression of
hepatocellular carcinoma. World J Gastroenterol. 19:8880–8886.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee HC, Huang KH, Yeh TS and Chi CW:
Somatic alterations in mitochondrial DNA and mitochondrial
dysfunction in gastric cancer progression. World J Gastroenterol.
20:3950–3959. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang Y, Huang J, Zhang J, Wang J, Qiao F,
Chen HM and Hong ZP: Detecting the somatic mutations spectrum of
Chinese lung cancer by analyzing the whole mitochondrial DNA
genomes. Mitochondrial DNA. 26:56–60. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sultana GN, Rahman A, Shahinuzzaman AD,
Begum RA and Hossain CF: Mitochondrial DNA mutations-candidate
biomarkers for breast cancer diagnosis in Bangladesh. Chin J
Cancer. 31:449–454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tipirisetti NR, Govatati S, Pullari P,
Malempati S, Thupurani MK, Perugu S, Guruvaiah P, Rao KL, Digumarti
RR, Nallanchakravarthula V, et al: Mitochondrial control region
alterations and breast cancer risk: A study in South Indian
population. PLoS One. 9:e853632014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mondal R, Ghosh SK, Talukdar FR and Laskar
RS: Association of mitochondrial D-loop mutations with GSTM1 and
GSTT1 polymorphisms in oral carcinoma: A case control study from
northeast India. Oral Oncol. 49:345–353. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kirches E, Krause G, Warich-Kirches M,
Weis S, Schneider T, Meyer-Puttlitz B, Mawrin C and Dietzmann K:
High frequency of mitochondrial DNA mutations in glioblastoma
multiforme identified by direct sequence comparison to blood
samples. Int J Cancer. 93:534–538. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vega A, Salas A, Gamborino E, Sobrido MJ,
Macaulay V and Carracedo A: mtDNA mutations in tumors of the
central nervous system reflect the neutral evolution of mtDNA in
populations. Oncogene. 23:1314–1320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mohamed Yusoff AA: Role of mitochondrial
DNA mutations in brain tumors: A mini-review. J Cancer Res Ther.
11:535–544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lueth M, Wronski L, Giese A,
Kirschner-Schwabe R, Pietsch T, von Deimling A, Henze G, Kurtz A
and Driever PH: Somatic mitochondrial mutations in pilocytic
astrocytoma. Cancer Genet Cytogenet. 192:30–35. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fliss MS, Usadel H, Caballero OL, Wu L,
Buta MR, Eleff SM, Jen J and Sidransky D: Facile detection of
mitochondrial DNA mutations in tumors and bodily fluids. Science.
287:2017–2019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharawat SK, Bakhshi R, Vishnubhatla S and
Bakhshi S: Mitochondrial D-loop variations in paediatric acute
myeloid leukaemia: A potential prognostic marker. Br J Haematol.
149:391–398. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ashtiani ZO, Heidari M, Hasheminasab SM,
Ayati M and Rakhshani N: Mitochondrial D-Loop polymorphism and
microsatellite instability in prostate cancer and benign
hyperplasia patients. Asian Pac J Cancer Prev. 13:3863–3868. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hung WY, Wu CW, Yin PH, Chang CJ, Li AF,
Chi CW, Wei YH and Lee HC: Somatic mutations in mitochondrial
genome and their potential roles in the progression of human
gastric cancer. Biochim Biophys Acta. 1800:264–270. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cai FF, Kohler C, Zhang B, Chen WJ,
Barekati Z, Garritsen HS, Lenner P, Toniolo P, Zhang JJ and Zhong
XY: Mutations of mitochondrial DNA as potential biomarkers in
breast cancer. Anticancer Res. 31:4267–4271. 2011.PubMed/NCBI
|
|
38
|
Lin JC, Wang CC, Jiang RS, Wang WY and Liu
SA: Impact of somatic mutations in the D-loop of mitochondrial DNA
on the survival of oral squamous cell carcinoma patients. PLoS One.
10:e01243222015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kassem AM, El-Guendy N, Tantawy M,
Abdelhady H, El-Ghor A and Wahab Abdel AH: Mutational hotspots in
the mitochondrial D-loop region of cancerous and precancerous
colorectal lesions in Egyptian patients. DNA Cell Biol. 30:899–906.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bragoszewski P, Kupryjanczyk J, Bartnik E,
Rachinger A and Ostrowski J: Limited clinical relevance of
mitochondrial DNA mutation and gene expression analyses in ovarian
cancer. BMC Cancer. 8:2922008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rahmani B, Azimi C, Omranipour R, Raoofian
R, Zendehdel K, Saee-Rad S and Heidari M: Mutation screening in the
mitochondrial D-loop region of tumoral and non-tumoral breast
cancer in Iranian patients. Acta Med Iran. 50:447–453.
2012.PubMed/NCBI
|
|
42
|
Yacoub HA, Mahmoud WM, El-Baz HA, Eid OM,
ELfayoumi RI, Elhamidy SM and Mahmoud MM: Novel mutations in the
displacement loop of mitochondrial DNA are associated with acute
lymphoblastic leukemia: A genetic sequencing study. Asian Pac J
Cancer Prev. 15:9283–9289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pang LJ, Shao JY, Liang XM, Xia YF and
Zeng YX: Mitochondrial DNA somatic mutations are frequent in
nasopharyngeal carcinoma. Cancer Biol Ther. 7:198–207. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu M, Shi Y, Zhang F, Zhou Y, Yang Y, Wei
X, Zhang L and Niu R: Sequence variations of mitochondrial DNA
D-loop region are highly frequent events in familial breast cancer.
J Biomed Sci. 15:535–543. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen JZ, Gokden N, Greene GF, Mukunyadzi P
and Kadlubar FF: Extensive somatic mitochondrial mutations in
primary prostate cancer using laser capture microdissection. Cancer
Res. 62:6470–6474. 2002.PubMed/NCBI
|
|
46
|
Sharma H, Singh A, Sharma C, Jain SK and
Singh N: Mutations in the mitochondrial DNA D-loop region are
frequent in cervical cancer. Cancer Cell Int. 5:342005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Van Trappen PO, Cullup T, Troke R, Swann
D, Shepherd JH, Jacobs IJ, Gayther SA and Mein CA: Somatic
mitochondrial DNA mutations in primary and metastatic ovarian
cancer. Gynecol Oncol. 104:129–133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Prior SL, Griffiths AP, Baxter JM, Baxter
PW, Hodder SC, Silvester KC and Lewis PD: Mitochondrial DNA
mutations in oral squamous cell carcinoma. Carcinogenesis.
27:945–950. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tan DJ, Chang J, Chen WL, Agress LJ, Yeh
KT, Wang B and Wong LJ: Somatic mitochondrial DNA mutations in oral
cancer of betel quid chewers. Ann N Y Acad Sci. 1011:310–316. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo W, Yang D, Xu H, Zhang Y, Huang J,
Yang Z, Chen X and Huang Z: Mutations in the D-loop region and
increased copy number of mitochondrial DNA in human laryngeal
squamous cell carcinoma. Mol Biol Rep. 40:13–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Czarnecka AM and Bartnik E: The role of
the mitochondrial genome in ageing and carcinogenesis. J Aging Res.
2011:1364352011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wallace DC: Mitochondria and cancer. Nat
Rev Cancer. 12:685–698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Verschoor ML, Ungard R, Harbottle A,
Jakupciak JP, Parr RL and Singh G: Mitochondria and cancer: Past,
present, and future. Biomed Res Int. 2013:6123692013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Carew JS, Zhou Y, Albitar M, Carew JD,
Keating MJ and Huang P: Mitochondrial DNA mutations in primary
leukemia cells after chemotherapy: Clinical significance and
therapeutic implications. Leukemia. 17:1437–1447. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sanchez-Cespedes M, Parrella P, Nomoto S,
Cohen D, Xiao Y, Esteller M, Jeronimo C, Jordan RC, Nicol T, Koch
WM, et al: Identification of a mononucleotide repeat as a major
target for mitochondrial DNA alterations in human tumors. Cancer
Res. 61:7015–7019. 2001.PubMed/NCBI
|
|
56
|
Rosson D and Keshgegian AA: Frequent
mutations in the mitochondrial control region DNA in breast tissue.
Cancer Lett. 215:89–94. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lièvre A, Chapusot C, Bouvier AM,
Zinzindohoué F, Piard F, Roignot P, Arnould L, Beaune P, Faivre J
and Laurent-Puig P: Clinical value of mitochondrial mutations in
colorectal cancer. J Clin Oncol. 23:3517–3525. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Choi SJ, Kim SH, Kang HY, Lee J, Bhak JH,
Sohn I, Jung SH, Choi YS, Kim HK, Han J, et al: Mutational hotspots
in the mitochondrial genome of lung cancer. Biochem Biophys Res
Commun. 407:23–27. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Montanini L, Regna-Gladin C, Eoli M,
Albarosa R, Carrara F, Zeviani M, Bruzzone MG, Broggi G, Boiardi A
and Finocchiaro G: Instability of mitochondrial DNA and MRI and
clinical correlations in malignant gliomas. J Neurooncol. 74:87–89.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kurtz A, Lueth M, Kluwe L, Zhang T, Foster
R, Mautner VF, Hartmann M, Tan DJ, Martuza RL, Friedrich RE, et al:
Somatic mitochondrial DNA mutations in neurofibromatosis type
1-associated tumors. Mol Cancer Res. 2:433–441. 2004.PubMed/NCBI
|
|
61
|
Lueth M, von Deimling A, Pietsch T, Wong
LJ, Kurtz A, Henze G and Driever PH: Medulloblastoma harbor somatic
mitochondrial DNA mutations in the D-loop region. J Pediatr Hematol
Oncol. 32:156–159. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lutz S, Wittig H, Weisser HJ, Heizmann J,
Junge A, Dimo-Simonin N, Parson W, Edelmann J, Anslinger K, Jung S
and Augustin C: Is it possible to differentiate mtDNA by means of
HVIII in samples that cannot be distinguished by sequencing the HVI
and HVII regions? Forensic Sci Int. 113:97–101. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Imaizumi K, Parsons TJ, Yoshino M and
Holland MM: A new database of mitochondrial DNA hypervariable
regions I and II sequences from 162 Japanese individuals. Int J
Legal Med. 116:68–73. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Parrella P, Seripa D, Matera MG, Rabitti
C, Rinaldi M, Mazzarelli P, Gravina C, Gallucci M, Altomare V,
Flammia G, et al: Mutations of the D310 mitochondrial
mononucleotide repeat in primary tumors and cytological specimens.
Cancer Lett. 190:73–77. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Alhomidi MA, Vedicherla B, Movva S, Rao
PK, Ahuja YR and Hasan Q: Mitochondrial D310 instability in Asian
Indian breast cancer patients. Tumour Biol. 34:2427–2432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mamba E, Gao X, Cohen Y, Guo Z, Talalay P
and Sidransky D: Electrophile and oxidant damage of mitochondrial
DNA leading to rapid evolution of homoplasmic mutations. Proc Natl
Acad Sci USA. 100:1838–1843. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Graziewicz MA, Day BJ and Copeland WC: The
mitochondrial DNA polymerase as a target of oxidative damage.
Nucleic Acids Res. 30:2817–2824. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Loueslati Yacoubi B, Troudi W, Cherni L,
Rhomdhane KB and Mota-Vieira L: Germline HVR-II mitochondrial
polymorphisms associated with breast cancer in Tunisian women.
Genet Mol Res. 9:1690–1700. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Liu VW, Wang Y, Yang HJ, Tsang PC, Ng TY,
Wong LC, Nagley P and Ngan H: Mitochondrial DNA variant 16189T>C
is associated with susceptibility to endometrial cancer. Hum Mutat.
22:173–174. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Czarnecka AM, Klemba A, Semczuk A, Plak K,
Marzec B, Krawczyk T, Kofler B, Golik P and Bartnik E: Common
mitochondrial polymorphisms as risk factor for endometrial cancer.
Int Arch Med. 2:332009. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cho S, Lee YM, Choi YS, Yang HI, Jeon YE,
Lee KE, Lim K, Kim HY, Seo SK and Lee BS: Mitochondria DNA
polymorphisms are associated with susceptibility to endometriosis.
DNA Cell Biol. 31:317–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liou CW, Lin TK, Chen JB, Tiao MM, Weng
SW, Chen SD, Chuang YC, Chuang JH and Wang PW: Association between
a common mitochondrial DNA D-loop polycytosine variant and
alteration of mitochondrial copy number in human peripheral blood
cells. J Med Genet. 47:723–728. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Czarnecka AM, Krawczyk T, Plak K, Klemba
A, Zdrozny M, Arnold RS, Kofler B, Golik P, Szybinska A, Lubinski
J, et al: Mitochondrial genotype and breast cancer predisposition.
Oncol Rep. 24:1521–1534. 2010.PubMed/NCBI
|
|
74
|
Mueller EE, Eder W, Ebner S, Schwaiger E,
Santic D, Kreindl T, Stanger O, Paulweber B, Iglseder B, Oberkofler
H, et al: The mitochondrial T16189C polymorphism is associated with
coronary artery disease in Middle European populations. PLoS One.
6:e164552011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Vidone M, Clima R, Santorsola M, Calabrese
C, Girolimetti G, Kurelac I, Amato LB, Iommarini L, Trevisan E,
Leone M, et al: A comprehensive characterization of mitochondrial
DNA mutations in glioblastoma multiforme. Int J Biochem Cell Biol.
63:46–54. 2015. View Article : Google Scholar : PubMed/NCBI
|