Introduction
Liver cancer is the fifth most prevalent cancer and
the third leading cause of cancer-associated mortality, immediately
following lung, and colorectal cancer worldwide (1). Hepatocellular carcinoma (HCC) is the
most common form of adult liver cancer, representing >90% of all
cases of primary liver cancer (2).
Great advances in the treatment of liver cancer, relapse and
metastasis are frequently observed in the clinic, and the poor
5-year survival rate is attributed to late diagnosis, resistance to
treatment, tumor recurrence and metastasis, hence stressing the
importance of novel diagnostics and therapeutics (3). It is also necessary to identify
biological markers that can be used to screen high-risk patients in
order to obtain earlier HCC diagnosis, earlier intervention and
increase the likelihood of successful treatment (4).
Regarding diagnosis biomarkers, microRNAs (miRNAs)
have become a hot topic in the field of cancer biological research.
miRNAs are non-coding RNA molecules of 21–24 nucleotides that
regulate the expression of target genes in a post-transcriptional
manner. Evidence indicates that miRNA serve essential roles in
embryogenesis, cell differentiation and the pathogenesis of various
human diseases, including cancer (5,6).
Furthermore, their expression levels have been identified to be
dysregulated in numerous cancer types some of which have been
directly implicated in carcinogenetic mechanisms, and several
altered expressions of miRNAs have previously been described in rat
and human HCC (7–9).
Several previous studies have revealed that the
expression of miRNAs is dysregulated in human HCC in comparison
with matched non-neoplastic tissue (10). miR-221, miR-19b and miR-224 expression
levels were increased in hepatitis C virus recurrence samples,
while miR-129, and miR-335 were decreased compared with normal
liver tissue (5,11). From the comprehensive miRNAs
expression analysis of HCC tissues paired with adjacent
non-cancerous hepatic tissues, miR-221 has been identified to
repress endogenous histone deacetylase 6 expression in HCC cells
(10). The overexpression of miR-19b
was significantly correlated with better overall and disease-free
survival rates for patients with HCC presenting with vascular
invasion or multifocal disease following curative surgery (11). In addition, previous results
demonstrated that miR-129 and miR-335 can suppress tumorigenesis,
and progression, defining them as potential treatment targets for
HCC (5).
miR-125a has been previously reported to be altered
in various human cancer types (12,13).
However, little is known about the association between miR-125a
expression and the survival of patients with HCC. In the present
study, in silico analysis of differentially expressed miRNAs
was first performed on patients with HCC vs. controls using the
Gene Expression Omnibus (GEO) database data. Subsequently, miR-125a
expression was assessed in 27 normal liver and 98 HCC tissue
samples using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). The aim of the current study was to evaluate
the efficacy of miR-125a as a prognostic marker for patients with
HCC.
Materials and methods
miRNA expression of HCC from GEO
database
In order to investigate the association between
miRNAs and the development of liver cancer, the raw data GSE31383
from the GEO database was downloaded (http://www.ncbi.nlm.nih.gov/geo/). This dataset
includes miRNA expression data from 10 healthy liver and 9 HCC. In
addition, GSE20594 (including 10 normal controls and 89 HCC) and
GSE36915 (including 21 normal controls and 69 HCC) were downloaded
to identify differentially expressed miRNAs for HCC.
Tissue samples
Liver tissue samples from 27 healthy liver and tumor
tissues from 98 HCC patients (Table
I), who underwent surgical resection between January 2008 and
December 2012, were collected from the Tissue Bank, Jilin
University (Changchun, China). Fresh tissue samples were frozen
within the 30 min after surgery and stored in liquid nitrogen until
use. The inclusion criteria was ≤75 years of age with
histologically proven CRC, no severe major organ dysfunction, World
Health Organization (WHO) (14)
performance status of 0 or 1 and no prior cancer chemotherapy. The
exclusion criteria included an age of ≥76 years, severe major organ
dysfunction, WHO performance status of >1 or prior cancer
chemotherapy. Two experienced pathologists confirmed HCC diagnosis
independently according to the WHO criteria. The present study was
approved by the Ethics Committee of Shanghai Tenth People's
Hospital, Tongji University School of Medicine (SHSY-IEC-pap-15-18;
Shanghai, China) and the Ethics Committee of Jilin University
(20151101). Patients and/or their legal surrogates provided written
informed consent to the surgical procedures, and participation of
the current study by donating tissue specimens.
 | Table I.Univariate analysis of overall
survival based on patients stratified by clinical
characteristics. |
Table I.
Univariate analysis of overall
survival based on patients stratified by clinical
characteristics.
|
|
|
|
| Overall survival |
|---|
|
|
|
|
|
|
|---|
| Factor | No. of patients | miR-125a
expressiona | P-value | Months (median) | 95% CI (median) | P-value (Log-rank
test) |
|---|
| Age (years) |
|
|
|
|
|
|
| ≥60 | 52 | 1.62±0.35 | 0.822 | 81.19 | 67.14–95.24 | 0.468 |
|
<60 | 46 | 1.52±0.27 |
| 73.01 | 54.16–91.86 |
|
| Gender |
|
|
|
|
|
|
| Male | 80 | 1.45±0.19 | 0.274 | 75.81 | 60.89–90.73 | 0.277 |
|
Female | 18 | 2.06±0.78 |
| 90.17 | 68.62–111.72 |
|
| No. of lesions |
|
|
|
|
|
|
|
Single | 63 | 1.79±0.37 | 0.431 | 88.44 | 76.56–100.33 | 0.003 |
|
Multiple | 35 | 1.44±0.27 |
| 51.13 | 30.31–71.96 |
|
| Invasion to tumor
capsule |
|
|
|
|
|
|
|
Negative | 43 | 1.95±0.22 | 0.472 | 101.21 | 86.34–116.06 | 0.019 |
|
Positive | 41 | 1.59±0.44 |
| 45.37 | 34.27–56.47 |
|
|
Unknown | 14 | 1.32±0.61 |
| 30.79 | 23.07–38.51 |
|
| Tumor
differentiation |
|
|
|
|
|
|
|
Poorly | 18 | 1.72±0.28 | 0.391 | 40.21 | 35.48–51.93 | 0.062 |
|
Moderately | 62 | 1.06±0.43 |
| 49.36 | 41.25–58.76 |
|
|
Well | 8 | 1.03±0.41 |
| 45.45 | 32.47–55.61 |
|
|
Unknown | 10 | 1.55±0.62 |
| 20.26 | 16.74–32.93 |
|
| Ki67
expression |
|
|
|
|
|
|
|
Negative | 21 | 1.31±0.27 | 0.027 | 45.84 | 30.21–61.48 | 0.005 |
|
Positive | 35 | 0.60±0.17 |
| 55.52 | 37.04–74.01 |
|
|
Unknown | 42 | 0.71±0.31 |
| 80.15 | 66.89–93.39 |
|
| TNM stage |
|
|
|
|
|
|
|
I–II | 7 | 0.86±0.61 | 0.836 | 51.17 | 34.98–67.35 | 0.051 |
|
III–IV | 6 | 0.69±0.52 |
| 47.29 | 27.34–67.23 |
|
|
Unknown | 85 | 0.63±0.16 |
| 84.05 | 71.65–96.45 |
|
| Drinking
status |
|
|
|
|
|
|
|
Negative | 72 | 1.80±0.26 | 0.038 | 66.34 | 45.62–81.21 | 0.413 |
|
Positive | 18 | 0.68±0.21 |
| 55.26 | 43.79–74.38 |
|
|
Unknown | 8 | 1.13±0.52 |
| 75.39 | 64.11–84.95 |
|
| Tumor capsule |
|
|
|
|
|
|
|
Negative | 36 | 2.21±0.49 | 0.125 | 45.69 | 33.94–57.47 | 0.009 |
|
Positive | 48 | 1.45±0.19 |
| 93.57 | 76.06–111.09 |
|
|
Unknown | 14 | 1.75±0.48 |
| 30.79 | 23.07–38.51 |
|
| Tumor embolus |
|
|
|
|
|
|
|
Negative | 64 | 2.21±0.58 | 0.064 | 87.64 | 69.28–106.01 | 0.008 |
|
Positive | 30 | 1.33±0.18 |
| 43.44 | 31.03–55.85 |
|
|
Unknown | 4 | 1.81±0.55 |
| 92.75 | 48.19–137.31 |
|
| Diameter (cm) |
|
|
|
|
|
|
| ≥5 | 58 | 1.25±0.23 | 0.226 | 49.92 | 38.73–61.12 | 0.149 |
|
<5 | 40 | 1.78±0.32 |
| 92.28 | 77.67–106.89 |
|
Collection of patients' clinical and
follow-up data
Clinical information was recorded including the
patient's characteristics (gender, age, drinking status), tumor
characteristics [number of lesions, invasion to tumor capsule,
tumor differentiation, Ki67 expression, tumor node metastasis (TNM)
stage, tumor capsule, tumor embolus and diameter; Table I], overall survival time (OS),
disease-free survival time (DFS) and chemotherapy status. The last
follow-up was performed on July 30th 2015 by direct correspondence
or phone interview. The occasion of mortality or tumor relapse was
verified by patients or their relatives or from their medical
records or the social security records. OS was analyzed for the
months from the date of diagnosis to the time of mortality,
regardless of the cause. DFS was defined as the period from the
initial date of diagnosis to the time of tumor progression by
computed tomography scan or to the time of mortality due to the
disease.
RNA isolation and RT-qPCR
Total RNA from HCC and normal tissues was isolated
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol. RNA
concentration was measured using NanoDrop ND-1000 (Thermo Fisher
Scientific, Inc.) and the quality was assessed using
electrophoresis with 1.5% denaturing agarose gels. TaqMan
probe-based qPCR was performed using a commercial kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RT was performed using a miR-125a-specific
primer and ABI's TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). miR-125a expression
level was detected using a Taqman MicroRNA assay (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Reverse transcriptase
reactions were performed using avian myeloblastosis virus reverse
transcriptase (Takara Biotechnology Co., Ltd., Dalian, China) and
qPCR was performed using a standard TaqMan PCR kit protocol with
the Applied Biosystems 7900HT Sequence Detection system. U6 was
used as the internal control. The RT-qPCR thermocycling conditions
were as follows: 94°C for 30 sec (initial denaturation), 94°C for 5
sec (denaturation) and 55°C for 30 sec (annealing), for 40 cycles.
U6 expression was used as the internal control. The following
primers were used: miR-125a forward, 5′-GGTAAGTCACGCGGT-3′ and
reverse, 5′-CAGTGCGTCTCGTGGAGT-3′; U6 forward,
5′-CTGGTTAGTACTTGGACGGGAGAC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′.
miR-125a levels were quantified using the 2−ΔΔCq method
(15).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical significances between groups were determined using
two-tailed Student's t-tests. The χ2 was used to compare
the differences of categorical variables and the Student's t-test
was used for comparison of differences between two groups.
Kaplan-Meier survival curves and the log-rank test were used to
analyze the OS or DFS of patients with HCC. Multivariate Cox
proportional hazards regression models were performed to explore
the prognostic value of multiple variables in HCC. All statistical
analyses were performed using SPSS software (version 20.0; IBM
Corp., Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
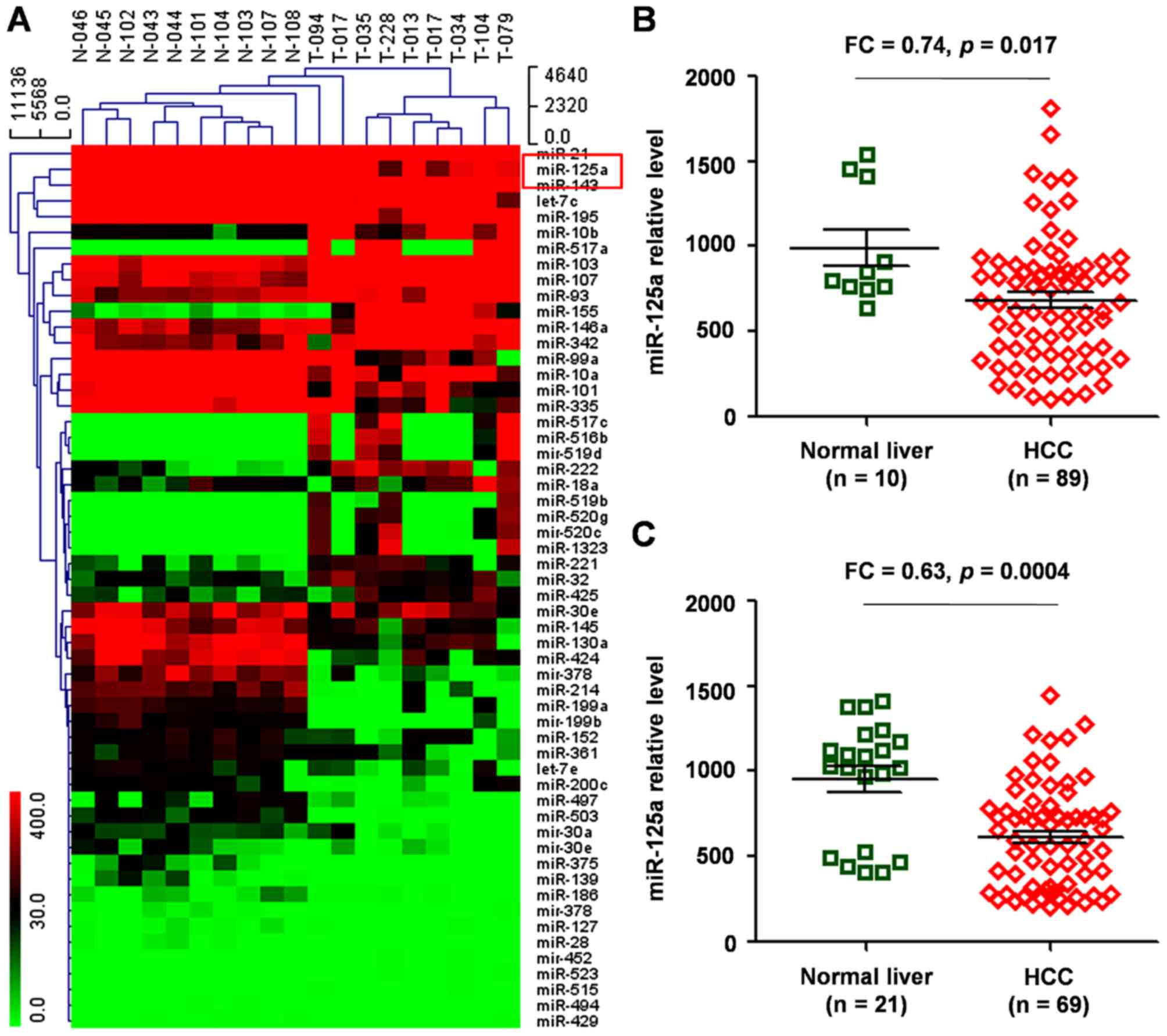
Expression of miR-125a using the GEO
database by clustering analysis
In order to identify the association between miRNA
expression and the prognosis of patients with HCC, in silico
analysis using GEO database data (GSE31383) was performed first.
There were 56 differentially expressed miRNAs identified between
normal controls and HCC (Fig. 1A),
which included specifically upregulated miRNAs (including miR-221,
miR-199a, let-7c/e, miR-10a, miR-21) that were reported previously
in liver cancer (10–12).
The prognostic value of the novel miRNAs identified
for patients with HCC was evaluated, one of which was miR-125a that
was identified to be significantly downregulated in HCC compared
with normal liver tissue [fold change (FC), 0.64; P=0.039].
Two datasets (GSE20594 and GSE36915) of HCC vs.
noncancerous tissue samples were used to validate the
aforementioned findings, and it was demonstrated that miR-125a
expression was significantly reduced in HCC compared with that in
normal controls (P=0.017, Fig. 1B;
P=0.0004, Fig. 1C).
miR-125a expression in HCC and
adjacent non-cancerous tissues
RT-qPCR was subsequently performed to quantify
miR-125a levels in 98 HCC specimens and 27 non-cancerous tissues.
The results of the qPCR analysis revealed that miR-125a levels were
significantly lower in HCC tissues compared with that in 27 paired
adjacent non-cancerous tissues (FC, 0.59; P=0.045; Fig. 2A). Furthermore, the level of miR-125a
expression was significantly lower in all 98 HCC biopsies compared
with that in 27 adjacent non-cancerous tissues (FC, 0.38; P=0.036;
Fig. 2B).
Cox regression model analysis for
prognosis based on various clinical characteristics in patients
with HCC
In addition, the association between miR-125a
expression in HCC samples and various clinical characteristics of
patients was analyzed (including age, gender, number of lesions,
invasion to tumor capsule, tumor differentiation, Ki67 expression,
TNM stage, drinking status, tumor capsule, tumor embolus and
diameter).
As presented in Table
I, miR-125a expression was positively correlated with Ki67
expression and drinking status (P<0.05). However, no significant
association was identified between miR-125a expression and other
clinical characteristics, including age, gender, and tumor
differentiation (P>0.05).
Association between clinical
characteristics and HCC prognosis
In order to further analyze the prognostic value of
other clinical factors, including age, gender, number of lesions,
invasion to tumor capsule, tumor differentiation, Ki67 expression,
drinking status, tumor capsule, tumor embolus, diameter and TNM
stage, Kaplan-Meier survival curves were plotted, and comparisons
were made using log-rank tests (Table
I). It was demonstrated that the number of lesions was
significantly associated with diminished OS (P=0.003) in patients
with HCC. In addition, invasion to tumor capsule was significantly
associated with decreased OS (P=0.019). Similar results were
obtained regarding Ki67 expression, tumor capsule, tumor embolus
and OS (P=0.005, 0.009 and 0.008, respectively).
As presented in Table
II, univariate analysis using the Cox regression model revealed
that miR-125a expression levels [hazard ratio (HR), 0.479;
confidence interval (CI), 0.25–0.92; P=0.027], number of lesions
(HR, 2.291; CI, 1.326–3.678; P=0.005), invasion to tumor capsule
(HR, 1.575; CI, 1.006–2.465; P=0.047), Ki67 expression (HR, 1.745;
CI, 1.183–2.577; P=0.005), tumor capsule (HR, 0.543; CI,
0.316–0.935; P=0.027) and tumor embolus (HR, 1.569; CI,
0.976–2.522; P=0.063) were positively associated with poor
prognosis (P<0.05). However, no significant association was
identified between HCC prognosis and clinicopathological
characteristics, including age, gender, tumor differentiation, TNM
stage, drinking status, and diameter exhibited (P>0.05).
 | Table II.Cox regression model analysis for
prognosis based on various clinical characteristics in patients
with HCC. |
Table II.
Cox regression model analysis for
prognosis based on various clinical characteristics in patients
with HCC.
|
| miR-125a univariate
analysis | miR-125a
multivariate analysis |
|---|
|
|
|
|
|---|
| Factor | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (years) | 1.259 | 0.670–2.364 | 0.474 |
|
|
|
| Gender | 0.600 | 0.235–1.536 | 0.287 |
|
|
|
| No. of lesions | 2.291 | 1.326–3.678 | 0.005 |
|
|
|
| Invasion to tumor
capsule | 1.575 | 1.006–2.465 | 0.047 |
|
|
|
| Tumor
differentiation | 0.843 | 0.511–1.391 | 0.504 |
|
|
|
| Ki67
expression | 1.745 | 1.183–2.577 | 0.005 | 2.561 | 1.578 - 4.375 | <0.001 |
| TNM stage | 1.121 | 0.627–2.005 | 0.701 |
|
|
|
| Drinking
status | 1.145 | 0.572–2.292 | 0.702 |
|
|
|
| Tumor capsule | 0.543 | 0.316–0.935 | 0.027 |
|
|
|
| Tumor embolus | 1.569 | 0.976–2.522 | 0.063 |
|
|
|
| Diameter | 1.575 | 0.840–2.951 | 0.156 |
|
|
|
| miR-125a | 0.479 | 0.250–0.920 | 0.027 |
|
|
|
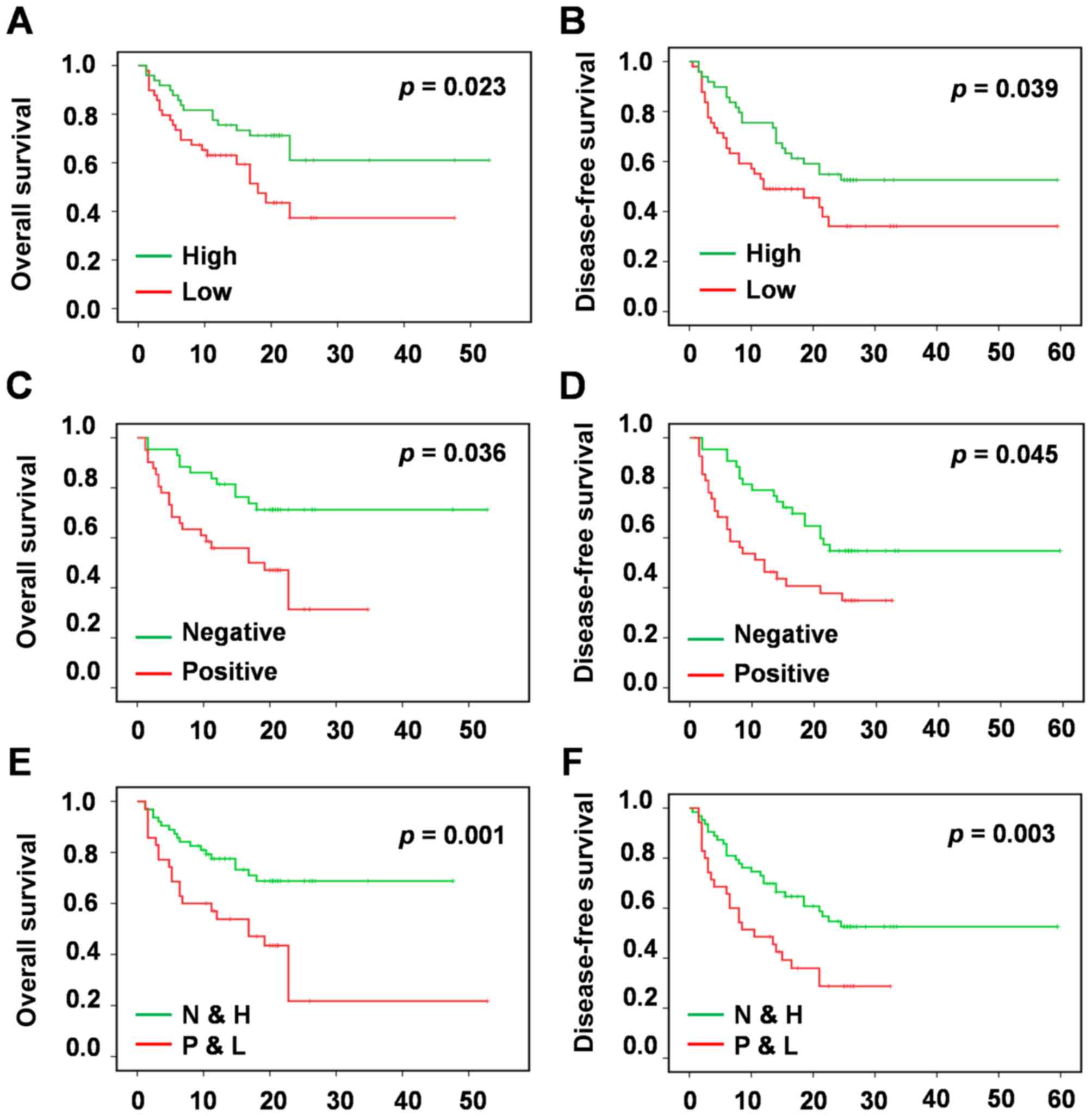
miR-125a downregulation is a
prognostic marker for survival in patients with HCC
To determine the prognostic value of miR-125a
expression in HCC, Kaplan-Meier survival analysis was used to
evaluate the association betweenmiR-125a expression, and OS and
DFS. The results revealed that low miR-125a expression associated
with poor OS, whereas high miR-125a mRNA levels were associated
with increased OS. Thus, reducedmiR-125a expression level was
significantly associated with poor OS (P=0.023; Fig. 3A) and DFS (P=0.039; Fig. 3B) in patients with HCC.
Considering that miR-125a expression was positively
associated with Ki67 expression (P<0.05; Table I), and miR-125a and Ki67 expression
levels exhibited associations with HCC prognosis (Table I and Fig.
3A-D), the prognostic value of miR-125a expression together
with Ki67 expression was further investigated. Multivariate
analysis of OS and DFS using Kaplan-Meier survival analysis
indicated that patients with HCC with low miR-125a expression and
high Ki67 expression had significantly decreased OS (P=0.001;
Fig. 3E), and DFS (P=0.003; Fig. 3F).
Discussion
HCC remains one of the most common types of solid
tumor malignancy worldwide, with Western Africa and China reporting
the highest incidence per capita (16). Management of advanced and metastatic
HCC continues to be challenging due to the high expression of drug
resistance genes, underlying cirrhosis, and poor liver function in
numerous patients (17).
It is now well established that miRNAs serve
essential roles in various biological processes, including
development, cellular proliferation, apoptosis and oncogenesis
(18,19). The role of miRNAs as oncogenes or
tumor suppressors in human cancer has been established. Various
studies have also begun to elucidate the molecular functional
associations between abnormal miRNA expression and the hallmarks of
malignant transformation: Aberrant cell growth, cell death,
differentiation, angiogenesis, invasion and metastasis (20).
Molecular biomarkers serve an important role in the
therapeutic decision making process, as they can be an indicator of
the response patients have to individual chemotherapeutic
interventions. In HCC, miRNAs exhibit aberrant processing and
expression profiles, in addition, the profile of circulating miRNAs
is also affected, which renders them potential biomarkers, with
possible applications in diagnosis, particularly for early,
pre-symptomatic disease, and prognosis of HCC. For instance,
research findings collectively demonstrate a tumor suppressor role
of miR-188-5p in HCC progression via targeting fibroblast growth
factor 5, suggesting that miR-188-5p may serve as a potential
prognostic biomarker and therapeutic target for HCC (21,22). The
expression of Rho associated coiled-coil containing protein kinase
1 (ROCK1) was decreased significantly following overexpression of
miR-335, indicating that ROCK1 is a target gene for miR-335, and
miR-335 can inhibit the proliferation and migration invasion of HCC
cells via regulating ROCK1, suggesting that miR-335 may be a
therapeutic biomarker of HCC in the future (23).
miR-125a has previously been reported to inhibit
breast cancer cell proliferation, invasion and migration (24). Furthermore, miR-125a was validated to
prevent the cancer cell invasion in different cancer types,
including ovarian (25), glioma
(26), gastric (27) and lung cancer (28). miR-125a has been identified to be
involved in hepatitis B virus duplication and the progression of
associated liver diseases caused by hepatitis b virus (29). Bi et al (30) investigated a molecular mechanism that
has been associated with the tumor-suppressive role of miR-125a. It
was demonstrated that miR-125a directly targets matrix
metalloproteinase 11 and vascular endothelial growth factor to
inhibit the proliferation, and migration of liver cancer cells
(30). Tang et al (31) identified that the expression of
miR-125a was significantly reduced in highly lung-invasive HCC-LM3
cells, which suggests that miR-125a may be associated with
conferring the invasive and migratory abilities of liver cancer
cells. In addition, this suggests that miR-125a may be used as a
marker for predicting the prognosis of patients with liver
cancer.
In the present study, it was revealed that miR-125a
was significantly lower in liver cancer and reduced miR-125a levels
in HCC tissues were associated with a shorter overall, and
disease-free survival of patients with HCC. In addition, it was
demonstrated that miR-125a was a predictor for shorter OS times of
patients with HCC. Furthermore, miR-125a expression was
significantly negatively associated with alcohol drinking status of
patients. Thus, further studies are warranted to investigate the
prognostic value of miR-125a in HCC. The evidence presented in the
current study and previous studies suggests that miR-125a may
function as a tumor suppressor gene in HCC. In addition, there is
sufficient evidence indicating that alcoholic beverages are
carcinogenic in humans. Therefore, it may be noteworthy to
investigate the association between miR-125a expression levels and
alcohol consumption in the livers of healthy people, individuals
with fatty liver disease, alcoholic hepatitis, alcoholic cirrhosis,
or HCC, and to illustrate its possible role and mechanism of
miR-125a in alcoholic liver diseases.
miR-125a expression was identified to be negatively
associated with Ki67 expression, and miR-125a and Ki67 expression
levels exhibited associations with HCC prognosis. Notably, patients
with HCC with low miR-125a expression and high Ki67expression
exhibited significantly decreased OS.
In conclusion, the results of the present study
provide the first evidence that reduced miR-125a expression is
associated with progression and poor prognosis in patients with
HCC. This suggests that miR-125a possesses potential prognostic
value as a tumor biomarker for the prognosis of patients with
HCC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81472501, 81201535,
81472202 and 81302065) and Shanghai Health and Family Planning
Commission Projects (grant no. 201540228).
References
|
1
|
Ma YS, Wu TM, Lv ZW, Lu GX, Cong XL, Xie
RT, Yang HQ, Chang ZY, Sun R, Chai L, et al: High expression of
miR-105-1 positively correlates with clinical prognosis of
hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget.
8:11896–11905. 2017.PubMed/NCBI
|
|
2
|
Wu SD, Ma YS, Fang Y, Liu LL, Fu D and
Shen XZ: Role of the microenvironment in hepatocellular carcinoma
development and progression. Cancer Treat Rev. 38:218–225. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ching RHH, Sze KMF, Lau EYT, Chiu YT, Lee
JMF, Ng IOL and Lee TKW: C-terminal truncated hepatitis B virus X
protein regulates tumorigenicity, self-renewal and drug resistance
via STAT3/Nanog signaling pathway. Oncotarget. 8:23507–23516.
2017.PubMed/NCBI
|
|
4
|
Kindrat I, Tryndyak V, de Conti A,
Shpyleva S, Mudalige TK, Kobets T, Erstenyuk AM, Beland FA and
Pogribny IP: MicroRNA-152-mediated dysregulation of hepatic
transferrin receptor 1 in liver carcinogenesis. Oncotarget.
7:1276–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Okajima W, Komatsu S, Ichikawa D, Miyamae
M, Kawaguchi T, Hirajima S, Ohashi T, Imamura T, Kiuchi J, Arita T,
et al: Circulating microRNA profiles in plasma: Identification of
miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma
independent of hepatic function. Oncotarget. 7:53820–53836. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng H, Zou AE, Saad MA, Wang XQ, Kwok
JG, Korrapati A, Li P, Kisseleva T, Wang-Rodriguez J and Ongkeko
WM: Alcohol-dysregulated microRNAs in hepatitis B virus-related
hepatocellular carcinoma. PLoS One. 12:e01785472017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kutay H, Bai S, Datta J, Motiwala T,
Pogribny I, Frankel W, Jacob ST and Ghoshal K: Downregulation of
miR-122 in the rodent and human hepatocellular carcinomas. J Cell
Biochem. 99:671–678. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang S, Tan G, Jiang X, Han P, Zhai B,
Dong X, Qiao H, Jiang H and Sun X: An artificial lncRNA targeting
multiple miRNAs overcomes sorafenib resistance in hepatocellular
carcinoma cells. Oncotarget. 7:73257–73269. 2016.PubMed/NCBI
|
|
9
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shi KQ, Lin Z, Chen XJ, Song M, Wang YQ,
Cai YJ, Yang NB, Zheng MH, Dong JZ, Zhang L and Chen YP:
Hepatocellular carcinoma associated microRNA expression signature:
Integrated bioinformatics analysis, experimental validation and
clinical significance. Oncotarget. 6:25093–25108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hung CL, Yen CS, Tsai HW, Su YC and Yen
CJ: Upregulation of microRNA-19b predicts good prognosis in
patients with hepatocellular carcinoma presenting with vascular
invasion or multifocal disease. BMC Cancer. 15:6652015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng J, Zhou Z, Xu Z, Li G, Dong P, Chen
Z, Lin D, Chen B and Yu F: Serum microRNA-125a-5p, a useful
biomarker in liver diseases, correlates with disease progression.
Mol Med Rep. 12:1584–1590. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jin L, Zhang Z, Li Y, He T, Hu J, Liu J,
Chen M, Gui Y, Chen Y and Lai Y: miR-125b is associated with renal
cell carcinoma cell migration, invasion and apoptosis. Oncol Lett.
13:4512–4520. 2017.PubMed/NCBI
|
|
14
|
Edge SB and Compton CC: The American Joint
Committee On Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McGlynn KA, Tsao L, Hsing AW, Devesa SS
and Fraumeni JF Jr: International trends and patterns of primary
liver cancer. Int J Cancer. 94:290–296. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dou JP, Yu J, Yang XH, Cheng ZG, Han ZY,
Liu FY, Yu XL and Liang P: Outcomes of microwave ablation for
hepatocellular carcinoma adjacent to large vessels: A propensity
score analysis. Oncotarget. 8:28758–28768. 2017.PubMed/NCBI
|
|
18
|
Chang L, Wang Y, Zhang J and Guo T: The
best strategy for HCC patients at each BCLC stage: A network
meta-analysis of observational studies. Oncotarget. 8:20418–20427.
2017.PubMed/NCBI
|
|
19
|
Xiang ZL, Zhao XM, Zhang L, Yang P, Fan J,
Tang ZY and Zeng ZC: MicroRNA-34a expression levels in serum and
intratumoral tissue can predict bone metastasis in patients with
hepatocellular carcinoma. Oncotarget. 7:87246–87256.
2016.PubMed/NCBI
|
|
20
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xue HY, Liu Y, Liao JZ, Lin JS, Li B, Yuan
WG, Lee RJ, Li L, Xu CR and He XX: Gold nanoparticles delivered
miR-375 for treatment of hepatocellular carcinoma. Oncotarget.
7:86675–86686. 2016.PubMed/NCBI
|
|
22
|
Fang F, Chang RM, Yu L, Lei X, Xiao S,
Yang H and Yang LY: MicroRNA-188-5p suppresses tumor cell
proliferation and metastasis by directly targeting FGF5 in
hepatocellular carcinoma. J Hepatol. 63:874–885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Li W, Chen C, Pei Y and Long X:
miR-335 acts as a potential tumor suppressor miRNA via
downregulating ROCK1 expression in hepatocellular carcinoma. Tumor
Biol. 36:6313–6319. 2015. View Article : Google Scholar
|
|
24
|
Scott GK, Goga A, Bhaumik D, Berger CE,
Sullivan CS and Benz CC: Coordinate suppression of ERBB2 and ERBB3
by enforced expression of micro-RNA miR-125a or miR-125b. J Biol
Chem. 282:1479–1486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dahl Cowden KD, Dahl R, Kruichak JN and
Hudson LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cortez MA, Nicoloso MS, Shimizu M, Rossi
S, Gopisetty G, Molina JR, Carlotti C Jr, Tirapelli D, Neder L,
Brassesco MS, et al: miR-29b and miR-125a regulate podoplanin and
suppress invasion in glioblastoma. Genes Chromosomes Cancer.
49:981–990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang L, Huang Q, Zhang S, Zhang Q, Chang
J, Qiu X and Wang E: Hsa-miR-125a-3p and hsa-miR-125a-5p are
downregulated in non-small cell lung cancer and have inverse
effects on invasion and migration of lung cancer cells. BMC Cancer.
10:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hashiguchi Y, Nishida N, Mimori K, Sudo T,
Tanaka F, Shibata K, Ishii H, Mochizuki H, Hase K, Doki Y and Mori
M: Down-regulation of miR-125a-3p in human gastric cancer and its
clinicopathological significance. Int J Oncol. 40:1477–1482.
2012.PubMed/NCBI
|
|
29
|
Potenza N, Papa U, Mosca N, Zerbini F,
Nobile V and Russo A: Human microRNA hsa-miR-125a-5p interferes
with expression of hepatitis B virus surface antigen. Nucleic Acids
Res. 39:5157–5163. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bi Q, Tang S, Xia L, Du R, Fan R, Gao L,
Jin J, Liang S, Chen Z and Xu G: Ectopic expression of miR-125a
inhibits the proliferation and metastasis of hepatocellular
carcinoma by targeting MMP11 and VEGF. PLoS One. 7:e401692012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang H, Li RP, Liang P, Zhou YL and Wang
GW: miR-125a inhibits the migration and invasion of liver cancer
cells via suppression of the PI3K/AKT/mTOR signaling pathway. Oncol
Lett. 10:681–686. 2015.PubMed/NCBI
|