Previous studies have provided support for the
hypothesis that breast cancer (BC) development is driven by a
subpopulation of cells that exhibit stem cell characteristics, such
as a capability for self-renewal, differentiation, metastasis,
tumorigenicity and intrinsic resistance to chemotherapy (1). This subpopulation of cells is recognized
as breast cancer stem cells (BCSCs), which are essential for BC
progression (2,3). MicroRNAs (miRNAs/miRs) are small
non-coding RNAs that regulate multiple signaling pathways and
affect cancer progression through targeting associated genes.
miRNAs may induce degradation or restrain translation of their
target mRNAs by binding to the 3′ untranslated region (UTR)
(4,5).
miRNAs have been implicated in tumor progression and therapeutic
resistance; however, the molecular mechanisms that define this
state remain unclear (6).
Dysregulation of miRNAs participating in BC progression, inlcuding
oncogenesis, apoptosis, proliferation, metastasis, invasion and
even drug resistance (7). Increasing
evidence suggests that miRNAs may participate in BC progression
through altering the stemness of BCSCs, which primarily involves
tumor formation, self-renewal, differentiation, metastasis,
tumorigenicity and chemotherapy resistance (8,9).
Therefore, BCSCs may be potential targets for miR-based
therapy.
BCSCs may be identified and isolated according to
their cell surface markers, including the phenotype of
CD44+CD24− and ALDH1+ (10,11).
miRNAs are involved in tumor biology by regulating associated
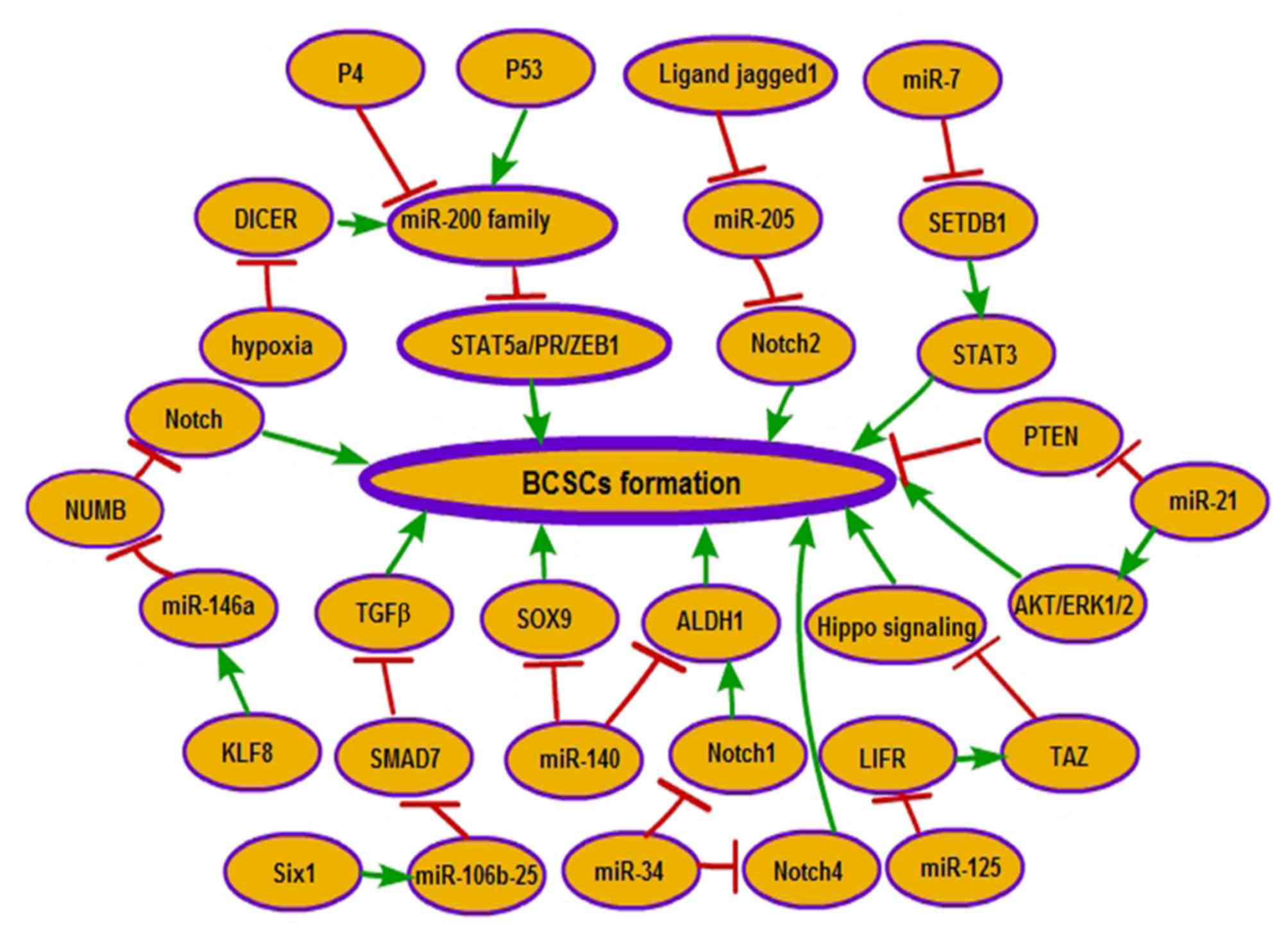
genes, and their roles in BCSC formation are becoming known: Tumor
suppressor tumor protein p53 (p53) transactivates miR-200c and
serves a role in reducing the CD44+CD24− stem
cell population through directly binding to the miR-200c promoter
and increasing expression of miR-200c (12). Endoribonuclease dicer (DICER), an
enzyme involved in microRNA processing, is suppressed by hypoxia
through silencing of the expression of the DICER promoter (13). Subsequently, decreased miRNA
processing leads to expression of the miR-200 target zinc finger
E-box binding homeobox 1 (ZEB1), which in turn causes an
epithelial-mesenchymal transition (EMT)-driven acquisition of stem
cell properties in BC (13). Sine
oculis homeobox homolog 1 (Six1), a metastatic regulator, was
suggested to activate the tumor-promotional arm of transforming
growth factor-β (TGF-β) signaling via increasing the expression of
the miR-106b-25 cluster. Upregulated miR-106b-25 by Six1 promotes
TGF-β-mediated upregulation of CD44+CD24−
BCSCs through targeting the inhibitory mothers against
decapentaplegic homolog (Smad) 7 protein of TGF-β signaling
(14). The miR-140/aldehyde
dehydrogenase 1 family member 1A (ALDH1)/sex determining region
Y-Box (SOX)9 axis also serves an important role in BCSC formation
in vivo. miR-140 is downregulated in ductal carcinoma in
situ (DCIS) stem-like cells, and inhibits CSC formation in
basal-like early-stage BC. miR-140 reduces BCSC formation by
targeting SOX9 and ALDH1, which have the highest level of activated
CSC factors in DCIS stem-like cells (15). mir-34a suppresses BCSC characteristics
at least partly through inhibiting Notch1 expression. Notch1
expression decreased by mir-34a was identified to decrease the
percentage of CD44+CD24− cells and the
expression of ALDH1 (16). Ligand
jagged1 is secreted from the tumor stroma to promote the BCSC
phenotype through repressing the expression of miR-205. Hairy and
enhancer of split-1, as a transcription repressor activated by
Jagged1-Notch1 signaling, is involved in the inhibition of miR-205
expression (17). Decreased miR-205
increases the BCSC population ratio through significantly promoting
the proportion of the CSCs population that exhibits the
CD44+CD24− phenotype (17). In addition, Notch2, as a target of
miR-205 and also activated by loss of miR-205, is involved in CSC
stemness through increasing the CD44+CD24−
cell population (15). Ectopic
expression of miR-7 significantly decreases the percentage of
CD44+CD24− cells in MDA-MB-231 cells. miR-7
decreases the BCSC population in BC partly by the downregulation of
the signal transducer and activator of transcription 3 (STAT3)
pathway via inhibiting the expression of SET domain bifurcated 1
(SETDB1) (18). Krüppel-like factor
(KLF) 8-induced expression of miR-146a was suggested to account for
the acquisition of BCSC traits, due to its effect on increasing the
CD44+CD24− and ALDH+ expression
levels. miR-146a mediates KLF8-induced CSC features by inhibiting
the expression of the Numb homolog (NUMB), a Notch signaling
inhibitor (19). miR-21 was
identified to increase the proportion of BCSCs that expressed the
CSC surface biomarkers CD44+CD242− and
ALDH1+ (20). miR-21
induces the BCSC phenotype through the depletion of phosphatase and
tensin homolog and the activation of protein kinase B (AKT) and
extracellular signal-related kinase 1/2 (20). miRNA-125a-targeted leukemia inhibitory
factor receptor changes the activity of transcriptional
co-activator with PDZ-binging motif, an effector molecule in the
Hippo pathway, through which miRNA-125a increases the percentage of
stem cells in MCF7 cells (21).
Increased miR-34c inhibits the development of
CD44+CD24− and ALDH+ cells in the
BC cell population by targeting Notch4 (22). Progesterone (P4) contributes to the
expansion of stem-like breast cancer cells through decreasing the
level of miR-141, a member of the miR-200 family of tumor
suppressors, which directly targets STAT5A and progesterone
receptor (PR) (Fig. 1) (23).
As a characteristic feature of stem cells,
self-renewal ensures that BCSCs survive for long periods of time.
miRNAs, including miR-145, miR-128b, miR-15/16 (miR-16, miR-15b),
and the miR-103/107 (miR-103, miR-107) and miR-200 (miR-200b,
miR-200a, miR-429, miR-200c) families, were identified to be
involved in the mammosphere formation of BCSCs. Individual
upregulation of these miRNAs restrains the formation of
mammospheres by at least 50%. The miR-200 family directly targets
the stem cell transcription factor KLF4, enhancer of zeste 2
polycomb repressive complex 2 subunit (EZH2) and polycomb complex
protein BMI1 (BMI1) (24).
Additionally, miR-200 also targets and inhibits the suppressor of
zeste 12 (SUZ12) (25) and BMI1
(26), which, respectively, are are
subunits of the polycomb repressive complex (PRC) 2 and PRC1 that
repress transcription. miR-200 target genes may also be regulated
by other miRNAs that are reduced in BCSCs and essential for BCSC
formation (27). For example, ZEB2
and KLF4 are putative targets of miR-145, BMI1 is a putative target
of miR-128b, and SUZ12 is a putative target of the miR-103/107 and
miR-15b/16 families (24). Thus, we
can conclude that the expression of the CSC-modulating miRNAs,
including miR-200b, miR-15b, miR-128b, miR-107 and miR-145, is
inhibited by ZEB1 and ZEB2. In addition, TGF-β expression
synergizes with RAC-α serine/threonine-protein kinase-knockdown in
promoting mammosphere formation through a decrease in the abundance
of miR-200 (28). miR-16 inhibits the
mammosphere-forming ability of mammary tumor cells by regulating
wild-type p53-induced phosphatase 1 (WIP1) induction in the DNA
damage response through targeting the 3′UTR of WIP1 (29). Pleckstrin homology-like domain, family
A, member1 (PHLDA1) in mammospheres, inhibited by miR-181a/b, leads
to attenuated mammosphere formation in estrogen receptor
(ER)+BC. Additionally, crosstalk between ER and the
nuclear factor κ-light-chain-enhancer of activated B cells pathway
contributes to the upregulation of PHLDA1, directly through the
increased transcription and indirectly through the inhibition of
miR-181a/b (30). Estrogen (E2) was
identified to enhance breast tumor-initiating cell survival by
downregulating miR-140, which targets SOX2 (31). Concomitantly, the transcription of
miR-140 was also inhibited by estrogen receptor α (ERα), which
binds to the promoter of miR-140; reduced miR-140 increases breast
tumor-initiating cell renewal via targeting SOX2 (32). In addition, miR-140 serves a critical
role in regulating stem cell signaling in basal-like DCIS. miR-140
overexpression reduces stem cell renewal and tumor growth in
vivo through directly targeting ALDH1 and SOX9, the stem-cell
factors with the highest expression level in basal-like DCIS stem
cells (15). Upregulated mir-93
inhibits several stem cell regulatory genes, including STAT3, Janus
kinase 1, high mobility group AT-hook 2 (HMGA2), enhancer of zeste
1 polycomb repressive complex 2 subunit, SOX4 and RAC-γ
serine/threonine-protein kinase, through which miR-93 results in
the depletion of BCSCs (33). Side
population (SP) cells exhibit characteristics similar to CSCs
(34). It was suggested that miR-99a
reduces the self-renewal capacities of BC SP cells in vivo
through activating mammalian target of rapamycin (mTOR), a
downstream effector of the AKT/phosphoinositide 3-kinase (PI3K)
signaling pathway (35). Ectopic
expression of miR-34c inhibits the self-renewal of BCSCs and
suppresses tumor growth by targeting and silencing expression of
Notch4 (21). Cyclo-oxygenase (COX)-2
promotes the BCSC phenotype by increasing the expression of
miR-526b, owing to the activation of the prostaglandin E2 receptor
EP4 and downstream PI3K/AKT and protein kinase A signaling pathways
(36). Ectopic miR-526b increases the
number and size of spheroids, which suggests that upregulated
miR-526b is associated with the stimulation of BCSCs (36).
The balance between self-renewal and differentiation
is an additionally important characteristic of BCSCs, and multiple
miRNAs have been suggested to participate in regulating this
balance. In the CD44+ cell population, miR-29 members
are downregulated by P4, which promotes the expansion of stem-like
cancer cells in ER+ and PR+ BC. Concurrently,
downregulated miR-29 members also enhance the expansion of
CD44+ and CK5+ cells in response to P4
(37). The reprogramming of
differentiated cells into pluripotent stem cells and the
maintenance of BCSCs are inhibited by miR-29 members that target
KLF4 (37). Induced expression of
miR-200c promotes differentiation of claudin-low tumors in
vivo by increasing the expression of basal and luminal markers,
specifically keratins K14 and K8 (27). Notably, the differentiation is more
similar to the differentiated basal-like tumors compared with the
undifferentiated claudin-low tumors from which they originated
(27). miR-200c alters the
functionality of BCSCs through exhibiting the expression of stem
cell-associated genes BMI1 and EZH2, and increases the levels of
differentiation markers GATA binding protein and E74-like factor 5,
which results in a more differentiated status of claudin-low tumors
in vivo (27). miR-200c may
also induce differentiation of BCSCs by targeting BMI1 (38). miR-100 serves a pivotal role in
modulating differentiation of patient-derived basal-like BCSCs
(39). Upregulated miR-100 interferes
with the properties of BCSCs, and alters the basal-like phenotype
into a more differentiated luminal phenotype, via inhibiting
polo-like kinase 1 (Plk1), SWItch/sucrose non-fermentable-related,
matrix-associated, actin-dependent regulator of chromatin, and the
Wnt/β-catenin signaling pathway (39).
Invasion and metastasis remain the most complex and
challenging problems of BC treatment and prognosis. EMT, which is
assessed by the decreased expression of epithelial cell markers
[keratins and epithelial (E)-cadherin] and the increased expression
of mesenchymal cell markers [α-smooth muscle actin (α-SMA),
vimentin and N-cadherin], contributes to invasion and metastasis in
BC and is significantly associated with the acquisition of BCSC
characteristics (40). Previous
evidence has demonstrated that multiple miRNAs are also involved in
the metastasis process of BC through inhibiting BCSC functionality.
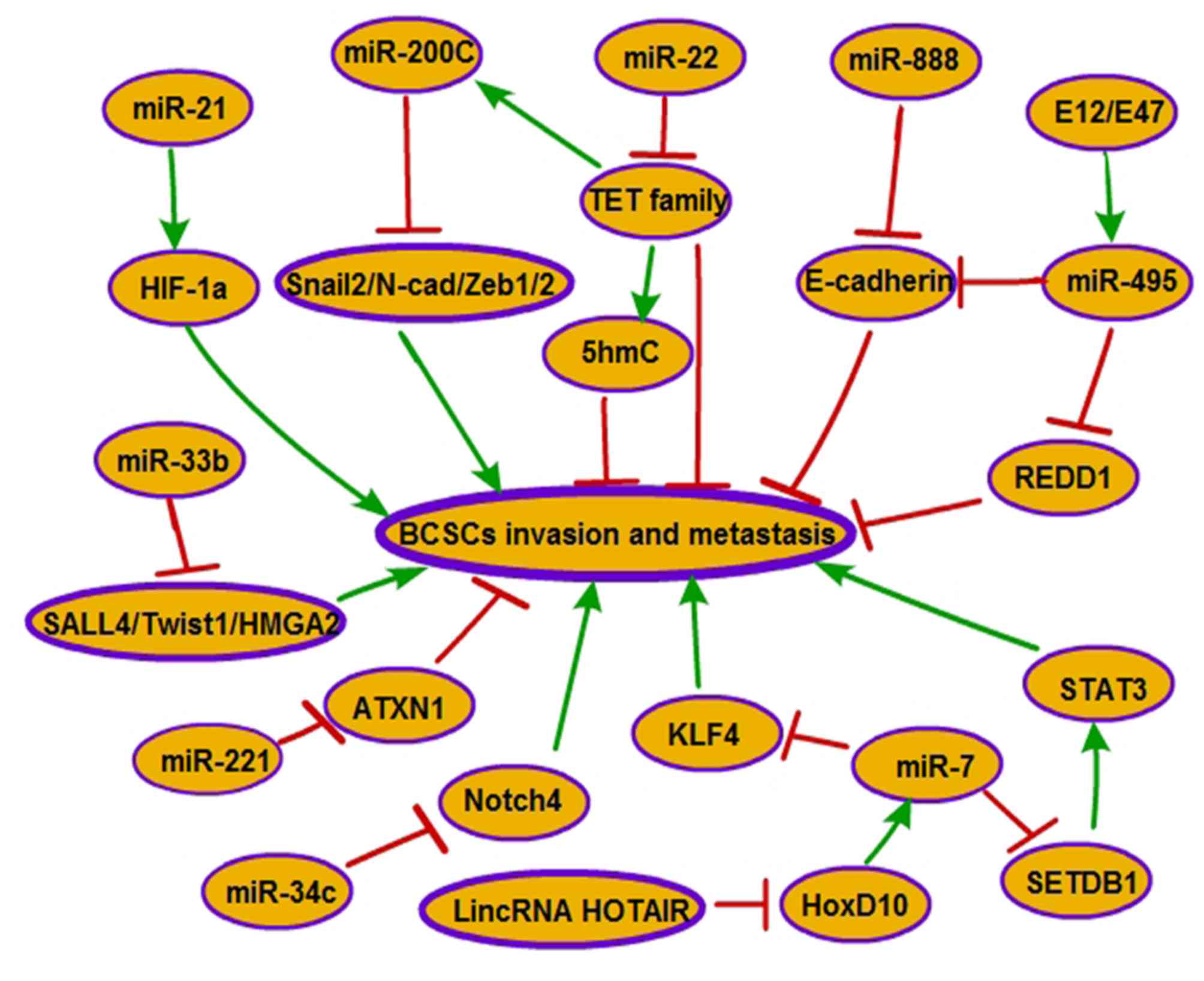
Han et al revealed that miR-21 and hypoxia-inducible
factor-1α (HIF-1α) are upregulated in the third-sphere forming
(3-S) CSC-like cells, which are isolated from MCF-7 parental cells
and exhibit high levels of CSC surface markers
(CD44+/CD24−/low and ALDH1+).
Antagonism of miR-21 reverses EMT and impedes invasion and
migration in the 3-S CSC-like cells via HIF-1α downregulation
(41). In addition, miR-21
re-expression promotes the process of migration and invasion by
enhancing the characteristics of CSCs and activating the EMT
process in BC MCF-7 cells (42). As
an important regulator of EMT, the upregulation of the miR-200
family reverses EMT and reduces metastatic potential in claudin-low
breast cancer, which is significantly enriched in BCSCs, via the
downregulation of ZEB1/2, zinc finger protein SNAI2, N-cadherin and
transcriptional repressors of E-cadherin (27). It was demonstrated that miR-22 expands
BCSC in size, and enhances cell invasion and metastasis in a BC
mouse xenograft through its ability to repress the expression of
miR-200 and 5-hydroxymethylcytosine (5hmC) by directly targeting
members of the ten-eleven translocation (TET) family (43). miR-33b acts as a negative regulator of
BC stem-like cell self-renewal, migration and invasion in highly
metastatic BC cells, and represses lung metastasis in vivo
by targeting its downstream targets, including sal-like protein 4,
twist-related protein 1 and HMGA2 (44). miR-888 was identified to act as a
repressor of the adherens junction pathway and serve a critical
role in maintaining SP properties and regulating EMT, invasion and
metastasis in MCF-7 SP cells via directly targeting E-cadherin
(45). Increases in levels of miR-495
enriched in PROCR+/ESA+ and
CD44+/CD24−/low BCSC subpopulations are
upregulated by E12/E47 (46). miR-495
overexpression maintains BCSC properties such as promotion of
metastasis and invasion via suppressing E-cadherin and DNA
damage-inducible transcript 4 protein (REDD1) (46). The overexpression of miR-221 is able
to stimulate stem-like properties in the luminal type of BC cells
and induce EMT in BC cells through downregulating ataxin-1
(47). miR-34c reduction via DNA
methylation in breast tumor-initiating cells (BT-Ics) promotes
self-renewal, EMT and migration of BT-ICs by targeting Notch4
(22). miR-7 suppresses brain
metastasis of BCSCs in vivo by downregulating the critical
downstream target KLF4, an induced pluripotent stem cell gene that
is important for the maintenance of stemness of progenitor cells
(48). Additionally, miR-7 was also
demonstrated to reduce the size of the BCSC population, partially
reverse EMT in MDA-MB-231 cells and repress the metastasis of BCSCs
in adrenal glands, kidneys and lungs in non-obese diabetic/severe
combined immune deficiency (NOD/SCID) mice by directly targeting
the 3′UTR of SETDB1, which serves a key role in activating the
STAT3 pathway. In addition, long intergenic non-coding RNA homeobox
(HOX) transcript antisense RNA indirectly inhibits miR-7 via
downregulating the expression of homeobox D10 (18) (Fig.
2).
miRNAs are considered to be potential biomarkers or
therapeutic targets of BC, due to their capability of modulating
stem cell biology, including clonogenicity and tumorigenicity.
miR-526b, a COX-2-upregulated oncogene, promoted tumorsphere
formation in BC cells and lung colony formation in an experimental
metastasis model, relying on EP4 receptor activity and cyclic
adenosine monophosphate (CAMP) and downstream PI3K/AKT signaling
pathways (36). In addition, miR-495
that is upregulated by transcription factor E2A immunoglobulin
enhancer-binding factors E12/E47, directly represses E-cadherin and
REDD1, and contribute to an increase in BCSC traits and hypoxia
resistance, which then promotes colony formation in BC cells and
tumorigenesis in mice (46).
Progestins significantly increase mammosphere formation in
vitro and enhance the tumor-initiating capability in
hormone-responsive breast cancer via repressing miR-29, to augment
the PR-mediated upregulation of KLF4 (15). The glabridin (GLA)/miR-148a/SMAD2 axis
serves a critical role in modulating CSC-like properties, such as
the formation of mammospheres and colonies. GLA-upregulated
miR-148a results in a repression of clone formation, as miR-148a is
able to inhibit endogenous TGF-β/SMAD2 signaling in BC cells
(49). It was identified that miR-99a
directly inhibits the mTOR signaling pathway in breast cancer SP
cells, which results in the suppression of tumorigenicity in
vivo (35). In addition, miR-200c
that targets BMI1, suppresses clonogenicity and tumorigenicity of
BCSCs in NOD/SCID mice due to the inhibition of self-renewal and
proliferation of BCSCs (38).
Conversely, miR-22, an oncogene, is able to promote tumorigenesis
in transgenic mice through expanding the BCSCs in size (43). The overexpression of miR-22 represses
the expression of miR-200 s and 5hmC by targeting members of the
TET family (43). miR-128-2, embedded
in the intron of the CAMP-regulated phosphoprotein 21 gene at
chromosome 3p22.3, serves critical roles in the modulation of
oncogenic transformation and progression in mammary epithelial
cells (50). miR-128-2 is
downregulated by TGF-β through the phosphorylation of TGF-β1
receptor to enhance a specific SNAIL protein expression. In
addition, miR-128-2, downregulated by SNAIL, promotes mammary
epithelial oncogenic transformation via expressing a group of
direct targets (colony-stimulating factor 1, BMI1, Lin-28 homology
A, nanog homeobox and KLF4), which together act to activate the
STAT3 and PI3K/AKT signaling pathways (50).
Chemotherapy resistance in BC is one of the major
obstacles for clinical intervention, and one of the hallmarks of
BCSCs. An increasing number of studies have suggested the key role
of miRNAs in chemoresistance by regulating BCSC traits (51). Cross-talk between miR-200c and BMI1,
modulated by p53, and BMI1 repression in breast cancer cells
promotes the sensitivity of BC to 5-fluorouracil through reducing
the proportion of CD44+/CD24− cells in the
BCSC population, and inducing susceptive apoptosis (52). The differentiation process, triggered
by miR-100, which attenuates BCSC properties and promotes the basal
like phenotype into a more differentiated luminal phenotype in
patient-derived basal-like BCSCs, induces the expression of ER and
sensitizes basal-like BCSCs to hormonal therapy via downregulating
PLK1 (39). KLF8 serves a critical
role in regulating the induction and maintenance of BCSC traits,
which contributes to the resistance of cells to the cytotoxic
effect of paclitaxel in MCF-10A cells via targeting miR-146a that
binds to the 3′-UTR of NUMB and inhibits NUMB expression (19). It was confirmed that histone
deacetylase (HDAC)1 and HDAC7 are downstream targets of miR-34a and
are upregulated in the CD44+CD24−
subpopulation (53). Deacetylation of
acetyl-heat shock protein 70 (HSP70; K246) by HDAC7 and HDAC1
increases resistance to therapeutics [paclitaxel (PTX), doxorubicin
and cisplatin] through the inhibition of autophagy in MCF-7 cells
expressing wild-type HSP70 (53).
Furthermore, the overexpression of miR-34a that targets Notch1 also
enhances chemosensitivity to PTX by suppressing the proliferation
of BCSCs (16). Metformin, the
anti-type II diabetes (T2D) drug, was identified to decrease the
generation of SPs in BC cells, leading to an attenuation in
chemoresistance to docetaxel and tumor-seeding ability through
miR-27b-mediated inhibition of ectonucleotide
pyrophosphatase/phosphodiesterase 1 (ENPP1). Uninhibited ENPP1
enhances the generation of SPs via upregulating the adenosine
5′-triphosphate (ATP) cassette sub-family G member 2 transporter
(54). miR-125b, as a positive
regulator of SP and CSC properties in BC cell lines and primary BC
cells, contributes to chemoresistance to paclitaxel (55). Additionally, ectopic overexpression of
miR-205 or miR-125b and silencing miR-424 expression are sufficient
to induce a subpopulation of cells that exhibit stem-like
characteristics, which were identified to confer aromatase
inhibitor (AI) resistance by activating the AKT/mTOR pathway in 2
AI-resistant cell lines (Res-Let cells and Res-Ana cells) (56). The activation of Akt, induced by
miR-125b, enhances sensitivity to letrozole and overcomes letrozole
resistance in Res-Let cells (56).
Downregulated miR-128 results in chemotherapeutic resistance to
doxorubicin, through enhancing cell viability and reducing
apoptosis and DNA damage in BT-ICs via the modulation of two
independent targets, BMI1 and ATP-binding cassette subfamily C
member 5 (57). miR-16 has been
revealed to be downregulated in BCSCs and to suppress BCSC
properties. The overexpression of miR-16 sensitizes MCF-7 cells to
doxorubicin by inhibiting Wip1 (Table
I) (29).
An increasing number of studies have demonstrated
that miRNAs participate in regulating BCSC characteristics via
targeting associated genes. miRNAs activate or inactivate multiple
signaling pathways by targeting associated genes to effect BCSC
formation, self-renewal, differentiation, invasion, metastasis,
clonogenicity, tumorigenicity and chemotherapy resistance. BCSCs,
as essential drivers of BC metastasis, chemotherapy resistance,
relapse and poor prognosis, may be effective therapeutic targets in
BC. miRNAs act as critical regulators of BCSC characteristics,
which may provide a novel therapeutic strategy for the treatment of
BC. In BCSCs, decreased expression of onco-miRNAs (miR-106b-25,
miR-146a, miR-21, miR125, miR-526b, miR-22 and miR-888) or
increased expression of anti-onco-miRNAs (miR-140, miR-34, miR-7,
miR-16, miR-93, miR-99a and the miR-200 family) may inhibit BC
progression by reducing the levels of expression of oncogenes,
while enhancing the levels of expression of anti-oncogenes.
Therefore, BCSCs may be potential targets for the miR-based therapy
of BC.
The present review focused on the complicated
associations between miRNAs and BCSCs in BC progression. miRNAs, as
oncogenes or tumor suppressor genes, may serve pivotal roles in BC
progression by regulating BCSCs, which are a subpopulation of cells
that exhibit significant potential for self-renewal, invasion,
metastasis and chemoresistance in BC. Several regulatory pathways
have been identified, and future studies should be performed to
investigate the effects of these regulatory pathways. A
comprehensive understanding of the association between BCSCs and
miRNAs may provide novel and safer therapeutic strategies for
BC.
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no.
8157101910).
|
1
|
Jeong H, Kim J, Lee Y, Seo JH, Hong SR and
Kim A: Neuregulin-1 induces cancer stem cell characteristics in
breast cancer cell lines. Oncol Rep. 32:1218–1224. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Visvader JE and Lindeman GJ: Cancer stem
cells: Current status and evolving complexities. Cell Stem Cell.
10:717–728. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Fan XM, Mao L, Zhang JY, Li J, Wu
JZ and Tang JH: MicroRNA-224: As a potential target for miR-based
therapy of cancer. Tumour Biol. 36:6645–6652. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang D, Zhou P, Wang W, Wang X, Li J, Sun
X and Zhang L: MicroRNA-616 promotes the migration, invasion and
epithelial-mesenchymal transition of HCC by targeting PTEN. Oncol
Rep. 35:366–374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
El Helou R, Pinna G, Cabaud O, Wicinski J,
Bhajun R, Guyon L, Rioualen C, Finetti P, Gros A, Mari B, et al:
miR-600 acts as a bimodal switch that regulates breast cancer stem
cell fate through WNT signaling. Cell Rep. 18:2256–2268. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Zhang J, Sun X, Su Q and You C:
Down-regulation of miR-29b in carcinoma associated fibroblasts
promotes cell growth and metastasis of breast cancer. Oncotarget.
8:39559–39570. 2017.PubMed/NCBI
|
|
8
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A MicroRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang Y, Xiang J, Chen Z, Gu X, Li Z, Tang
F and Zhou Z: miRNA expression profile of colon cancer stem cells
compared to non-stem cells using the SW1116 cell line. Oncol Rep.
28:2115–2124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dontu G, Abdallah WM, Foley JM, Jackson
KW, Clarke MF, Kawamura MJ and Wicha MS: In vitro propagation and
transcriptional profiling of human mammary stem/progenitor cells.
Genes Dev. 17:1253–1270. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chang CJ, Chao CH, Xia W, Yang JY, Xiong
Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al: p53 regulates
epithelial-mesenchymal transition and stem cell properties through
modulating miRNAs. Nat Cell Biol. 13:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van den Beucken T, Koch E, Chu K,
Rupaimoole R, Prickaerts P, Adriaens M, Voncken JW, Harris AL,
Buffa FM, Haider S, et al: Hypoxia promotes stem cell phenotypes
and poor prognosis through epigenetic regulation of DICER. Nat
Commun. 5:52032014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smith AL, Iwanaga R, Drasin DJ, Micalizzi
DS, Vartuli RL, Tan AC and Ford HL: The miR-106b-25 cluster targets
Smad7, activates TGF-β signaling, and induces EMT and tumor
initiating cell characteristics downstream of Six1 in human breast
cancer. Oncogene. 31:5162–5171. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li Q, Yao Y, Eades G, Liu Z, Zhang Y and
Zhou Q: Downregulation of miR-140 promotes cancer stem cell
formation in basal-like early stage breast cancer. Oncogene.
33:2589–2600. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang L, Mao J, Tao Y, Song B, Ma W, Lu Y,
Zhao L, Li J, Yang B and Li L: MicroRNA-34a suppresses the breast
cancer stem cell-like characteristics by downregulating Notch1
pathway. Cancer Sci. 106:700–708. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chao CH, Chang CC, Wu MJ, Ko HW, Wang D,
Hung MC, Yang JY and Chang CJ: MicroRNA-205 signaling regulates
mammary stem cell fate and tumorigenesis. J Clin Invest.
124:3093–3106. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang X, Lu H, Li T, Yu L, Liu G, Peng X
and Zhao J: Krüppel-like factor 8 promotes tumorigenic mammary stem
cell induction by targeting miR-146a. Am J Cancer Res. 3:356–373.
2013.PubMed/NCBI
|
|
20
|
Han M, Liu M, Wang Y, Chen X, Xu J, Sun Y,
Zhao L, Qu H, Fan Y and Wu C: Antagonism of miR-21 reverses
epithelial-mesenchymal transition and cancer stem cell phenotype
through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One.
7:e395202012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nandy SB, Arumugam A, Subramani R, Pedroza
D, Hernandez K, Saltzstein E and Lakshmanaswamy R: MicroRNA-125a
influences breast cancer stem cells by targeting leukemia
inhibitory factor receptor which regulates the Hippo signaling
pathway. Oncotarget. 6:17366–17378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu F, Jiao Y, Zhu Y, Wang Y, Zhu J, Cui X,
Liu Y, He Y, Park EY, Zhang H, et al: MicroRNA 34c gene
down-regulation via DNA methylation promotes self-renewal and
epithelial-mesenchymal transition in breast tumor-initiating cells.
J Biol Chem. 287:465–473. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Finlay-Schultz J, Cittelly DM, Hendricks
P, Patel P, Kabos P, Jacobsen BM, Richer JK and Sartorius CA:
Progesterone downregulation of miR-141 contributes to expansion of
stem-like breast cancer cells through maintenance of progesterone
receptor and Stat5a. Oncogene. 34:3676–3687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Polytarchou C, Iliopoulos D and Struhl K:
An integrated transcriptional regulatory circuit that reinforces
the breast cancer stem cell state. Proc Natl Acad Sci USA.
109:14470–14475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iliopoulos D, Lindahl-Allen M, Polytarchou
C, Hirsch HA, Tsichlis PN and Struhl K: Loss of miR-200 inhibition
of Suz12 leads to polycomb-mediated repression required for the
formation and maintenance of cancer stem cells. Mol Cell.
39:761–772. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wellner U, Schubert J, Burk UC,
Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D,
zur Hausen A, et al: The EMT-activator ZEB1 promotes tumorigenicity
by repressing stemness-inhibiting microRNAs. Nat Cell Biol.
11:1487–1495. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Knezevic J, Pfefferle AD, Petrovic I,
Greene SB, Perou CM and Rosen JM: Expression of miR-200c in
claudin-low breast cancer alters stem cell functionality, enhances
chemosensitivity and reduces metastatic potential. Oncogene.
34:5997–6006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Iliopoulos D, Polytarchou C,
Hatziapostolou M, Kottakis F, Maroulakou IG, Struhl K and Tsichlis
PN: MicroRNAs differentially regulated by Akt isoforms control EMT
and stem cell renewal in cancer cells. Sci Signal. 2:ra622009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang X, Wan G, Mlotshwa S, Vance V,
Berger FG, Chen H and Lu X: Oncogenic Wip1 phosphatase is inhibited
by miR-16 in the DNA damage signaling pathway. Cancer Res.
70:7176–7186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kastrati I, Canestrari E and Frasor J:
PHLDA1 expression is controlled by an estrogen
receptor-NFκB-miR-181 regulatory loop and is essential for
formation of ER+ mammospheres. Oncogene. 34:2309–2316. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vazquez-Martin A, Cufí S, López-Bonet E,
Corominas-Faja B, Cuyàs E, Vellon L, Iglesias JM, Leis O, Martín AG
and Menendez JA: Reprogramming of non-genomic estrogen signaling by
the stemness factor SOX2 enhances the tumor-initiating capacity of
breast cancer cells. Cell Cycle. 12:3471–3477. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang Y, Eades G, Yao Y, Li Q and Zhou Q:
Estrogen receptor α signaling regulates breast tumor-initiating
cells by down-regulating miR-140 which targets the transcription
factor SOX2. J Biol Chem. 287:41514–41522. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu S, Patel SH, Ginestier C, Ibarra I,
Martin-Trevino R, Bai S, McDermott SP, Shang L, Ke J, Ou SJ, et al:
MicroRNA93 regulates proliferation and differentiation of normal
and malignant breast stem cells. PLoS Genet. 8:e10027512012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liao J, Liu PP, Hou G, Shao J, Yang J, Liu
K, Lu W, Wen S, Hu Y and Huang P: Regulation of stem-like cancer
cells by glutamine through β-catenin pathway mediated by redox
signaling. Mol Cancer. 16:512017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Z, Han Y, Cheng K, Zhang G and Wang
X: miR-99a directly targets the mTOR signalling pathway in breast
cancer side population cells. Cell Prolif. 47:587–595. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Majumder M, Landman E, Liu L, Hess D and
Lala PK: COX-2 elevates oncogenic miR-526b in breast cancer by EP4
activation. Mol Cancer Res. 13:1022–1033. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cittelly DM, Finlay-Schultz J, Howe EN,
Spoelstra NS, Axlund SD, Hendricks P, Jacobsen BM, Sartorius CA and
Richer JK: Progestin suppression of miR-29 potentiates
dedifferentiation of breast cancer cells via KLF4. Oncogene.
32:2555–2564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Petrelli A, Carollo R, Cargnelutti M,
Iovino F, Callari M, Cimino D, Todaro M, Mangiapane LR, Giammona A,
Cordova A, et al: By promoting cell differentiation, miR-100
sensitizes basal-like breast cancer stem cells to hormonal therapy.
Oncotarget. 6:2315–2330. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chiotaki R, Polioudaki H and
Theodoropoulos PA: Cancer stem cells in solid and liquid tissues of
breast cancer patients: Characterization and therapeutic
perspectives. Curr Cancer Drug Targets. 15:256–269. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han M, Wang Y, Liu M, Bi X, Bao J, Zeng N,
Zhu Z, Mo Z, Wu C and Chen X: MiR-21 regulates
epithelial-mesenchymal transition phenotype and hypoxia-inducible
factor-1α expression in third-sphere forming breast cancer stem
cell-like cells. Cancer Sci. 103:1058–1064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Han M, Liu M, Wang Y, Mo Z, Bi X, Liu Z,
Fan Y, Chen X and Wu C: Re-expression of miR-21 contributes to
migration and invasion by inducing epithelial-mesenchymal
transition consistent with cancer stem cell characteristics in
MCF-7 cells. Mol Cell Biochem. 363:427–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Song SJ, Poliseno L, Song MS, Ala U,
Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC, et al:
MicroRNA-antagonism regulates breast cancer stemness and metastasis
via TET-family-dependent chromatin remodeling. Cell. 154:311–324.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin Y, Liu AY, Fan C, Zheng H, Li Y, Zhang
C, Wu S, Yu D, Huang Z, Liu F, et al: MicroRNA-33b inhibits breast
cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci Rep.
5:99952015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang S, Cai M, Zheng Y, Zhou L, Wang Q
and Chen L: miR-888 in MCF-7 side population sphere cells directly
targets E-cadherin. J Genet Genomics. 41:35–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hwang-Verslues WW, Chang PH, Wei PC, Yang
CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY and Lee WH: miR-495
is upregulated by E12/E47 in breast cancer stem cells, and promotes
oncogenesis and hypoxia resistance via downregulation of E-cadherin
and REDD1. Oncogene. 30:2463–2474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ke J, Zhao Z, Hong SH, Bai S, He Z, Malik
F, Xu J, Zhou L, Chen W, Martin-Trevino R, et al: Role of
microRNA221 in regulating normal mammary epithelial hierarchy and
breast cancer stem-like cells. Oncotarget. 6:3709–3721. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Okuda H, Xing F, Pandey PR, Sharma S,
Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, et al:
miR-7 suppresses brain metastasis of breast cancer stem-like cells
by modulating KLF4. Cancer Res. 73:1434–1444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang F, Li Y, Mu J, Hu C, Zhou M, Wang X,
Si L, Ning S and Li Z: Glabridin inhibits cancer stem cell-like
properties of human breast cancer cells: An epigenetic regulation
of miR-148a/SMAd2 signaling. Mol Carcinog. 55:929–940. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qian P, Banerjee A, Wu ZS, Zhang X, Wang
H, Pandey V, Zhang WJ, Lv XF, Tan S, Lobie PE and Zhu T: Loss of
SNAIL regulated miR-128-2 on chromosome 3p22.3 targets multiple
stem cell factors to promote transformation of mammary epithelial
cells. Cancer Res. 72:6036–6050. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen W, Zhou S, Mao L, Zhang H, Sun D,
Zhang J, Li J and Tang JH: Crosstalk between TGF-β signaling and
miRNAs in breast cancer metastasis. Tumour Biol. 37:10011–10019.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yin J, Zheng G, Jia X, Zhang Z, Zhang W,
Song Y, Xiong Y and He Z: A Bmi1-miRNAs cross-talk modulates
chemotherapy response to 5-fluorouracil in breast cancer cells.
PLoS One. 8:e732682013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wu MY, Fu J, Xiao X, Wu J and Wu RC:
MiR-34a regulates therapy resistance by targeting HDAC1 and HDAC7
in breast cancer. Cancer Lett. 354:311–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Takahashi RU, Miyazaki H, Takeshita F,
Yamamoto Y, Minoura K, Ono M, Kodaira M, Tamura K, Mori M and
Ochiya T: Loss of microRNA-27b contributes to breast cancer stem
cell generation by activating ENPP1. Nat Commun. 6:73182015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang HJ, Guo YQ, Tan G, Dong L, Cheng L,
Li KJ, Wang ZY and Luo HF: miR-125b regulates side population in
breast cancer and confers a chemoresistant phenotype. J Cell
Biochem. 114:2248–2257. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Vilquin P, Donini CF, Villedieu M, Grisard
E, Corbo L, Bachelot T, Vendrell JA and Cohen PA: MicroRNA-125b
upregulation confers aromatase inhibitor resistance and is a novel
marker of poor prognosis in breast cancer. Breast Cancer Res.
17:132015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhu Y, Yu F, Jiao Y, Feng J, Tang W, Yao
H, Gong C, Chen J, Su F, Zhang Y and Song E: Reduced miR-128 in
breast tumor-initiating cells induces chemotherapeutic resistance
via Bmi-1 and ABCC5. Clin Cancer Res. 17:7105–7115. 2011.
View Article : Google Scholar : PubMed/NCBI
|