Introduction
Lung cancer is the most common type of cancer among
men worldwide, accounting for ~16.7% of all estimated new cancer
cases (1). In Thailand, lung cancer
is the second leading cancer in men with an age-standardized
incidence rate of 27.1/100,000 (2).
Approximately 85% of patients with lung cancer are diagnosed with
non-small cell lung cancer (NSCLC). The survival of patients with
lung cancer primarily depends on the stage of disease at the time
of diagnosis. The 5-year survival rate of patients with early stage
NSCLC is between 25 and 52%, while the rate is <4% for those
with advanced stages (3). Although
several advanced therapeutic modalities are available, the
mortality rate remains high. Thus, identifying biological markers
that are able to detect the disease at the early stage, or predict
treatment response or prognosis is important for improving patient
survival.
14-3-3 proteins are small acidic polypeptides that
are 28–33 kDa in size, and consist of at least seven isoforms, β,
ε, γ, η, σ, τ/θ, and ξ, in mammalian cells. The proteins are
spontaneously self-assembled to form homodimers or heterodimers and
bind to various cellular proteins (4). The interaction between 14-3-3 proteins
and other proteins has been demonstrated in a number of signaling
pathways, including cell cycle progression, signal transduction,
and apoptosis (5,6). Among the various isoforms, 14-3-3σ is
the most common isoform reported to be involved in carcinogenesis
via a tumor suppression manner (7).
Loss of 14-3-3σ expression has been demonstrated in a variety of
cancer types, particularly in adenocarcinoma-type tumors, including
breast carcinoma (8) and gastric
carcinoma (9). Subsequently, a number
of these studies have demonstrated that epigenetic silencing
through CpG methylation is responsible for the loss or reduction of
14-3-4σ expression (10). With
regards to prognosis, loss of expression of this protein has been
reported to be associated with poor prognosis in ovarian and
nasopharyngeal carcinoma (11,12). By
contrast, poor overall survival in patients with high expression of
14-3-3σ has been revealed in colorectal cancer, oral squamous cell
carcinoma and gastric cancer (13–15).
In lung cancer, 14-3-3σ expression has been
demonstrated to be abundantly expressed in cancerous tissue samples
compared with normal lung tissues (16). In contrast with the results identified
in breast cancer (8), it has been
reported that 14-3-3σ expression is observed in the majority of
lung adenocarcinoma (17) or NSCLC
(18) tissues, and methylation is
more frequently observed in small cell lung cancer compared with
NSCLC (18). However, these findings
have been reported in a few studies with a limited number of cases.
In addition, there is little evidence to suggest that peripheral
DNA sources reflect 14-3-3σ methylation in NSCLC tissue. In the
present study, the association of 14-3-3σ methylation between tumor
tissues and matched serum was investigated, and the prognostic
value of 14-3-3σ expression was evaluated.
Materials and methods
Patients and specimens
Tissues and matched serum samples were obtained from
36 NSCLC patients who had not received any previous treatment. The
obtained tissues were frozen immediately at −80°C for DNA
extraction. For the matched serum samples, 10 ml peripheral blood
was collected from the fore arm vein and kept at room temperature
for 2 h. Serum was separated by double centrifugation at 1,600 × g
for 10 min and kept at −80°C until analysis. All patients were
diagnosed at Songklanagarind Hospital (Hat-Yai, Thailand), a
university hospital in Southern Thailand, between May 2012 and
April 2013. Fresh tumor tissue for methylation analysis was
obtained via bronchial biopsy through bronchoscopy simultaneously
when tissue was obtained for pathological diagnosis. Normal serum
samples (n=7) were collected from healthy blood donors. Written
informed consent was obtained from all patients. The mean age was
61 years (range, 32–83 years). Twenty-one patients were males and
15 were females. Seventeen cases were adenocarcinoma (ADC) and 15
cases were squamous cell carcinoma (SCC). The specific subtype was
not specified in 4 cases, which were recorded as
NSCLC-unclassified.
For the evaluation of prognostic significance of
14-3-3σ expression, 167 patients with stage I–IV of NSCLC who were
diagnosed, and treated at Songklanagarind Hospital between January
2006 and December 2008 were included. The clinicopathological data,
including the clinical stage were retrieved from the hospital
registry data. Clinical staging was based on the Tumor Node
Metastasis staging system of the International Union against Cancer
(7th Edition) (19). The histological
diagnosis was performed according to the WHO classification of lung
and pleural tumors (2004) (20). The
patients were followed up until September 2012. Data associated
with the mortality of patients was obtained from the provincial
nationwide-linked register of mortalities, where the law requires
all mortalities that have occurred in Thailand to be registered
within 24 h of occurrence. The present study was approved by the
Ethics Committee on Human Research, Faculty of Medicine, and Prince
of Songkla University (EC, 54-273-04-2-3 and 55-020-04-1-2).
Methylation-specific polymerase chain
reaction (MSP)
Genomic DNA was extracted from frozen tissue and
serum samples using standard proteinase K/phenol/chloroform methods
(21). The structural integrity of
DNA was confirmed using 1% agarose gel electrophoresis and
quantified with a spectrophotometer. The genomic DNA (1 µg) was
subjected to sodium bisulfite modification using the EZ DNA
Methylation-Gold kit (Zymo Research, Irvine, CA, USA) according to
the manufacturer's protocol. Modified DNA was resuspended in 15 µl
of nuclease-free water, quantified using a spectrophotometer and
stored at −70°C. For MSP analysis, the modified DNA (50 ng) was
amplified using methylation or unmethylation primers spanning the
region between CpG dinucleotides 3 and 9 of the 14-3-3σ gene. The
primers were designed according to a previous report by Ferguson
et al (8). Primers sequence
were as follows: Methylation forward,
5′-GATATGGTAGTTTTTATGAAAGGCGTCG-3′ and reverse,
5′-CCTCTAACCGCCCACCACG-3′; unmethylation forward,
5′-GATATGGTAGTTTTTATGAAAGGTGTTGTG-3′ and reverse,
5′-CCCTCTAACCACCCACCACA-3′. The MSP conditions maintained were as
follows: 1 cycle at 94°C for 3 min; 35 cycles at 94°C for 30 sec,
64°C (methylated reaction) or 59°C (unmethylated reaction) for 30
sec, 72°C for 45 sec; and 1 cycle at 72°C 10 min. The MSP products
were 108 and 109 bp for methylation, and unmethylation primers,
respectively. Universal human methylated and unmethylated DNA
strands (Zymo Research) were used as a positive control for each
primer. Following amplification, the MSP products were separated on
a 10% polyacrylamide gel, stained with ethidium bromide for 10 min
at room temperature, visualized as bands under ultraviolet
illumination and imaged using Gel Doc™ XR (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The density of bands was measured using
ImageJ software (National Institutes of Health, Bethesda, MD, USA).
The relative density of each methylated and unmethylated products
were obtained by dividing their values by the density of the
corresponding positive control. The 14-3-3σ methylation level
percentage was calculated as follows: Relative density of
methylated products/(relative density of methylated products +
relative density of unmethylated products).
Immunohistochemistry
Sections 4 µm thick were cut from a
paraffin-embedded block, deparaffinized with xylene and rehydrated
with ethanol. Antigen retrieval was enhanced by rapid heating in a
microwave in a citrate buffer (10 mM, pH 6.0) for 10 min.
Endogenous peroxidase activity was blocked at room temperature by
incubation with 3% hydrogen peroxide in methanol for 10 min. The
slides were then incubated with 10% normal goat serum (Santa Cruz
Biotechnology, Dallas, TX, USA) at room temperature for 20 min and
incubated with monoclonal antibody against 14-3-3σ (5D7,
sc-100,638; Santa Cruz Biotechnology) at a dilution of 1:800
overnight at 4°C in a humidified chamber. After washing with PBS
(pH 7.4), the slides were incubated with a biotinylated goat
anti-mouse IgG-B (sc-2039; Santa Cruz Biotechnology) at a dilution
of 1:300 for 40 min at room temperature. Antigen-antibody complexes
were detected using the avidin-biotin complex staining kit (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and a diaminobenzidine
solution (Merck KGaA, Darmstadt, Germany) as a substrate for 5 min
at room temperature. Finally, the slides were counterstained with
hematoxylin for 5 min at room temperature (Santa Cruz
Biotechnology), cover slipped and examined under a light microscope
at ×200. Oral squamous carcinoma tissue from a patient with oral
cancer was used as a positive control. Negative controls using the
same tissue without primary antibody were run in parallel.
Evaluation of immunohistochemical
staining
Immunoreactivity was qualitatively and
quantitatively evaluated in terms of intensity, and percentage of
positively stained cells, respectively. The intensity was scored as
follows: 0, no staining; 1, weak; 2, moderate; and 3, intense. The
percentage of positive cells was scored as follows: 0, ≤10%; 1,
11–30%; 2, 31–60%; and 3, ≥61%. Final scores (0–9) were then
obtained through multiplication of both scores. Four expression
groups were assigned as follows: No expression, final score 0; weak
expression, final score 1–3; moderate expression, final score 4–6;
and strong expression, final score 7–9. The expression of 14-3-3σ
was dichotomized to give negative expression (final score 0) and
positive expression (final score 1–9). Immunostaining was evaluated
by two independent pathologists, and discordant cases was
reevaluated and scored on the basis of consensus
interpretation.
Statistical analysis
Methylation levels are presented as the mean ±
standard deviation. The differences and correlation of methylation
level between tumor, and matched serum were analyzed using a paired
t-test and the Spearman correlation, respectively. The associations
between 14-3-3σ expression and clinicopathological variables were
analyzed using the chi-squared test. The survival rates according
to 14-3-3σ expression status and other variables were examined
using Kaplan-Meier analysis, and compared using the log-rank test.
Cancer-associated mortality was considered to be the end event. The
Cox multivariate proportional hazards model was used to identify
independent prognostic variables. P<0.05 was considered to
indicate a statistically significant difference. Statistical
analysis was performed using STATA software version 12.1 (StataCorp
LP, College Station, TX, USA).
Results
14-3-3σ methylation in tumor and
serum
Methylation of 14-3-3σ gene was identified in all
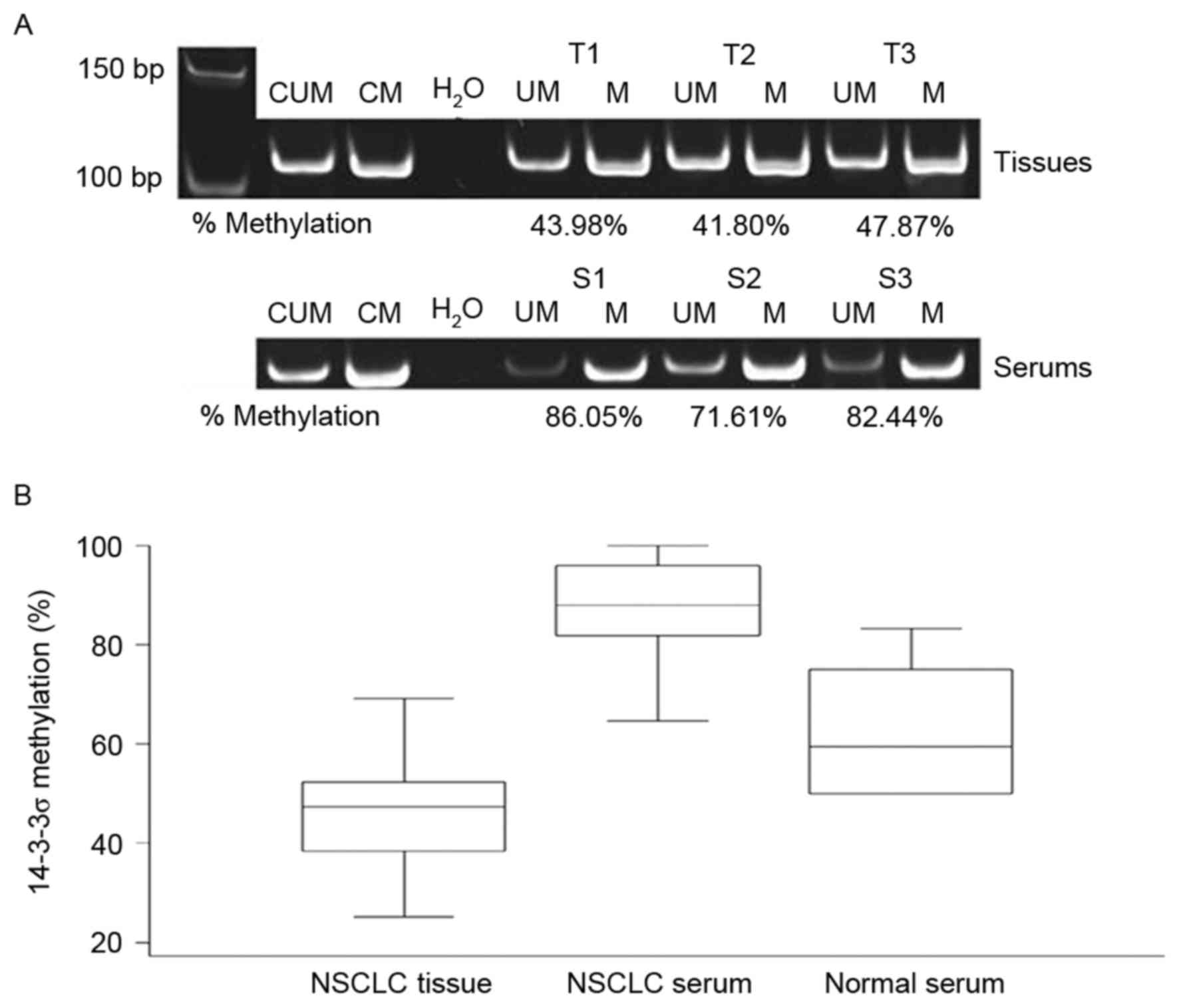
samples. Representative methylated and unmethylated products of the
samples run on the 10% polyacrylamide gel are presented in Fig. 1. The mean methylation level across all
tumor tissues was 46.7% (range, 25.3–69.2%). The methylation level
in ADC (mean, 43.6%; range, 25.3–56.3%) and SCC (mean, 48.6; range,
32.7–68.6%) samples were comparable. The mean methylation level in
patient sera was ~2 times higher compared with that of the primary
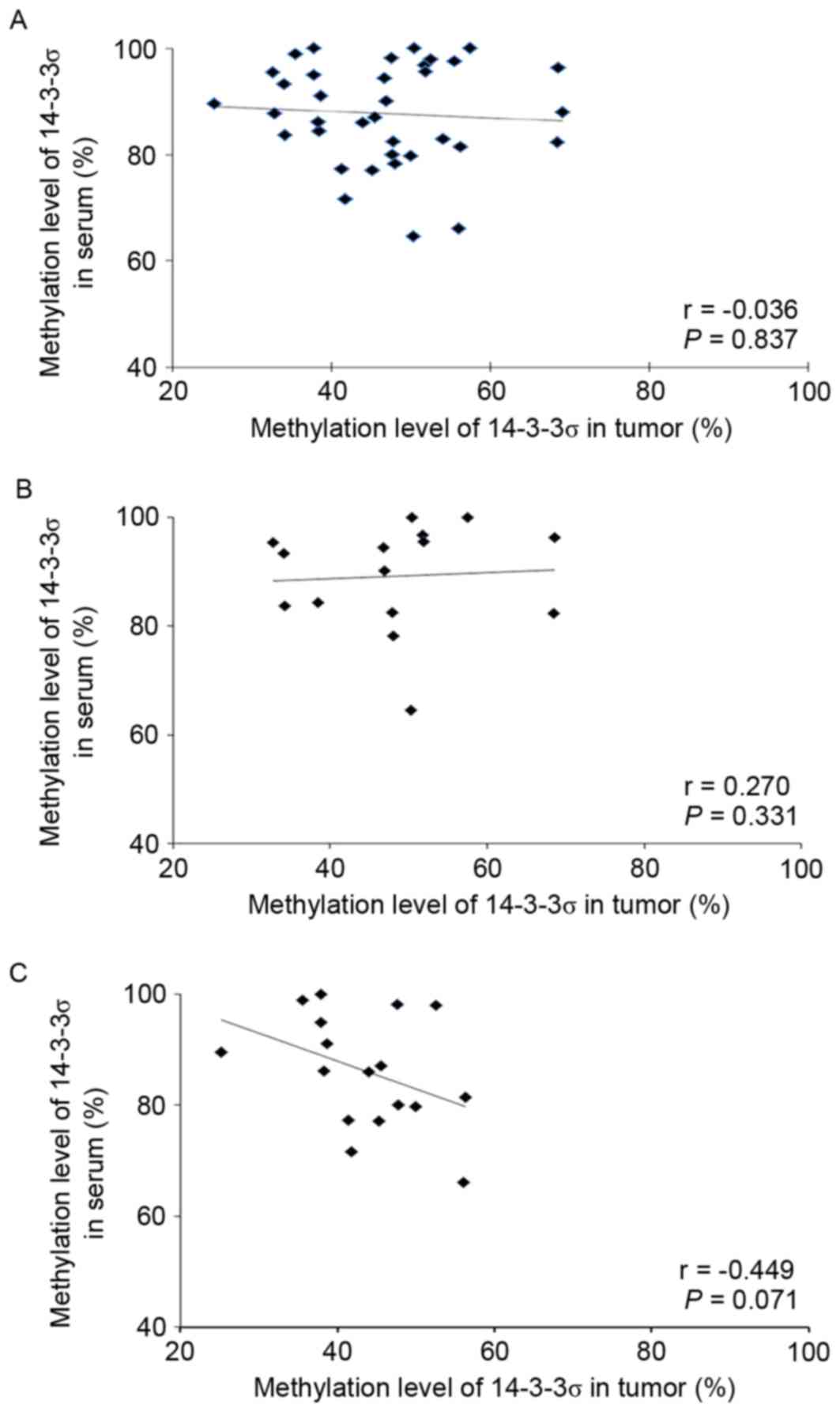
tumors with a mean value of 87.7% (range, 64.6–100%). However, the
methylation levels in tissues and serums were not linearly
correlated [Spearman's correlation (r), −0.036; P=0.837; Fig. 2]. The methylation level in normal
serum (mean, 60.2%; range, 50.0–75.0%) was lower compared with in
patient sera.
Correlation between 14-3-3σ
methylation and protein expression
The correlation between 14-3-3σ methylation and
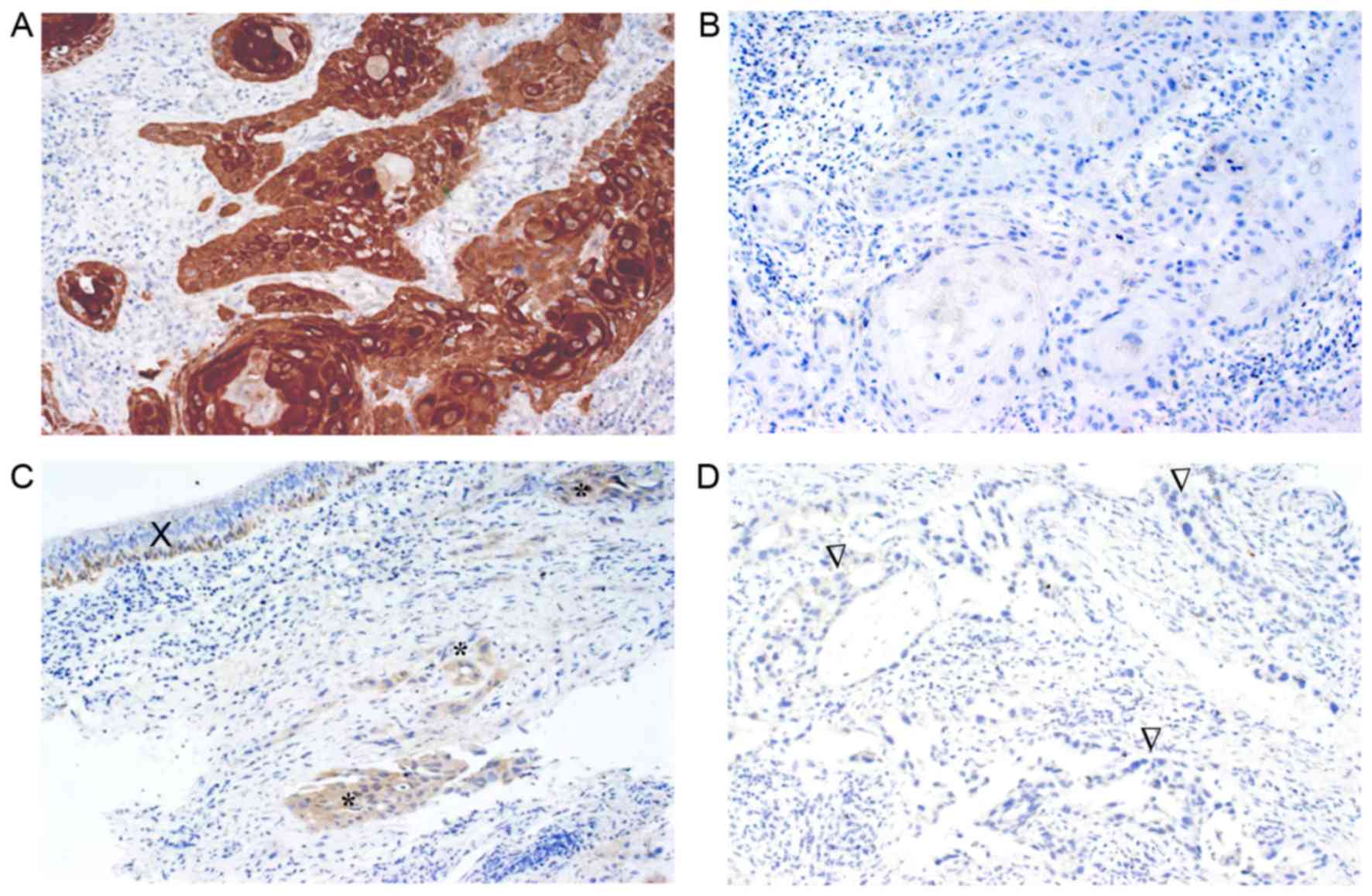
immunohistochemical protein expression was evaluated in 32 cases.
The 14-3-3σ protein was primarily observed in the cytoplasm
(Fig. 3) and 18 cases (56.2%)
exhibited no expression. The remaining cases exhibited weak
expression (7 cases, 21.9%) and moderate to strong expression (7
cases, 21.9%). No significant correlation was observed between
immunohistochemical expression and the methylation level (r, 0.153;
P=0.402).
Association between 14-3-3σ expression
and clinicopathological variables
The immunohistochemical expression of 14-3-3σ
protein in relation to clinicopathological characteristics and
prognosis was evaluated in 167 patients. The patients had a mean
age of 64 years (range, 37–93 years; Table I). The majority of patients exhibited
the advanced stages of the disease (89.8%). The majority of the
cases (140 cases, 83.8%) revealed no expression, whereas 19 (11.4%)
and 8 (4.8%) cases demonstrated weak expression, and
moderate/strong expression, respectively. Patients in the ADC group
had a significantly higher frequency of no expression (91.4%)
compared with SCC (70.20) (P=0.002). In the further analysis, the
weak to strong expression samples were grouped as positive
expression. Sex and histological type were identified to be
significantly associated with the expression status. In addition,
tumors in males had a significantly higher frequency of positive
expression compared with that of females (Table I).
 | Table I.Correlation between 14-3-3σ expression
and clinicopathological variables. |
Table I.
Correlation between 14-3-3σ expression
and clinicopathological variables.
|
|
| 14-3-3σ expression
(%) |
|
|---|
|
|
|
|
|
|---|
| Variable | No. of cases | Negative | Positive | P-value |
|---|
| Sex |
|
|
| 0.03 |
| Male | 124 | 99 (79.8) | 25 (20.2) |
|
|
Female | 43 | 41 (95.3) | 2 (4.7) |
|
| Age, years |
|
|
| 0.46 |
|
<60 | 63 | 55 (87.3) | 8 (12.7) |
|
|
≥60 | 104 | 85 (81.7) | 19 (18.3) |
|
| Histological
type |
|
|
| 0.002 |
|
ADC | 105 | 96 (91.4) | 9 (8.6) |
|
|
SCC | 57 | 40 (70.2) | 17 (29.8) |
|
|
NSCLC-UC |
5 | 4 (80.0) | 1 (20.0) |
|
| Clinical stage |
|
|
| 0.38 |
| I | 10 | 8 (80) | 2 (20) |
|
| II |
5 | 3 (60) | 2 (40) |
|
|
III | 59 | 51 (86.4) | 8 (13.6) |
|
| IV |
9 | 77 (84.6) | 14 (15.4) |
|
|
Unknown |
2 | 1 (50) | 1 (50) |
|
| LN metastasis |
|
|
| 0.24 |
| No | 72 | 64 (88.9) | 8 (11.1) |
|
|
Yes | 95 | 76 (80) | 19 (20) |
|
| Distant
metastasis |
|
|
| 0.93 |
| No | 76 | 63 (82.9) | 13 (17.1) |
|
|
Yes | 91 | 77 (84.6) | 14 (15.4) |
|
| Surgery |
|
|
| 0.92 |
| No | 157 | 132 (84.1) | 25 (15.9) |
|
|
Yes | 10 | 8 (80) | 2 (20) |
|
| Chemotherapy |
|
|
| 0.22 |
| No | 87 | 70 (80.5) | 17 (19.5) |
|
|
Yes | 80 | 70 (87.5) | 10 (12.5) |
|
| Radiotherapy |
|
|
| 0.14 |
| No | 116 | 101 (87.1) | 15 (12.9) |
|
|
Yes | 51 | 39 (76.5) | 12 (23.5) |
|
Prognostic significance of 14-3-3σ
expression
The patients had a median survival time of 5.7
months. The Kaplan-Meier estimates revealed no significant
difference in overall survival according to 14-3-3σ expression
status. Furthermore, no significant difference was identified in
survival for ADC and SCC groups with P=0.13 and P=0.60,
respectively (data not shown). Clinical stage, surgery,
chemotherapy and histological type were associated with survival
rates in the univariate analysis, but only age, and treatments were
significant independent prognostic parameters in the multivariate
analysis (Table II). 14-3-3σ
expression did not exhibit prognostic significance.
 | Table II.Univariate and multivariate analysis
of clinicopathological variables for overall survival. |
Table II.
Univariate and multivariate analysis
of clinicopathological variables for overall survival.
|
| Univariate
analysis | Multivariable
analysis |
|---|
|
|
|
|
|---|
| Variable | Risk ratio (95%
CI) | P-value | Risk ratio (95%
CI) | P-value |
|---|
| Sex |
| 0.339 |
|
|
|
Male | 1 |
|
|
|
|
Female | 0.85
(0.61–1.19) |
|
|
|
| Age, years |
| 0.609 |
| 0.031 |
|
<60 | 1 |
|
|
|
|
≥60 | 1.08
(0.79–1.48) |
| 0.69
(0.50–0.97) |
|
| Histological
type |
| 0.022 |
|
|
|
ADC | 1 |
|
|
|
|
SCC | 1.27
(0.93–1.74) |
|
|
|
|
NSCLC-UC | 3.9
(1.56–9.76) |
|
|
|
| Clinical stage |
| <0.001 |
|
|
| I | 1 |
|
|
|
| II | 6.19
(1.47–26.11) |
|
|
|
|
III | 9.02
(2.79–29.10) |
|
|
|
| IV | 9.62
(3.00–30.87) |
|
|
|
|
Unknown | 52.6
(10.17–272.16) |
|
|
|
| Surgery |
| <0.001 |
| <0.001 |
| No | 1 |
|
|
|
|
Yes | 0.09
(0.03–0.28) |
| 0.06
(0.02–0.20) |
|
| Chemotherapy |
| <0.001 |
| <0.001 |
| No | 1 |
|
|
|
|
Yes | 0.5
(0.37–0.68) |
| 0.47
(0.33–0.66) |
|
| Radiotherapy |
| 0.069 |
| 0.015 |
| No | 1 |
|
|
|
|
Yes | 0.75
(0.55–1.03) |
| 0.66
(0.47–0.92) |
|
| 14–3-3σ
expression |
| 0.248 |
|
|
|
Negative | 1 |
|
|
|
|
Positive | 1.44
(0.95–2.19) |
|
|
|
Discussion
In recent years, the aberrant expression levels of
the 14-3-3 protein family have been reported in various cancer
types and as potential novel biological markers (4). Among various isoforms, 14-3-3σ is the
most common isoform reported to be involved in carcinogenesis via a
tumor suppressive manner (7). Loss of
14-3-3σ expression has been reported in various types of epithelial
cancer and is reported to be associated with hypermethylation of
the promoter of the gene (8–10). In the present study, the methylation
status of the NSCLC tissue in relation to protein expression as
well as in relation to the methylation level in their match serum
was evaluated. The results revealed that all tumors harbored
certain levels of methylation; however, it was not correlated with
the level of protein expression. In addition, it was demonstrated
that methylation level in serum was significantly higher compared
with in primary tumor samples.
Hypermethylation of CpG islands is a well-known
epigenetic mechanism for inactivating tumor suppressor genes, thus
contributing significantly to tumor development (10,22).
Methylation in the promoter region of the 14-3-3σ gene has been
demonstrated in a high proportion of breast (90%) (23), nasopharynx (84%) (24), ovary, endometrium and prostate
(11) carcinoma. The results of the
present study demonstrated that all NSCLC tumor samples harbored
methylation in the promoter of 14-3-3σ gene (relative methylation
level, 25.3–69.2%). The methylation status may be reported as
partial methylation as methylated and unmethylated products were
identified. These results are consistent with that of Shiba-Ishii
and Noguchi (25) where
invasive adenocarcinoma harbored partial methylation. SCC, in the
present study, also revealed a comparable methylation level with
ADC. However, these results were inconsistent with the study of
Osada et al (18), whereby
hypermethylation was identified to be frequent in small cell
carcinoma cell lines, but rare in NSCLC cell lines.
It is well known that circulating cell-free DNA is
released into the blood of patients with cancer, with increasing
levels compared with normal healthy individuals (26), thus allowing for the detection of gene
alternation of the primary tumor. Detection of hypermethylation in
the promoter regions of certain tumor suppressor genes in the serum
of patients with NSCLC was first reported by Esteller et al
(27). Later, Ramirez et al
(28) detected methylation in the
sera of one-third of 115 advanced-stage patients with NSCLC. In the
present study, a higher methylation level (mean, 87.7%) was
observed in the serum of the patients compared with normal serum
(mean, 60.2%). In addition, the serum methylation was level was two
to three times higher compared with the matched primary tumor
samples and was not linearly correlated. The possible explanation
is that the circulating DNA is contaminated by other sources,
including inflammatory cells reacting to the tumor. The
inflammatory process has been demonstrated to serve a role in the
pathogenesis of NSCLC and the majority of lung cancer cases coexist
with inflammatory reactions (29). In
addition, lysis of peripheral blood lymphocytes during serum
separation may cause an artificial increase in DNA (30). However, this risk was minimized by
performing centrifugation of the collected serum within 2 h.
Methylation of the promoter region of genes is
typically associated with decreased or a loss of protein
expression. Shiba-Ishii and Noguchi (25) identified an inverse correlation
between the level of the 14-3-3σ transcript and methylation level
in lung adenocarcinoma tissue. By contrast, no significant
correlation was identified in present study. Similarly, no
significant correlation between the methylation of the 14-3-3σ gene
in the tumor and protein expression was noted in the study of Osada
et al (18). The authors
demonstrated that certain SCLC tissues exhibited almost complete
unmethylation of the 14-3-3σ gene as indicated by the loss of
protein expression. This may indicate that 14-3-3σ protein
expression is affected by additional mechanisms. Furthermore,
clinical tissue specimen may be contaminated by other cells/tissue,
including stromal cells or inflammatory cells as reported by Osada
et al (18), whereby it was
demonstrated that microdissected stromal tissue also harbored
14-3-3σ hypermethylation.
Previous studies regarding the expression of 14-3-3σ
in NSCLC are conflicting. Osada et al (18) reported immunohistochemical expression
of 14-3-3σ in 21/22 NSCLC specimens and Shiba-Ishii et al
(17) observed immunopositive
staining in 95% of ADC. By contrast, Liu et al (31) observed the downregulation of 14-3-3σ
in NSCLC cell lines. The present study demonstrated that the
majority of NSCLC (84%) demonstrated no expression of 14-3-3σ
protein following immunohistochemistry, which is consistent with
the results of studies on other cancer types, in particular breast
(8) and prostate (32) cancer. The number of specimens examined
may contribute to the contradictory results in lung cancer.
Regarding the prognostic role, the decreased
expression of 14-3-3σ has been reported to be correlated with a
short survival rate in esophageal squamous cell carcinoma (33) and ovarian cancer (33,34), and a
good survival rate in gastric cancer (35). However, the present study did not
identify prognostic significance of 14-3-3σ expression in NSCLC.
This may possibly be due to the small numbers of patients with a
positive expression. In addition, the majority of the patients had
stage III–IV cancer, thus the insignificance may also be due to the
homogeneity of cases regarding of stage of disease.
In conclusion, the results of the present study have
demonstrated that NSCLC harbored partial 14-3-3σ methylation and
may, in part, contribute to the loss of protein expression in the
tumor. The serum of patients with advanced NSCLC exhibited a high
level of 14-3-3σ methylation, but its clinical value remains to be
elucidated. The prognostic significance of immunohistochemical
expression of the protein was not demonstrated, possibly due to the
small number of cases with positive expression and homogeneity of
advanced cases.
Acknowledgements
The present study was supported by the Prince of
Songkla University (grant no. MED540677S) and Faculty of Medicine,
Songkhla, Thailand (grant no. 540200412). The Excellent Research
Laboratory of Cancer Molecular Biology was acknowledged for
research facilities.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
LN
|
lymph node
|
|
ADC
|
adenocarcinoma
|
|
SCC
|
squamous cell carcinoma
|
|
MSP
|
methylation-specific polymerase chain
reaction
|
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Attasara P and Sriplung H: Cancer
incidence in Thailand. In: Cancer in Thailand Volume VI,
2004–2006Khuhaprema T, Attasara P, Sriplung H, Wiangnon S,
Sumitsawan Y and Sangrajrang S: National Cancer Institute
(Thailand); Bangkok: pp. 3–68. 2012
|
|
3
|
National Cancer Institute: SEER Cancer
Statistics Review. 1975–2012 http://seer.cancer.gov/csr/1975_2012/Accessed.
April;2015.
|
|
4
|
Aitken A: 14-3-3 proteins: A historic
overview. Semin Cancer Biol. 16:162–172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galan JA, Geraghty KM, Lavoie G, Kanshin
E, Tcherkezian J, Calabrese V, Jeschke GR, Turk BE, Ballif BA,
Blenis J, et al: Phosphoproteomic analysis identifies the tumor
suppressor PDCD4 as a RSK substrate negatively regulated by 14-3-3.
Proc Natl Acad Sci USA. 111:E2918–E2927. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dar A, Wu D, Lee N, Shibata E and Dutta A:
14-3-3 proteins play a role in the cell cycle by shielding cdt2
from ubiquitin-mediated degradation. Mol Cell Biol. 34:4049–4061.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hermeking H, Lengauer C, Polyak K, He TC,
Zhang L, Thiagalingam S, Kinzler KW and Vogelstein B: 14-3-3 sigma
is a p53-regulated inhibitor of G2/M progression. Mol Cell. 1:3–11.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferguson AT, Evron E, Umbricht CB, Pandita
TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA,
Stampfer MR and Sukumar S: High frequency of hypermethylation at
the 14-3-3 sigma locus leads to gene silencing in breast cancer.
Proc Natl Acad Sci USA. 97:6049–6054. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suzuki H, Itoh F, Toyota M, Kikuchi T,
Kakiuchi H and Imai K: Inactivation of the 14-3-3 sigma gene is
associated with 5′ CpG island hypermethylation in human cancers.
Cancer Res. 60:4353–4357. 2000.PubMed/NCBI
|
|
10
|
Lodygin D and Hermeking H: The role of
epigenetic inactivation of 14-3-3sigma in human cancer. Cell Res.
15:237–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mhawech P, Greloz V, Assaly M and Herrmann
F: Immunohistochemical expression of 14-3-3 sigma protein in human
urological and gynecological tumors using a multi-tumor microarray
analysis. Pathol Int. 55:77–82. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Perathoner A, Pirkebner D, Brandacher G,
Spizzo G, Stadlmann S, Obrist P, Margreiter R and Amberger A:
14-3-3sigma expression is an independent prognostic parameter for
poor survival in colorectal carcinoma patients. Clin Cancer Res.
11:3274–3279. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Laimer K, Blassnig N, Spizzo G, Kloss F,
Rasse M, Obrist P, Schäfer G, Perathoner A, Margreiter R and
Amberger A: Prognostic significance of 14-3-3sigma expression in
oral squamous cell carcinoma (OSCC). Oral Oncol. 45:127–134. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou WH, Tang F, Xu J, Wu X, Feng ZY, Li
HG, Lin DJ, Shao CK and Liu Q: Aberrant upregulation of 14-3-3o
expression serves as an inferior prognostic biomarker for gastric
cancer. BMC Cancer. 11:3972011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qi W, Liu X, Qiao D and Martinez JD:
Isoform-specific expression of 14-3-3 proteins in human lung cancer
tissues. Int J Cancer. 113:359–63. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shiba-Ishii A, Kano J, Morishita Y, Sato
Y, Minami Y and Noguchi M: High expression of stratifin is a
universal abnormality during the course of malignant progression of
early-stage lung adenocarcinoma. Int J Cancer. 129:2445–2453. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Osada H, Tatematsu Y, Yatabe Y, Nakagawa
T, Konishi H, Harano T, Tezel E, Takada M and Takahashi T: Frequent
and histological type-specific inactivation of 14-3-3sigma in human
lung cancers. Oncogene. 21:2418–2424. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
International Union Against Cancer (UICC):
TNM classification of malignant tumours. Sobin LH, Gospodarowicz MK
and Wittekind CH: 7th. Wiley-Blackwell; Hoboken, NJ: 2009
|
|
20
|
Travis WD, Brambilla E, Müller-Hermelink
HK and Harris CC: World Health Organization Classification of
TumoursPathology and Genetics of Tumours of the Lung, Pleura,
Thymus and Heart. 3rd. IARC Press; Lyon: pp. 145–975. 2004
|
|
21
|
Green MR and Sambrook J: Molecular
cloningA laboratory manual. 4th. Cold Spring Harbor Laboratory
Press; Cold Spring Harbor, NY: 2012
|
|
22
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
23
|
Luo J, Feng J, Lu J, Wang Y, Tang X, Xie F
and Li W: Aberrant methylation profile of 14-3-3 sigma and its
reduced transcription/expression levels in Chinese sporadic female
breast carcinogenesis. Med Oncol. 27:791–797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yi B, Tan SX, Tang CE, Huang WG, Cheng AL,
Li C, Zhang PF, Li MY, Li JL, Yi H, et al: Inactivation of 14-3-3
sigma by promoter methylation correlates with metastasis in
nasopharyngeal carcinoma. J Cell Biochem. 106:858–866. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shiba-Ishii A and Noguchi M: Aberrant
stratifin overexpression is regulated by tumor-associated CpG
demethylation in lung adenocarcinoma. Am J Pathol. 180:1653–1662.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gormally E, Hainaut P, Caboux E, Airoldi
L, Autrup H, Malaveille C, Dunning A, Garte S, Matullo G, Overvad
K, et al: Amount of DNA in plasma and cancer risk: A prospective
study. Int J Cancer. 111:746–749. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Esteller M, Sanchez-Cespedes M, Rosell R,
Sidransky D, Baylin SB and Herman JG: Detection of aberrant
promoter hypermethylation of tumor suppressor genes in serum DNA
from non-small cell lung cancer patients. Cancer Res. 59:67–70.
1999.PubMed/NCBI
|
|
28
|
Ramirez JL, Rosell R, Taron M,
Sanchez-Ronco M, Alberola V, de Las Peñas R, Sanchez JM, Moran T,
Camps C, Massuti B, et al: 14-3-3sigma methylation in pretreatment
serum circulating DNA of cisplatin-plus-gemcitabine-treated
advanced non-small-cell lung cancer patients predicts survival: The
Spanish Lung Cancer Group. J Clin Oncol. 23:9105–9112. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jylhävä J, Jylhä M, Lehtimäki T, Hervonen
A and Hurme M: Circulating cell-free DNA is associated with
mortality and inflammatory markers in nonagenarians: The Vitality
90+ Study. Exp Gerontol. 47:372–378. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Umetani N, Kim J, Hiramatsu S, Reber HA,
Hines OJ, Bilchik AJ and Hoon DS: Increased integrity of free
circulating DNA in sera of patients with colorectal or
periampullary cancer: Direct quantitative PCR for ALU repeats. Clin
Chem. 52:1062–1069. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu Y, Chen Q and Zhang JT: Tumor
suppressor gene 14-3-3sigma is down-regulated whereas the
proto-oncogene translation elongation factor 1delta is up-regulated
in non-small cell lung cancers as identified by proteomic
profiling. J Proteome Res. 3:728–735. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cheng L, Pan CX, Zhang JT, Zhang S, Kinch
MS, Li L, Baldridge LA, Wade C, Hu Z, Koch MO, et al: Loss of
14-3-3sigma in prostate cancer and its precursors. Clin Cancer Res.
10:3064–3068. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ren HZ, Pan GQ, Wang JS, Wen JF, Wang KS,
Luo GQ and Shan XZ: Reduced stratifin expression can serve as an
independent prognostic factor for poor survival in patients with
esophageal squamous cell carcinoma. Dig Dis Sci. 55:2552–2560.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Akahira J, Sugihashi Y, Suzuki T, Ito K,
Niikura H, Moriya T, Nitta M, Okamura H, Inoue S, Sasano H, et al:
Decreased expression of 14-3-3 sigma is associated with advanced
disease in human epithelial ovarian cancer: Its correlation with
aberrant DNA methylation. Clin Cancer Res. 10:2687–3793. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li YL, Liu L, Xiao Y, Zeng T and Zeng C:
14-3-3σ is an independent prognostic biomarker for gastric cancer
and is associated with apoptosis and proliferation in gastric
cancer. Oncol Lett. 9:290–294. 2015.PubMed/NCBI
|