Introduction
Bladder cancer is one of the most frequent
malignancies in the world. Despite the significant advances in the
development of diagnostic and therapeutic methods in recent years,
patients with invasive bladder cancer still have a poor 5-year
survival rate of less than 60% (1,2).
Epithelial-mesenchymal transition (EMT), which is characterized by
loss of cell adhesion and gain of migratory and invasive
capability, is an important mechanism for metastatic potential and
the initial stage of cancer metastasis, including bladder cancer
(3,4).
Long non-coding RNAs (lncRNAs) are a class of
noncoding regulatory RNA molecules that are longer than 200
nucleotides. Previous studies have focused on the regulatory roles
of lncRNAs in several biological processes, such as chromosome
inactivation, genomic imprinting and development (5–7). Recently,
increasing reports have revealed that lncRNAs are frequently
dysregulated in various human cancers, including bladder cancer
(8,9).
Of note, many dysregulated lncRNAs are involved in EMT and
metastasis in human cancer. For example, high expression of the
lncRNA PVT1 promotes invasion by inducing EMT in esophageal cancer
(10), and increased lncRNA PANDAR
expression indicates a poor prognosis for colorectal cancer and
promotes metastasis by the EMT pathway (11). LncRNAs can also function as a
competing endogenous RNA to sponge microRNAs (12–14). For
example, the lncRNA H19 promotes EMT by functioning as a miRNA
sponge to miR-138 and miR-200a in colorectal cancer (15). Therefore, by interacting with specific
miRNAs, lncRNAs can play an important role in cancer development
and progression.
UCA1 was first identified as an oncogenic lncRNA in
bladder cancer (16). UCA1 is highly
expressed in diverse cancer types, such as breast cancer (17), pancreatic cancer (18), gastric cancer (19), and colorectal cancer (20,21),
suggesting that high expression of UCA1 might serve as a molecular
marker for predicting metastasis and prognosis in these cancers
(22). Several studies also
demonstrated that upregulation of UCA1 is associated with EMT in
breast cancer (23) and bladder
cancer (24), but the underlying
mechanism remains largely unknown.
HMGB1, a member of the high mobility group box
(HMG-box) subfamily, is involved in many cancers (25,26). HMGB1
also functions as an inducer of EMT in human cancer cells (27–31). HMGB1
expression was much higher in bladder cancer cells than normal
urethra epithelial cells and was associated with cell invasion
(32,33). However, the mechanism of HMGB1 in
bladder cancer is largely unknown.
MicroRNAs (miRNAs) play important roles during
tumorigenesis, and dysregulated miRNAs are also involved in EMT in
human cancer. In the past decades, the involvement of miRNAs in
human bladder cancer has been widely studied. We previously
reported that miR-143 acts as a tumor suppressor in human bladder
cancer (34).
In the present study, UCA1 and HMGB1 were
upregulated and miR-143 was downregulated in bladder cancer
specimens. Bioinformatics found that binding sites of the tumor
suppressive miR-143 within UCA1 and the 3′UTR of HMGB1. And taken
the functional examinations together, we hypothesis that UCA1 may
promote the invasion and EMT of bladder cancer cells by regulating
the miR-143/HMGB1 pathway.
Materials and methods
Tissue specimens
A total of 81 bladder tissue specimens, including 52
tumor tissues and 29 adjacent noncancerous tissues (at least 2.5 cm
away from the tumor), were obtained from our hospital. All tissues
were collected from bladder cancer at the time of radical
cystectomy, snap-frozen in liquid nitrogen and stored at −70°C
until use. All patients gave informed consent prior to collection
of specimens according to institutional guidelines. The study was
approved by the Institute Research Ethics Committee at Peking
University Shenzhen Hospital.
Cell culture
The human bladder cancer cell lines (T24, 5637, J82,
RT4 and HT1376) were purchased from China Academia Sinica Cell
Repository (Shanghai, China) and cultured in DMEM (Invitrogen,
Carlsbad, CA, USA) or RPMI 1640 (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal calf serum at
37°C in a 5% CO2 incubator.
RNA isolation and qRT-PCR
Total RNA was extracted by TRIzol reagent
(Invitrogen) and reverse transcribed using the Primer Script RT
Reagent kit (Takara Bio, Tokyo, Japan). UCA1 and miR-381 expression
levels were determined using the SYBR-Green qRT-PCR assay (BioRad,
Berkeley, CA, USA) and mirVana qRT-PCR miRNA Detection kit (Ambion,
Austin, TX, USA), respectively. GAPDH and U6 snRNA were used as
endogenous controls for UCA1 and miR-143 relative expression,
respectively. Primers used are as follows: UCA1-F,
5′-CTCTCCATTGGGTTCACCATTC-3′, UCA1-R, 5′-GCGGCAGGTCTTAAGAGATGAG-3′;
HMGB1-F, 5′-ATCCCAATGCACCCAAGAGGCCT-3′, HMGB1-R,
5′-TTCGCAACATCACCAATGGACAGG-3′; β-actin-F,
5′-AGGGGCCGGACTCGTCATACT-3′, β-actin-R,
5′-GGCGGCACCACCATGTACCCT-3′; miR-143-F, 5′-CCTGGCCTGAGATGAAGCAC-3′,
miR-143-R, 5′-CAGTGCTGGGTCCGAGTGA-3′; and U6-F,
5′-CTCGCTTCGGCAGCACA-3′, U6-R, 5′-AACGCTTCACGAATTTGCGT-3′. The
relative levels were measured in triplicate using the
2−∆∆Cq method (35).
Luciferase reporter assay
Fragments of the 3′UTR of HMGB1 and UCA1 containing
miR-143 binding sites were amplified and cloned into the psiCHECK-2
reporter vector (Promega, Shanghai, China). The mutated versions of
the 3′UTR of HMGB1 and UCA1 (with substitutions of four bases in
the miR-143 seed binding sites) were generated using the
QuikChange™ Site-Directed Mutagenesis kit (Stratagene, La Jolla,
CA, USA). T24 cells were co-transfected with the reporter vector
(0.5 µg) and miR-143 mimics or scrambled mimics (50 nM) using
Lipofectamine 2000 (Invitrogen). After 48 h, the Dual Luciferase
Reporter assay system (Promega, Fitchburg, WI, USA) was used to
examine luciferase activity. MiR-143, scramble mimics and miR-143
inhibitor mimics were purchased from RiboBio (Guangzhou,
China).
Vector construction and siRNA
To construct the UCA1 expression vector, we
amplified UCA1 cDNA with flanking sequences (forward primer,
5′-ATGCACCTTGTGACTCCCTCCTCT-3′; reverse primer,
5′-CACCTCATCAGACTGCCTTTGG-3′) and cloned it into the pcDNA3.1
vector in BamHI/XhoI sites (Invitrogen). We used the following
siRNAs for knockdown experiments: si-UCA1,
5′-GAGCCGAUCAGACAAACAAUU-3′ (S), 5′-UUGUUUGUCUGAUCGGCUCUU-3′ (AS);
si-HMGB1, 5′-CCCGUUAUGAAAGAGAAAUUU-3′ (S),
5′-AUUUCUCUUUCAUAAUGGGUU-3′ (AS); si-NC (control),
5′-UUCGUCUGUACUCCACAUATT-3′ (S), 5′-GAUGUCUUCUACAGUCCGATT-3′ (AS).
We used Lipofectamine 2000 Reagent (Invitrogen) for cell
transfection.
Cell invasion assays
Cell invasion was assessed by Transwell assays with
matrigel (BD Bioscience, San Diego, CA, USA). Briefly,
5×104 cells in 200 µl of serum-free media were added
into the upper chamber and 500 µl media with 10% fetal bovine serum
were added to the bottom chamber. After 24 h, the cells that
invaded through the membrane were fixed with 3% formaldehyde,
stained with 0.5% crystal violet, and imaged under an inverted
microscope (Olympus, Tokyo, Japan).
Western blot analysis
Total cellular extracts were prepared using lysis
buffer (Sigma-Aldrich, St. Louis, MO, USA). Approximately 50 ug of
total protein was separated by 10% SDS-PAGE and transferred to a
PVDF membrane. Membranes were incubated with anti-E-cadherin
(1:500), anti-N-cadherin (1:1,000) and anti-Vimentin (1:500)
antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) followed
by the HRP-conjugated secondary antibody (Santa Cruz
Biotechnology). Signals were visualized using the ECL detection
system (SuperSignal West Femto, Pierce, USA).
Statistical analysis
Statistical analyses were carried out using the SPSS
19.0 software (IBM SPSS, Armonk, NY, USA). Student's t-test was
used to analyze experimental significance. Spearman's correlation
analysis was used to analyze the correlation between miR-143 and
UCA1 and HMGB1 expression.
Results
UCA1 modulated the invasion and EMT of
bladder cancer cells
Previous studies revealed that UCA1 expression is
associated with enhanced invasion ability of bladder cancer cells
(24,36,37). We
first investigated the relationship between the expression of UCA1
and the invasion ability of five bladder cancer cell lines.
Comparison of Transwell assay results and qRT-PCR analysis of UCA1
expression showed that UCA1 expression was positively correlated
with cell invasion ability (Fig.
1A-C). T24 cells showed stronger invasion capability and higher
UCA1 expression levels, while RT4 cells showed relatively weaker
invasion capability and lower UCA1 expression levels among the five
tested bladder cancer cell lines. Knockdown of endogenous UCA1 in
T24 cells by siRNA significantly (P<0.05) reduced the number of
invading cells, while UCA1 overexpression in RT4 cells increased
the number of invading cells significantly (P<0.05) (Fig. 1D and E), suggesting that UCA1 is
closely involved in the invasion ability of bladder cancer
cells.
EMT is an important mechanism of enhanced invasion
of bladder cancer cells. Therefore, we further investigated whether
UCA1 modulated EMT in bladder cancer cells. The epithelial marker
E-cadherin and mesenchymal markers N-cadherin and vimentin were
examined by western blot analysis (Fig.
1F). UCA1 knockdown in T24 cells led to increased E-cadherin
and decreased N-cadherin and vimentin, while UCA1 overexpression in
RT4 cells inhibited E-cadherin and enhanced N-cadherin and Vimentin
protein expression. Together these results suggest that UCA1 may
modulate invasion and EMT of bladder cancer cells.
UCA1 promoted the invasion and EMT of
bladder cancer cells by regulating HMGB1
To investigate the potential mechanism by which UCA1
regulates EMT in bladder cancer cells, we examined the effect of
UCA1 on the expression of HMGB1, which is reported to be associated
with the invasion ability of many types of human cancers including
bladder cancer (24). qRT-PCR showed
that HMGB1 mRNA expression was significantly (P<0.05) reduced by
knockdown of UCA1 in T24 cells and increased by overexpression of
UCA1 in RT4 cells (Fig. 2A). In
addition, Transwell assays revealed that knockdown of HMGB1 by
specific siRNA significantly inhibited the invasion of RT4 cells
(Fig. 2B and C), similar to the
effects of UCA1 knockdown. Of note, we found that knockdown of
HMGB1 and overexpression of UCA1 significantly attenuated the
effect of UCA1 on invasion and EMT of T24 cells (Fig. 2C and D). These results illustrated
that UCA1 promoted the invasion and EMT of bladder cancer cells at
least in part by regulating HMGB1.
UCA1 induced HMGB1 expression through
directly interacting with miR-143
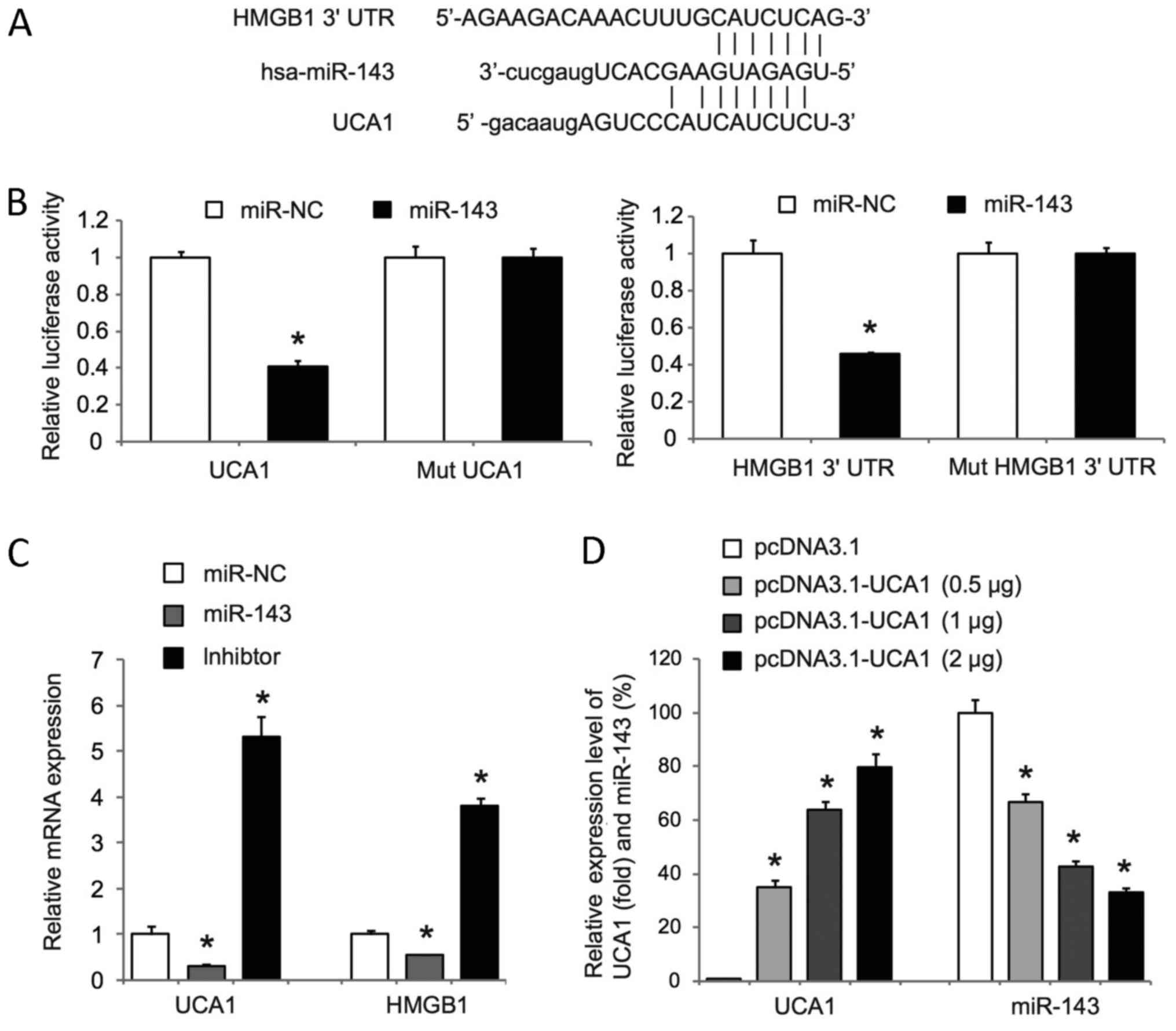
Bioinformatics revealed putative binding sites for
the tumor suppressive miR-143 within UCA1 and the 3′UTR of HMGB1
(Fig. 3A). Luciferase reporter assay,
which was a standard way to prove the direct interaction between
microRNAs and target genes (38), was
used to determine whether miR-143 targets UCA1 and the 3′UTR of
HMGB1. We cloned the full-length UCA1 and a fragment of the HMGB1
3′UTR containing miR-143 binding sites into luciferase reporter
plasmids (psiCheck-2) and co-transfected the reporter plasmids with
miR-143 mimics or scrambled mimics (miR-NC) into T24 cells.
Luciferase assays showed that miR-143 mimics significantly reduced
the activity of psiCheck-2-UCA1 and psiCheck-2-HMGB1 (Fig. 3B). Of note, mutations of the miR-143
binding sites in psiCheck-2-UCA1 and psiCheck-2-HMGB1 significantly
abolished the effects on luciferase activity mediated by miR-143
mimics (Fig. 3B). In addition,
miR-143 mimics significantly decreased UCA1 and HMGB1 expression,
whereas miR-143 inhibitor mimics significantly increased UCA1 and
HMGB1 expression in T24 cells (Fig.
3C). These data revealed that miR-143 could directly target and
inhibit UCA1 and HMGB1 in T24 cells. We also observed that UCA1
overexpression in RT4 cells significantly repressed miR-143
expression in a dose-dependent manner (Fig. 3D). Taken together, our data suggest
that UCA1 may act as a competing endogenous RNA of miR-143 and
inhibit miR-143 to upregulate HMGB1 in bladder cancer cells.
UCA1 and HMGB1 were upregulated and
miR-143 was downregulated in bladder tumors
To examine the clinical significance of UCA1, HMGB1
and miR-143 in human bladder cancer development, qRT-PCR was
performed to evaluate the expression levels of UCA1, miR-143 and
HMGB1 mRNAs in a cohort of 81 bladder tissue specimens including 52
tumor tissues and 29 adjacent noncancerous tissues. As shown in
Fig. 4A, UCA1 and HMGB1 were
significantly upregulated while miR-143 was significantly
downregulated in tumors compared with adjacent noncancerous
tissues. In addition, correlation analysis revealed that UCA1 and
HMGB1 expression levels had a positive correlation, but were both
inversely correlated with miR-143 expression (Fig. 4B). Taken together, our observations
suggest that dysregulation of the UCA1/miR-143/HMGB1 axis may be
associated with the development and progression of bladder
cancer.
Discussion
Increasing reports have revealed that dysregulated
expression of lncRNAs is closely involved in the pathological
development of a wide variety of human cancers including bladder
cancer (8,9). UCA1 is a sensitive and specific
oncogenic lncRNA in bladder cancer (16). Consistent with previous observations
that high UCA1 expression was involved in cell invasion in many
cancers as well as bladder cancer (24,36,37), our
results clearly showed that cells with stronger invasion capability
had higher UCA1 expression levels in bladder cancer cell lines
in vitro. EMT is a well-characterized process that
facilitates invasion and metastatic dissemination of human cancers
(3,4).
Therefore, we further investigated whether UCA1 could modulate EMT
of bladder cancer cells. Gain and loss of function analysis showed
that UCA1 knockdown increased E-cadherin and decreased N-cadherin
and vimentin, while UCA1 overexpression led to the opposite
results. These data suggest that UCA1 may modulate cell invasion by
promoting EMT in bladder cancer cells.
The HMGB family could be divided into 3 subgroups,
which are HMGB1, HMGB2 and HMGB3 (39). HMGB1 is widely distributed in the
lymphoid tissue, brain, liver, lung, heart, spleen, kidney tissues,
and exists in most organizations in the nucleus (40); The HMGB2/3 distribution is limited.
HMGB2 is mainly distributed in the testicular and lymphoid tissues
(41,42), while HMGB3 is found mainly in embryos
(43). This is why we chose HMGB1 as
our candidate.
To investigate the potential mechanism by which UCA1
regulates EMT in bladder cancer cells, we examined the effect of
UCA1 on the expression of HMGB1. We found that HMGB1 mRNA
expression was significantly reduced by knockdown of UCA1 in T24
cells and increased by overexpression of UCA1 in RT4 cells,
suggesting that UCA1 can positively regulate HMGB1 mRNA expression.
Cellular function assays further revealed that knockdown of HMGB1
significantly inhibited the invasion of RT4 cells and attenuated
the effect of UCA1 on invasion and EMT of T24 cells. These results
revealed that UCA1 promoted the invasion and EMT of bladder cancer
cells, at least in part by inducing HMGB1. And in order to look
into the biological function of HMGB1 in bladder cancer, we might
pursue another research on how the protein level of HMGB1 affected
by UCA1.
LncRNAs interact with miRNAs to function as miRNA
sponges or inhibitors and also modulate the repression of miRNA
targets. Increasing reports have revealed that UCA1 functions as an
oncogenic lncRNA by interacting with and inhibiting tumor
suppressive miRNAs such as miR-507 (44), miR-145 (24), miR-16 (45) and miR-216b (46). Based on bioinformatics and luciferase
reporter assays, we confirmed binding sites of the tumor
suppressive miR-143 within UCA1 and the 3′UTR of HMGB1. In
addition, we observed that UCA1 overexpression significantly
repressed miR-143 expression in a dose-dependent manner in RT4
cells. Our observation was closely consistent with a recently
published report demonstrating that UCA1 promoted bladder cancer
cell migration and invasion by the hsa-mir145-ZEB1/2-FSCN1 pathway
(24). Similar to miR-143, miR-145
was also reported to be a tumor suppressive gene and a negative
regulator of EMT (47–49). Thus, UCA1 might function as an
oncogene by sponging EMT-related miRNAs.
miR-143 was already known to act as a tumor
suppressor in multiple cancers, including bladder cancer (34). According to the literature found in
Pubmed, miR-143 was found to suppress EMT and cell invasion in
multiple cancers, including spinal glioblastoma (50), esophageal squamous cell carcinoma
(51), and breast cancer (52). Our research showed that UCA1 may act
as a competing endogenous RNA of miR-143 and inhibit miR-143 to
upregulate HMGB1, result in promoting the invasion and EMT of
bladder cancer cells. Taken together, we believe that miR-143 might
also affect EMT in bladder cancer. Further studies were needed to
look into the function of miR-143 on EMT and invasion in bladder
cancer.
Expression analysis on clinical bladder cancer
specimens also revealed that UCA1 and HMGB1 expression levels were
positively correlated but were both inversely correlated with
miR-143 expression. Taken together, our data suggest that UCA1 may
act as a miRNA sponge to miR-143 and inhibit miR-143 to upregulate
HMGB1 in bladder cancer cells. The limitation in this research was
that this study only included in vitro models. Our future
studies will pursue these findings in an in vivo model.
In conclusion, our study provided evidence that UCA1
promotes the invasion and EMT of bladder cancer by regulating the
miR-143/HMGB1 pathway, which may play an important regulatory role
in the pathology of bladder cancer.
Acknowledgements
The authors would like to thank the Guangdong and
Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics,
Institute of Urology, Peking University Shenzhen Hospital. The
present study was supported by Shenzhen Health and Family Planning
System Scientific Research Project (grant no. 201401033, for Junhua
Luo), the Science and Technology Development Fund Project of
Shenzhen (grant no JCYJ20150403091443304, for Shangqi Yang), the
fund of ‘San-ming’ Project of Medicine in Shenzhen and the fund of
the Guangdong Key Medical Subject.
References
|
1
|
Dancik GM and Theodorescu D:
Pharmacogenomics in bladder cancer. Urol Oncol. 32:16–22. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ploussard G, Shariat SF, Dragomir A, Kluth
LA, Xylinas E, Masson-Lecomte A, Rieken M, Rink M, Matsumoto K,
Kikuchi E, et al: Conditional survival after radical cystectomy for
bladder cancer: Evidence for a patient changing risk profile over
time. Eur Urol. 66:361–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yun SJ and Kim WJ: Role of the
epithelial-mesenchymal transition in bladder cancer: From prognosis
to therapeutic target. Korean J Urol. 54:645–650. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Autuoro JM, Pirnie SP and Carmichael GG:
Long noncoding RNAs in imprinting and X chromosome inactivation.
Biomolecules. 4:76–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Caley DP, Pink RC, Trujillano D and Carter
DR: Long noncoding RNAs, chromatin, and development. Scientific
World Journal. 10:90–102. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long noncoding RNA MEG3 affects
cell proliferation and predicts a poor prognosis in patients with
colorectal cancer. Tumour Biol. 36:4851–4859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Q, Su M, Lu G and Wang J: The
complexity of bladder cancer: Long noncoding RNAs are on the stage.
Mol Cancer. 12:1012013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng X, Hu H and Li S: High expression of
lncRNA PVT1 promotes invasion by inducing epithelial-to-mesenchymal
transition in esophageal cancer. Oncol Lett. 12:2357–2362.
2016.PubMed/NCBI
|
|
11
|
Lu M, Liu Z, Li B, Wang G, Li D and Zhu Y:
The high expression of long non-coding RNA PANDAR indicates a poor
prognosis for colorectal cancer and promotes metastasis by EMT
pathway. J Cancer Res Clin Oncol. 143:71–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dey BK, Pfeifer K and Dutta A: The H19
long noncoding RNA gives rise to microRNAs miR-675-3p and
miR-675-5p to promote skeletal muscle differentiation and
regeneration. Genes Dev. 28:491–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW,
Li MQ, Chen YC, Qian XP, Lu TJ, Yu LZ, et al: Rapid identification
of UCA1 as a very sensitive and specific unique marker for human
bladder carcinoma. Clin Cancer Res. 12:4851–4858. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu
M and Mo YY: Long non-coding RNA UCA1 promotes breast tumor growth
by suppression of p27 (Kip1). Cell Death Dis. 5:e10082014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen P, Wan D, Zheng D, Zheng Q, Wu F and
Zhi Q: Long non-coding RNA UCA1 promotes the tumorigenesis in
pancreatic cancer. Biomed Pharmacother. 83:1220–1226. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C,
Ye H, Zhou B, Chen JJ and Chen P: Aberrant expression of UCA1 in
gastric cancer and its clinical significance. Clin Transl Oncol.
17:640–646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ni B, Yu X, Guo X, Fan X, Yang Z, Wu P,
Yuan Z, Deng Y, Wang J, Chen D and Wang L: Increased urothelial
cancer associated 1 is associated with tumor proliferation and
metastasis and predicts poor prognosis in colorectal cancer. Int J
Oncol. 47:1329–1338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang HM, Lu JH, Chen WY and Gu AQ:
Upregulated lncRNA-UCA1 contributes to progression of lung cancer
and is closely related to clinical diagnosis as a predictive
biomarker in plasma. Int J Clin Exp Med. 8:11824–11830.
2015.PubMed/NCBI
|
|
22
|
He A, Hu R, Chen Z, Liao X, Li J, Wang D,
Lv Z, Liu Y, Wang F and Mei H: Role of long noncoding RNA UCA1 as a
common molecular marker for lymph node metastasis and prognosis in
various cancers: A meta-analysis. Oncotarget. 8:1937–1943.
2017.PubMed/NCBI
|
|
23
|
Xiao C, Wu CH and Hu HZ: LncRNA UCA1
promotes epithelial-mesenchymal transition (EMT) of breast cancer
cells via enhancing Wnt/beta-catenin signaling pathway. Eur Rev Med
Pharmacol Sci. 20:2819–2824. 2016.PubMed/NCBI
|
|
24
|
Xue M, Pang H, Li X, Li H, Pan J and Chen
W: Long non-coding RNA urothelial cancer-associated 1 promotes
bladder cancer cell migration and invasion by way of the
hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107:18–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang W, Jiang H, Zhu H, Zhang H, Gong J,
Zhang L and Ding Q: Overexpression of high mobility group box 1 and
2 is associated with the progression and angiogenesis of human
bladder carcinoma. Oncol Lett. 5:884–888. 2013.PubMed/NCBI
|
|
26
|
Yang GL, Zhang LH, Bo JJ, Huo XJ, Chen HG,
Cao M, Liu DM and Huang YR: Huang, Increased expression of HMGB1 is
associated with poor prognosis in human bladder cancer. J Surg
Oncol. 106:57–61. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang BP, Wang DS, Xing JW, Yang SH, Chu Q
and Yu SY: miR-200c inhibits metastasis of breast cancer cells by
targeting HMGB1. J Huazhong Univ Sci Technolog Med Sci. 34:201–206.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen YC, Statt S, Wu R, Chang HT, Liao JW,
Wang CN, Shyu WC and Lee CC: High mobility group box 1-induced
epithelial mesenchymal transition in human airway epithelial cells.
Sci Rep. 6:188152016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng M, Liu H, Zhang D, Liu Y, Wang C,
Liu F and Chen J: HMGB1 enhances the AGE-induced expression of CTGF
and TGF-β via RAGE-dependent signaling in renal tubular epithelial
cells. Am J Nephrol. 41:257–266. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lynch J, Nolan S, Slattery C, Feighery R,
Ryan MP and McMorrow T: High-mobility group box protein 1: A novel
mediator of inflammatory-induced renal epithelial-mesenchymal
transition. Am J Nephrol. 32:590–602. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu L, Li X, Chen Y, Fang J and Ge Z:
High-mobility group box 1: A novel inducer of the
epithelial-mesenchymal transition in colorectal carcinoma. Cancer
Lett. 357:527–534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang Z, Zhong Z, Zhang L, Wang X, Xu R,
Zhu L, Wang Z, Hu S and Zhao X: Down-regulation of HMGB1 expression
by shRNA constructs inhibits the bioactivity of urothelial
carcinoma cell lines via the NF-κB pathway. Sci Rep. 5:128072015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liao H, Xiao Y, Hu Y, Xiao Y, Yin Z and
Liu L: Suppression of cellular proliferation and invasion by HMGB1
knockdown in bladder urothelial carcinoma cells. Oncol Res.
22:235–245. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin T, Dong W, Huang J, Pan Q, Fan X,
Zhang C and Huang L: MicroRNA-143 as a tumor suppressor for bladder
cancer. J Urol. 181:1372–1380. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang F, Li X, Xie X, Zhao L and Chen W:
UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma
and embryo, influencing cell growth and promoting invasion. FEBS
Lett. 582:1919–1927. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xue M, Li X, Li Z and Chen W: Urothelial
carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted
long noncoding RNA that enhances hypoxic bladder cancer cell
proliferation, migration, and invasion. Tumour Biol. 35:6901–6912.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vella MC, Reinert K and Slack FJ:
Architecture of a validated microRNA: Target interaction. Chem
Biol. 11:1619–1623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yanai H, Ban T, Wang Z, Choi MK, Kawamura
T, Negishi H, Nakasato M, Lu Y, Hangai S, Koshiba R, et al: HMGB
proteins function as universal sentinels for nucleic-acid-mediated
innate immune responses. Nature. 462:99–103. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bonaldi T, Talamo F, Scaffidi P, Ferrera
D, Porto A, Bachi A, Rubartelli A, Agresti A and Bianchi ME:
Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect
it towards secretion. EMBO J. 22:5551–5560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mosevitsky MI, Novitskaya VA, Iogannsen MG
and Zabezhinsky MA: Tissue specificity of nucleo-cytoplasmic
distribution of HMG1 and HMG2 proteins and their probable
functions. Eur J Biochem. 185:303–310. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bucci LR, Brock WA and Meistrich ML:
Heterogeneity of high-mobility-group protein 2. Enrichment of a
rapidly migrating form in testis. Biochem J. 229:233–240. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Moleri S, Cappellano G, Gaudenzi G,
Cermenati S, Cotelli F, Horner DS and Beltrame M: The HMGB protein
gene family in zebrafish: Evolution and embryonic expression
patterns. Gene Expr Patterns. 11:3–11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wei Y, Sun Q, Zhao L, Wu J, Chen X, Wang
Y, Zang W and Zhao G: LncRNA UCA1-miR-507-FOXM1 axis is involved in
cell proliferation, invasion and G0/G1 cell cycle arrest in
melanoma. Med Oncol. 33:882016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li HJ, Li X, Pang H, Pan JJ, Xie XJ and
Chen W: Long non-coding RNA UCA1 promotes glutamine metabolism by
targeting miR-16 in human bladder cancer. Jpn J Clin Oncol.
45:1055–1063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chiou GY, Cherng JY, Hsu HS, Wang ML, Tsai
CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, et al: Cationic
polyurethanes-short branch PEI-mediated delivery of Mir145
inhibited epithelial-mesenchymal transdifferentiation and cancer
stem-like properties and in lung adenocarcinoma. J Control Release.
159:240–250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ling DJ, Chen ZS, Zhang YD, Liao QD, Feng
JX, Zhang XY and Shi TS: MicroRNA-145 inhibits lung cancer cell
metastasis. Mol Med Rep. 11:3108–3114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ren D, Wang M, Guo W, Zhao X, Tu X, Huang
S, Zou X and Peng X: Wild-type p53 suppresses the
epithelial-mesenchymal transition and stemness in PC-3 prostate
cancer cells by modulating miR-145. Int J Oncol. 42:1473–1481.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yan Y, Wu J, Wu M, Xia Y, Tang W and Liao
Z: MiR-143 suppresses the epithelial-mesenchymal transition of
spinal glioblastoma through down-regulation of ERK5. Oncotarget.
2016.(Epub ahead of print). View Article : Google Scholar
|
|
51
|
Liu J, Mao Y, Zhang D, Hao S, Zhang Z, Li
Z and Li B: RETRACTED: MiR-143 inhibits tumor cell proliferation
and invasion by targeting STAT3 in esophageal squamous cell
carcinoma. Cancer Lett. 373:97–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhai L, Ma C, Li W, Yang S and Liu Z:
miR-143 suppresses epithelial-mesenchymal transition and inhibits
tumor growth of breast cancer through down-regulation of ERK5. Mol
Carcinog. 55:1990–2000. 2016. View Article : Google Scholar : PubMed/NCBI
|