Introduction
The increasing incidence of esophageal cancer has
led to its becoming one of the common digestive diseases (1), and in fact, was the sixth most common
cancer disease in China by the end of 2015 (2) causing the fourth highest mortality rates
among all cancer diseases (2).
Methylation plays an important role in the
development and progression of tumors. Previous studies showed that
the increased methylation rate of the tumor suppressor gene rate
and its promoter region can significantly increase the occurrence
of the tumor (3). The methylation
rate of CDH1 gene in esophageal cancer patients is
approximately 12.3%, compared to 73.4% in healthy individuals and
this reduced gene methylation can promote gene expression (4). Thus, the levels of related oncogene
proteins are increased. Previous findings have shown significantly
higher expression levels of RUNX3 and P53 in cancer cells compared
to those in healthy cells (5). The
main function of RUNX3 gene is to bind DNA to form a complex
to inhibit or promote the process of cell growth and
differentiation (6). P53 gene
is a common transcription factor (7)
and the expression of P53 in healthy cells is normally low, but
when the cells are stimulated by toxic substances or carcinogenic
factors, the expression rapidly increases and thus makes P53
closely related to the development of cancer. As a newly discovered
tumor inhibitor (8), RECK can inhibit
tumor cell infiltration.
In this study, we explored the relationship between
RECK, P53 and RUNX gene methylation and
esophageal cancer to reveal the interactions between them and to
provide the theoretical and the experimental basis for the
diagnosis and treatment of esophageal cancer.
Materials and methods
General information
In total, 58 esophageal cancer patients (28 males,
30 females) with an average age of 32.4±15.3 years were selected
during the period of February 2013 to February 2014 and 58 healthy
inidivduals (21 males, 21 females) with a mean age of 33.2±12.4
years were also considered as control group. All the patients
signed informed consent and the study was approved by the Ethics
Committee of the Tumor Hospital Affiliated to Xinjiang Medical
University (Xinjiang, China). Inclusion criteria for the study
were: a) suffering from esophageal cancer, and b) aged between 32
and 65 years. The exclusion criteria were a) suffering from other
tumors and cancer, b) suffering from digestive system diseases, and
c) <32 or >65 years of age, and d) other reasons.
Main reagents and instruments
The following main reagents were used: RNA
Extraction kit (Xinmai Biotechnology Co., Ltd., Shanghai, China),
RT-qPCR kit (Applied Biosystems, Foster City, CA, USA), rabbit
anti-human RECK, P53, and RUNX monoclonal primary antibody (Acris
Antibodies Inc., San Diego, CA, USA), mouse anti-rabbit polyclonal
secondary antibody (HRP-labeled) (Genewiz, Suzhou, China), primary
antibody and secondary antibody of GAPDH were purchased from Thermo
Fisher Scientific (Waltham, MA, USA), immunohistochemistry kit
(Roche, Indianapolis, IN, USA), ELISA kit (Takara, Dalian, China),
methylation determination kit (Kang Century Biotech Co., Ltd.,
Beijing, China) and other chemical reagents were from Sinopharm
Chemical Reagent Co., Ltd. (Shanghai, China). In addition, the
following main instruments were used: Fluorescence quantitative PCR
instrument (Applied Biosystems), microplate reader (Beijing Liuyi
Biotechnology, Beijing, China), protein electrophoresis (Beijing
Liuyi Biotechnology), gel imager (Bio-Rad, Hercules, CA, USA),
Olympus microscope X53, Mindrop micro-nucleic acid quantitative
instrument (Bio-Rad).
Methylation detection
The total DNA was extracted and processed by
methylation assay kit according to the instructions. The specific
methylation primers were designed according to the methylation
principle. Table I shows the primer
sequences.
 | Table I.Methylation primers. |
Table I.
Methylation primers.
| Primer name | Sequences |
|---|
| RECK-F |
ATCTACTATTCCTCTATCTATCCAC |
| RECK-R |
CTATCTATTCATCTTCTATCTACC |
| P53-F |
CTATCTTATCTATCTTCTCTATCTTC |
| P53-R |
CTATCTTATCTTCTCTCATCTCTAC |
| RUNX-F |
CTATCTTTATACTATCTTCTCTATC |
| RUNX-R |
TCTCTCTATTATCTTAAACTTCTAC |
RT-qPCR
RNA extraction
In order to extract RNA, the digestive tract samples
were collected from both patients and healthy people, then stored
in liquid nitrogen or in a refrigerator at −80°C. Then 0.5 g of the
sample was mixed with 300 µl RNA Plus by pipetting and was kept at
room temperature for 10 min, followed by centrifugation (10,000 ×
g) at 4°C for 5 min. The supernatant was collected and 250 µl
chloroform was added. After vigorous mixing, the mixture was kept
at room temperature for 5 min. After centrifugation (10,000 × g) at
4°C for 5 min, the supernatant was collected and then an equal
volume of isoamyl alcohol was added and mixed gently, followed by
centrifugation (10,000 × g) at 4°C for 10 min. Then the supernatant
was removed and the sample was washed with 75% ethanol 2 or 3
times. The samples were placed at room temperature until the
ethanol was completely removed. Then 50 µl of RNase-free ultra-pure
water was added and the mass of the RNA extracted was quantitated
by the Mindrop microarray and analyzed by electrophoresis (9).
Fluorescence quantitation
All the operations were performed according to the
instructions of Takara fluorescence quantitative PCR. Furthermore,
the PCR primers were synthesized by the Sangon Biotech (Shanghai,
China) (Table II).
 | Table II.q-RT-PCR primers. |
Table II.
q-RT-PCR primers.
| Primer name | Sequences |
|---|
| RECK-F |
AGCTGATGCATCGATCGATCGATC |
| RECK-R |
CGTATCGTGGTCAGTCGTACGTCAC |
| P53-F |
AGTCGATGCTAGCTAGCTAGCTAC |
| P53-R |
TGATCGATCGATGATAGTACACGC |
| RUNX-F |
TGATGCGCGCTAGCATGAAGTCGATCG |
| RUNX-R |
CTGATCGGAGATCAGTCAGCGATCAGCTG |
| GAPDH-F |
GTCGATGGCTAGTCGTAGCATCGAT |
| GAPDH-R |
TGCTAGCTGGCATGCCCGATCGATC |
Cell total protein extraction
The esophageal cancer samples were collected from
patients with esophageal cancer and from healthy subjects and then
preserved in liquid nitrogen. After melting ice, reaction solution
A was quickly added and the mixture was vigorously mixed for 2–3
min. Then reaction mixture B was added and mixed gently followed by
centrifugation (12,000 × g) at 4°C for 10 min. The supernatant was
kept.
Enzyme-linked immunosorbent assay
(ELISA)
The expression of different gene proteins in
different samples was detected by ELISA kit (Takara). The main
steps are as follows: the standard protein samples were first
diluted at 1:50 with dilution buffer in the kit and a standard
curve was drawn. The sample to be tested was then diluted at the
ratio of 1:100 with PBS (pH 7.2) and then 100 µl solution was added
to each well. Test solution (50 µl) was added and incubated at room
temperature for 2 h and TMB chromogenic substrate was added. The
absorbance values were measured at 495 nm, and the protein
concentration in each sample was calculated according to the
standard curve given previously (10).
Western blotting
In order to determine the protein expression of
different genes in different samples, western blotting was used.
The total protein extracted from the samples was quantified by
Coomassie blue staining. Processed sample (20 µl) was subjected to
SDS-PAGE electrophoresis and then protein was transferred to the
membrane by the semi-dry method. The membrane was blocked with 1%
skim milk for 2 h. The primary antibody was diluted with a ratio of
1:1,000, and then was added and incubated at room temperature for 2
h. Membranes were first incubated with primary rabbit monoclonal
Reck antibody (dilution: 1:500; cat. nos. ab115844), rabbit
monoclonal p53 antibody (dilution: 1:500; cat. nos. ab32049) and
rabbit polyclonal Runx antibody (dilution: 1:500; cat. nos.
ab23981) at 20°C for 2 h, and then incubated with secondary goat
anti-rabbit (HRP) IgG antibody (dilution: 1:2000; cat. nos.
ab6721). The antibodes were purchased from Abcam (Cambridge, MA,
USA). The membrane was washed 5 times (10 min each time) and color
development solution was added (11).
Immunohistochemistry
The digestive tract samples were collected from both
patients and healthy people and were stored in liquid nitrogen or
at −80°C. The samples were processed according to the experimental
procedure described by Shi et al (12,13). The
positive rate was determined by immunoassay kit according to the
instructions of the kit.
Statistical analysis
Data were analyzed using SPSS 19.0 software (IBM,
Armonk, NY, USA). The Chi-square test was used to compare the
statistical data considering the level of significance α=0.05.
Results
Methylation of different genes in
different samples
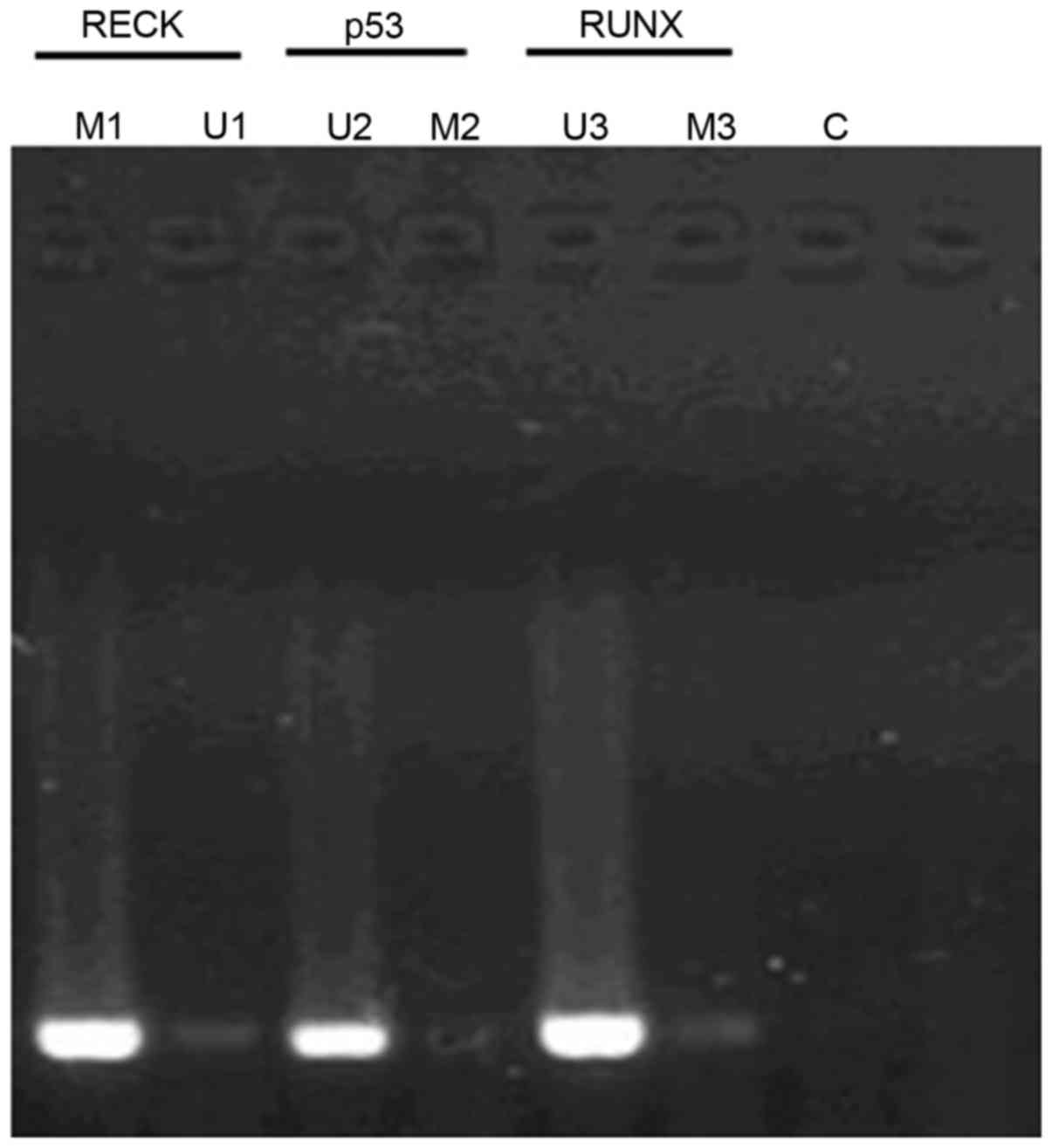
Fig. 1 shows the
methylation status of different genes in different samples by MSP
method (methylation-specific PCR). The methylation rates of
RECK, P53 and RUNX genes in patients with
esophageal cancer were 72.4% (42/58), 1.7% (1/58) and 3.4% (2/58),
respectively; while the methylation rates of RECK, P5 and RUNX in
healthy tissue were 7.1% (3/42), 90.5% (38/42) and 83.3% (35/42),
respectively. Furthermore, there were significant differences
between patients and healthy people (P<0.05).
mRNA levels of different genes in
patients with esophageal cancer and healthy people
The mRNA expression levels of different genes in
patients with esophageal cancer and healthy subjects were
determined by RT-qPCR, as shown in Fig.
2A-C. The mRNA level of RECK in patients with esophageal cancer
was found significantly lower (approximately 2.3% of the RECK mRNA
level in healthy subjects) than that of healthy subjects
(P<0.05) (Fig. 2A). Moreover, the
mRNA level of P53 in patients with esophageal cancer was
significantly higher than that of healthy subjects (P<0.05)
(Fig. 2B). Finally, the mRNA level of
RUNX in patients with esophageal cancer was 47.2 times higher than
that of healthy subjects and this difference was statistically
significant (P<0.05) (Fig.
2C).
Protein levels of different genes in
patients with esophageal cancer and healthy people
Herein, ELISA was used to detect the protein levels
of different genes in different samples and the results are shown
in Fig. 3A-C. The protein level of
RECK in patients with esophageal cancer (0.12±0.05 µg/l) was
significantly lower than that in healthy people (3.46±0.08 µg/l),
(P<0.05) (Fig. 3A) Moreover, the
protein levels of P53 and RUNX patients with esophageal cancer and
healthy people (6.43±0.12 µg/l and 4.32±0.14 µg/l) were
significantly higher than those of healthy subjects (0.64±0.06 µg/l
and 0.53±0.09 µg/l) (P<0.05) (Fig. 3B
and C).
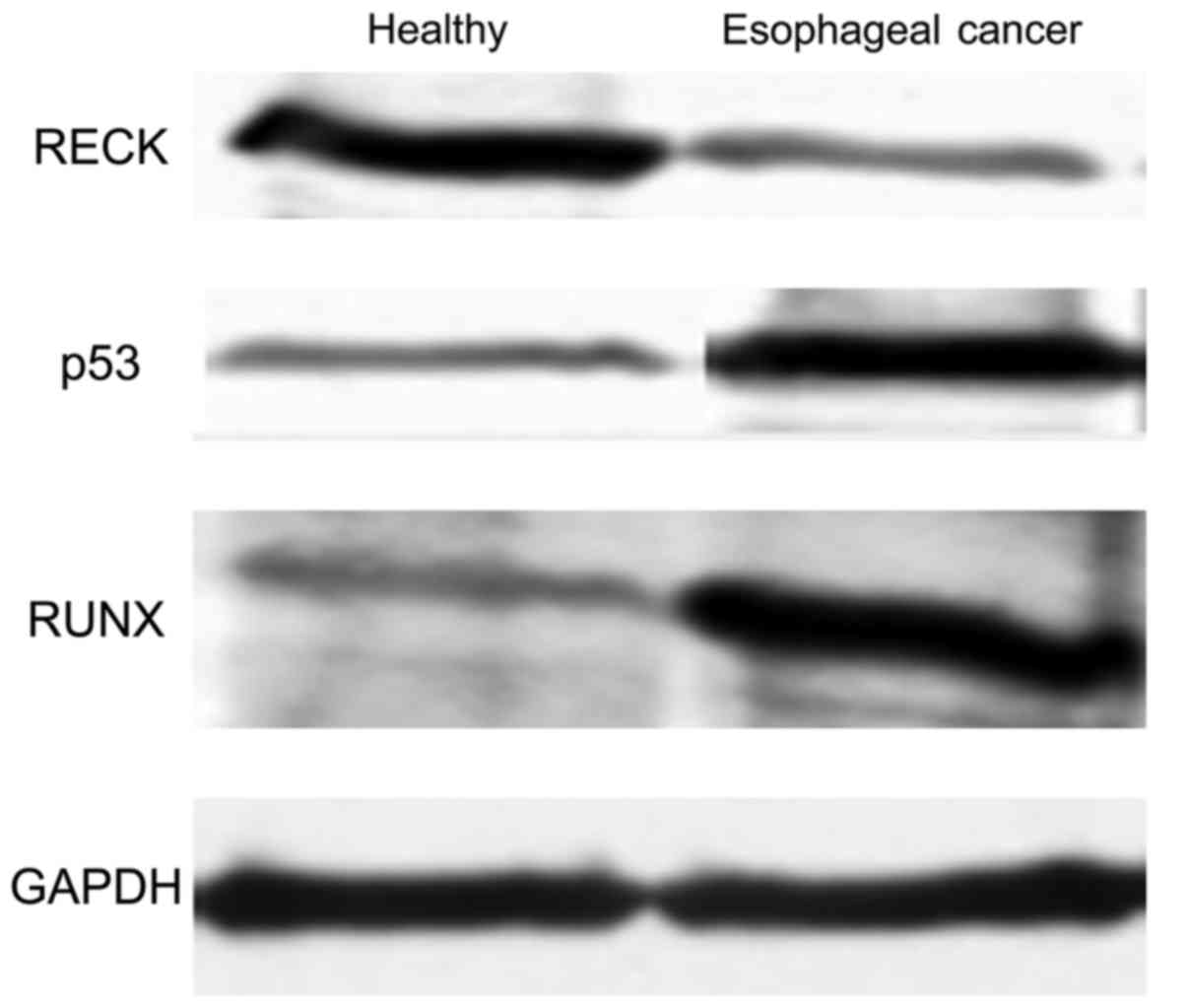
Protein levels of different genes in
different samples detected by western blotting
In order to determine further the differences in
protein expression between healthy individuals and patients with
esophageal cancer, different levels of genes in both groups were
determined by western blot. As shown in Fig. 4, the level of RECK protein was
significantly higher in the healthy people than that in the
patients with esophageal cancer (p<0.05), whereas protein levels
of P53 and RUNX in patients with esophageal cancer were
significantly higher than those in healthy people (p<0.05). In
addition, western blot results were consistent with ELISA
results.
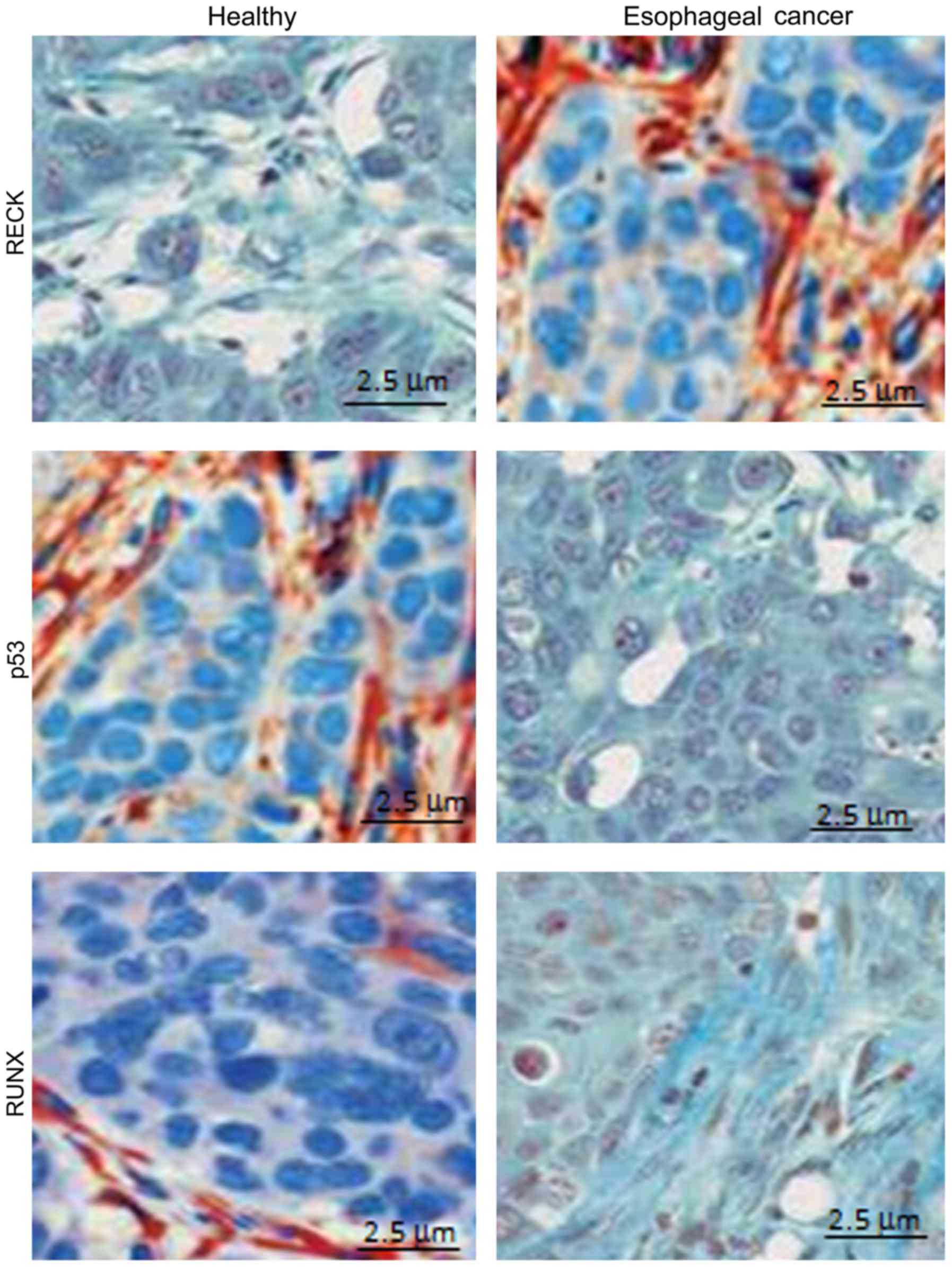
Positive expression of different genes
in different samples detected by immunohistochemical method
Using immunohistochemistry, we detected the positive
expression of different genes in different samples. The results
showed that RECK protein was mainly expressed in the cytoplasm, and
the positive expression rate of RECK in healthy subjects (82.3%)
was significantly higher than that in patients with esophageal
cancer (9.5%) (P<0.05) (Fig. 5A).
Moreover, P53 protein was mainly expressed in the cytoplasm, and
the positive expression rate of P53 in healthy subjects (11.1%) was
significantly lower than that in patients with esophageal cancer
(78.4%) (P<0.05) (Fig. 5B).
Furthermore, RUNX protein was mainly expressed in the cytoplasm,
and the positive expression rate of RUNX in healthy subjects
(9.06%) was significantly lower than that in patients with
esophageal cancer (87.3%) (P<0.05) (Fig. 5C) (Table
III).
 | Table III.A summary of the positive expression
rate of different genes in the samples. |
Table III.
A summary of the positive expression
rate of different genes in the samples.
| Group | Cell number (n) | Positive rate of RECK
(%) | Positive rate of P53
(%) | Positive rate of RUNX
(%) |
|---|
| Control group | 400 | 82.3 | 11.1 | 9.06 |
| Observation
group | 400 | 9.5 | 78.4 | 87.3 |
| P-value |
| <0.001 | <0.001 | <0.001 |
Discussion
Studies on different tumors and cancers have shown
that tumor cells and cancer cells are affected by a variety of
factors including environmental factors such as various toxic and
hazardous substances, radioactive carcinogens, and self-factors
such as autoimmune disorders, mutations in tumor suppressor gene
and oncogene activation (14). In
addition, the environmental factors can affect the expression of
intracellular genes to promote the occurrence of cancer cells and
tumor cells. Recent findings have identified many tumor suppressor
genes in human cells (15). Tumor
suppressor genes can also maintain body health by inhibiting the
expression of oncogenes. In addition, tumor suppressor genes can be
inactivated by mutations within them or the abnormal regulation
network, leading to the occurrence of cancer cells and tumor cells.
Ethanol and other chemicals can significantly increase the
methylation rate of the promoter of human aldehyde dehydrogenase
gene, while the accumulation of acetaldehyde can damage the
function of liver cells (16).
In previous studies, it was found that the
RECK gene, which is highly expressed in normal tissue, can
negatively regulate the expression of oncogenes (17). Moreover, the RECK can inhibit
angiogenesis. In the present study, we found that methylation rate
of RECK gene in patients with esophageal cancer was
significantly higher than that of normal people, indicating that
the methylation of RECK gene can promote the occurrence and
development of esophageal cancer. It is well-known that the
P53 gene, which is involved in the process of cell signal
transduction in cancer cells, can inhibit the occurrence of cancer
cells by interacting with carcinogenic factors (18,19). In
addition, the RUNX gene, which acts as a downstream
regulator of TGF-β, can promote abnormal cell proliferation and
tumor cell infiltration (20). Our
results showed that the methylation rates of P53 and
RUNX genes in patients with esophageal cancer were
significantly reduced, leading to the increased expression of the
two genes in esophageal cancer cells, which in turn promote the
proliferation and infiltration of esophageal cancer cells. In
conclusion, the increased methylation of RECK gene and the
decreased methylation of P53 and RUNX genes can
promote the occurrence of esophageal cancer.
References
|
1
|
Sottocornola R, Royer C, Vives V, Tordella
L, Zhong S, Wang Y, Ratnayaka I, Shipman M, Cheung A,
Gaston-Massuet C, et al: ASPP2 binds Par-3 and controls the
polarity and proliferation of neural progenitors during CNS
development. Dev Cell. 19:126–137. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xie T, Cui X, Zheng H, Chen D, He L and
Jiang B: Meta-analysis: Eradication of Helicobacter pylori
infection is associated with the development of endoscopic
gastroesophageal reflux disease. Eur J Gastroenterol Hepatol.
25:1195–1205. 2013.PubMed/NCBI
|
|
3
|
Mhaskar RS, Ricardo I, Azliyati A,
Laxminarayan R, Amol B, Santosh W and Boo K: Assessment of risk
factors of Helicobacter pylori infection and peptic ulcer
disease. J Glob Infect Dis. 5:60–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jarl J and Gerdtham UG: Time pattern of
reduction in risk of oesophageal cancer following alcohol cessation
- a meta-analysis. Addiction. 107:1234–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Venerito M, Kohrs S, Wex T, Adolf D,
Kuester D, Schubert D, Peitz U, Mönkemüller K and Malfertheiner P:
Helicobacter pylori infection and fundic gastric atrophy are
not associated with esophageal squamous cell carcinoma: A
case-control study. Eur J Gastroenterol Hepatol. 23:859–864. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu R, Jiang Y, Wu Q, Li Q, Cheng D, Xu L,
Zhang C, Zhang M and Ye L: Diagnostic value of microRNA-21 in the
diagnosis of lung cancer: Evidence from a meta-analysis involving
11 studies. Tumour Biol. 35:8829–8836. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu J, Cheng Y, Li Y, Jin Z, Pan Y, Liu G,
Fu S, Zhang Y, Feng K and Feng Y: microRNA-128 plays a critical
role in human non-small cell lung cancer tumourigenesis,
angiogenesis and lymphangiogenesis by directly targeting vascular
endothelial growth factor-C. Eur J Cancer. 50:2336–2350. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jiang T, Tang HM, Lu S, Yan DW, Yang YX
and Peng ZH: Up-regulation of tripartite motif-containing 29
promotes cancer cell proliferation and predicts poor survival in
colorectal cancer. Med Oncol. 30:7152013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Solomon H, Buganim Y, Kogan-Sakin I,
Pomeraniec L, Assia Y, Madar S, Goldstein I, Brosh R, Kalo E,
Beatus T, et al: Various p53 mutant proteins differently regulate
the Ras circuit to induce a cancer-related gene signature. J Cell
Sci. 125:3144–3152. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jia G1, Stormont RM, Gangahar DM and
Agrawal DK: Role of matrix Gla protein in angiotensin II-induced
exacerbation of vascular calcification. Am J Physiol Heart Circ
Physiol. 303:H523–H532. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Olesen M, Skov V, Mechta M, Mumm BH and
Rasmussen LM: No influence of OPG and its ligands, RANKL and TRAIL,
on proliferation and regulation of the calcification process in
primary human vascular smooth muscle cells. Mol Cell Endocrinol.
362:149–156. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi HJ, Wen JK, Miao SB, Liu Y and Zheng
B: KLF5 and hhLIM cooperatively promote proliferation of vascular
smooth muscle cells. Mol Cell Biochem. 367:185–194. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sehgal S, Kaul S, Gupta BB and Dhar MK:
Risk factors and survival analysis of the esophageal cancer in the
population of Jammu, India. Indian J Cancer. 49:245–250. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shanahan CM, Crouthamel MH, Kapustin A and
Giachelli CM: Arterial calcification in chronic kidney disease: Key
roles for calcium and phosphate. Circ Res. 109:697–711. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou JY, Chen X, Zhao J, Bao Z, Chen X,
Zhang P, Liu ZF and Zhou JY: MicroRNA-34a overcomes HGF-mediated
gefitinib resistance in EGFR mutant lung cancer cells partly by
targeting MET. Cancer Lett. 351:265–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Steinmetz CG, Xie P, Weiner H and Hurley
TD: Structure of mitochondrial aldehyde dehydrogenase: The genetic
component of ethanol aversion. Structure. 5:701–711. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kapustin AN, Davies JD, Reynolds JL,
McNair R, Jones GT, Sidibe A, Schurgers LJ, Skepper JN, Proudfoot
D, Mayr M, et al: Calcium regulates key components of vascular
smooth muscle cell-derived matrix vesicles to enhance
mineralization. Circ Res. 109:e1–e12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Toko H, Takahashi H, Kayama Y, Oka T,
Minamino T, Okada S, Morimoto S, Zhan DY, Terasaki F, Anderson ME,
et al: Ca2+/calmodulin-dependent kinase IIdelta causes
heart failure by accumulation of p53 in dilated cardiomyopathy.
Circulation. 122:891–899. 2012. View Article : Google Scholar
|
|
19
|
Kapustin AN, Davies JD, Reynolds JL,
McNair R, Jones GT, Sidibe A, Schurgers LJ, Skepper JN, Proudfoot
D, Mayr M, et al: Calcium regulates key components of vascular
smooth muscle cell-derived matrix vesicles to enhance
mineralization. Circ Res. 109:e1–e12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Speer MY, Li X, Hiremath PG and Giachelli
CM: Runx2/Cbfa1, but not loss of myocardin, is required for smooth
muscle cell lineage reprogramming toward osteochondrogenesis. J
Cell Biochem. 110:935–947. 2010. View Article : Google Scholar : PubMed/NCBI
|