Introduction
As the fifth leading cause of cancer-associated
mortalities resulting from gynecological malignancies, ovarian
cancer (OC) presented with the highest incidence rates in Europe
and North America, and lowest in Africa and Asia in 2008.
Additionally, an estimated 22,240 new cases and 14,030 mortalities
were reported in the United States and ~4,500 mortalities per year
in the UK (1,2). Although China still has the lowest rates
and improved overall survival among patients with OC, increasing
incidence rates and decreasing age of diagnosis was noted in
patients with OC in a population-based registry in Taiwan and Hong
Kong (3–5). In addition, patients suffering from OC
are usually diagnosed at advanced stages, since early diagnosis is
difficult due to the subtle and nonspecific initial symptoms; thus,
significant efforts are directed at understanding the molecular
mechanism and developing targeted therapy for OC (6). Deciphering of the signaling pathways
brings about novel advances in the understanding of the pathogenic
mechanism of ovarian carcinogenesis, and growing evidence has
demonstrated that microRNAs (miRs) are differentially expressed in
OC and may target molecular pathways involved in OC pathogenesis
(6,7).
Accumulating evidence revealed significant roles of
miRs in various human cancers by modulating several biological and
pathological processes, including differentiation, growth,
proliferation and apoptosis in cells, as well as angiogenesis,
invasion and metastasis in tumors (7–9). Putative
tumor suppressor miR-214 regulates invasion and metastasis in
hepatocellular carcinoma (HCC) and is able to directly or
indirectly target CTNNB1 to modulate the β-catenin signaling
pathway in HCC (9,10). The role of miR-214 in driving melanoma
metastasis has been demonstrated, and miR-214 may contribute to
melanoma tumor progression (11,12).
miR-214 has been reported to function as an onco-miR in
osteosarcoma, and promotes the proliferation and invasion of
osteosarcoma cells by directly suppressing leucine zipper putative
tumor suppressor 1 (13). Notably,
down-regulated miR-214 expression has been found in human cervical
cancer; upregulated miR-214 expression in HeLa cancer cells was
found to significantly reduce cell growth (14). It has been suggested that miR-214
serves a critical role in OC stem cells via regulation of the
p53-Nanog axis (14). Previous
studies presented evidence that miR-214 is frequently upregulated
in OC tissues compared with normal ovarian tissues, and miR-214
induces cell survival and cisplatin resistance primarily through
targeting the phosphatase and tensin homolog (PTEN)/Akt signaling
pathway, leading to downregulated PTEN protein and activation of
the Akt signaling pathway (15,16).
Studies have suggested that the phosphoinositide 3-kinase
(PI3K)/Akt/mechanistic target of rapamycin (mTOR) pathway is
frequently activated in OC and is a potential predictor of distinct
invasive and migratory capacities in human OC cell lines, and the
PTEN/PI3K/Akt/mTOR axis receives more attention in treatment of
various types of cancer (17–19). Therefore, the present study aimed to
evaluate the potential role of miR-214 in regulating the
PTEN-mediated PI3K/Akt signaling pathway and to highlight the
mechanisms of miR-214 in OC progression.
Materials and methods
Cell culture
The SK-OV-3 (human ovarian cancer) cell line was
provided by the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). SK-OV-3 cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life
Sciences, Logan, UT, USA), and incubated in a 5% CO2
incubator at 37°C. Cell subcultures were performed when cells were
grown to 90% confluence. The primary medium was subsequently
discarded. The cells were washed twice with phosphate-buffered
saline (PBS), digested with trypsin (Gibco; Thermo Fisher
Scientific, Inc.) and treated with the RPMI-1640 medium containing
10% FBS to form a single-cell suspension. Routine subculture was
performed. The cells in logarithmic phase were collected for
subsequent usage.
Luciferase reporter gene assay
DNA extraction was performed with the TIANamp
Genomic DNA kit (Tiangen Biotech Co., Beijing, China) according to
the manufacturer's protocol. In order to investigate whether
binding sites predicted by miR-214 induced changes in luciferase
activity, PTEN-mutation (PTEN-mut; 5′-TTA-TTT TAC TAG TTT TCA ATC
ATA ATA CCT GAC AT-3′) and wide-type PTEN (PTEN-wt; 5′-TTA-TTT TAC
TAG TTT TCA ATC ATA ATA CCT GCT GT-3′) both from Synbio
Technologies (Jiangshu, China), were designed and inserted into a
reporter plasmid, which was purchased from Miaolingbio, Inc.
(Hubei, China). PTEN and the corresponding miR-214 binding target
site were determined using a target prediction software (Target
Scan) (http://www.targetscan.org). Entering the
‘Target Scan’ and selecting Human in the column called ‘Select a
species’, miR-214 was input in the column called ‘Enter a microRNA
name (e.g., miR-9-5p)’, and the target site combined with PTEN were
found. The PTEN gene was digested by EcoRI and
SmalII. The digestion conditions were as follows: Following
recovery of purpose gene fragments using 30 µl KpnI, 2 µl
HindIII, 2 µl buffer and 5 µl ddH2O,
respectively, the fragments were sealed using T4 DNA ligase.
Subsequently the sealed fragments were incubated with
Escherichia coli DH5a from Weidi Biotechnology Co., Ltd.
(Shanghai, China) at 37°C overnight. Finally, luciferase reporter
gene pGL3 vectors were constructed. Luciferase activity was
determined with a Dual-Luciferase Reporter assay system (E1910;
Promega Corporation, Madison, WI, USA). Following transfection at
37°C for 48 h, the culture medium was removed. The cells were
washed with PBS twice. Passive lysis buffer (100 µl per well) was
added, and the cells were gently agitated for 15 min at room
temperature to obtain cell lysate. The program was set with a 2-sec
pre-read delay followed by a 10-sec measurement period, and
luciferase assay reagent II (LARII; 100 µl) and Stop &
Glo® reagent (100 µl) were added to each sample. The
prepared LARII and Stop & Glo® reagent and luminous
tubes or plates containing the cell lysate were placed into a
bioluminescence detection system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The program was run, and data were saved at the
end of reading.
Cell transfection
The cells were divided into four groups: Blank
(cells untransfected with any miR-214 sequence), negative control
(NC), miR-214 mimic and miR-214 inhibitor. The cells in the NC
group were transfected with a vector containing the miR-214 NC
sequence (5′CCU GAC AAU UAG UAU UU-3′; Shanghai GenePharma Co.,
Ltd., Shanghai, China) and the cells in the miR-214 mimic group
were transfected with a miR-214 mimic
(5′-ACAGGUAGCUGAACACUGGGUU-3′; Synbio Technologies). The cells in
the miR-214 inhibitor group were transfected with mirVana™ miRNA
inhibitor (5′-UCACAGUGCUCAUCAUGAAUAA-3′; Shanghai Bioleaf,
Shanghai, China). SK-OV-3 cells (200 µl/well) in the logarithmic
growth phase were seeded on a 6-well plate with antibiotic-free
complete medium. SK-OV-3 cells were transfected with Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) when cell density
reached 30–50%, according to the manufacturer's instructions. The
miR-214 NC vector, miR-214 mimic and a miR-214 inhibitor (100 pmol;
final concentration, 50 nM) were respectively diluted in 250 µl
serum-free medium Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.),
gently mixed until even and incubated for 5 min at room
temperature. Lipofectamine 2000 (5 µl) was diluted in 250 µl
serum-free medium Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.),
gently mixed until even, and incubated for 5 min at room
temperature. The diluted miR-214 NC vector, miR-214 mimic and
miR-214 inhibitor were evenly mixed with the diluted Lipofectamine
2000, respectively. The mixture was added into the well containing
cells following incubation for 20 min at room temperature and
gently mixed until even. The transfected cells were placed in a 5%
CO2 incubator at 37°C. The medium was replaced with
complete medium after 6–8 h incubation at 37°C, and the transfected
cells were incubated at 37°C for 24–48 h for subsequent
experiments.
MTT colorimetric assay
The cells (80% confluence) were washed twice with
PBS and digested with 0.25% trypsin (Gibco; Thermo Fisher
Scientific, Inc.) to form a single-cell suspension. The cells were
counted using Moxi Z mini automated cell counter (Bio Excellence
International Tech Co., Ltd., Beijing, China), according to the
manufacturer's instructions and were inoculated in a 96-well plate
(200 µl per well, 6 repeated wells) at a density of
3×103 to 6×103. Following culture at 37°C for
48 h, 5 mg/ml MTT solution (20 µl) was added to each well. After 4
h incubation at 37°C, the culture medium was discarded. Dimethyl
sulfoxide (150 µl; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
was added to each well and gently agitated for 10 min. Optical
density (OD) values were determined at a wavelength of 490 nm with
an enzyme-linked immunosorbent detector (De Tie Inc., Nanjing,
China) at 12, 24 and 48 h, respectively. Cell viability curves were
plotted with time as the x-axis and the OD value as the y-axis.
Apoptosis detected by Annexin V and PI
staining
Following a 48-h transfection at 37°C, the cells
were digested with the EDTA-free trypsin and collected in the flow
tube for centrifugation at 157 × g at 4°C for 5 min, and the
supernatant was discarded. Cells were washed three times with PBS,
and subsequently centrifuged at 157 × g at 4°C for 5 min for
removal of supernatant. According to the manufacturer's
instructions, 150 µl binding buffer and 5 µl (per tube) Annexin
V-FITC [Annexin V-fluorescein isothiocyanate (FITC) apoptosis
detection kit; Sigma-Aldrich; Merck KGaA] was added to the cells
and gently agitated. The cells were incubated in darkness for 15
min at room temperature, and an additional 100 µl binding buffer
and 5 µl propidium iodide (PI) dye (Sigma-Aldrich; Merck KGaA) was
added. The mixture was oscillated and mixed evenly. The proportion
of cells undergoing apoptosis was analyzed by CytoFLEX flow
cytometer (Beckman Coulter, Inc., Brea, CA, USA) and the results
were analyzed using CytExpert 2.0 software (Beckman Coulter,
Inc.).
PI staining for cell cycle
analysis
Following a 48-h transfection at 37°C, the cells
were collected, washed with cold PBS for three times and
centrifuged at 157 × g at 4°C for 5 min to remove the supernatant.
Cell concentration was adjusted to ~1×105/ml, and the
cells were fixed with 1 ml 75% ice-cold ethanol overnight at 4°C.
The cells were subsequently washed twice with PBS with supernatant
discarded prior to staining and 100 µl RNase A was then added in
darkness. The cells were incubated in a 37°C water bath for 30 min,
stained with 400 µl PI and mixed evenly. Subsequently, the samples
were incubated in the dark at 4°C for 30 min. Cell cycle was
analyzed by detecting red fluorescence at excitation wavelength of
488 nm with the CytoFLEX flow cytometer (Beckman Coulter,
Inc.).
Western blot analysis
After 48 h of transfection, the cells were
collected, lysed by Tissue Total Protein Lysis Buffer Solution
(Shanghai Yu Bo Biological Technology Co., Ltd., Shanghai, China)
and centrifuged. The bicinchoninic acid kit (Thermo Fisher
Scientific, Inc.) was used following the manufacturer's
instructions to measure total protein concentration. The total
protein (20 µg) was run on a 12% SDS-PAGE and electro-transferred
to polyvinylidene fluoride membranes. The membranes were blocked
with skimmed milk at room temperature for 1 h, incubated with
monoclonal antibodies GAPDH (GTX101025, 1:500), PTEN (GTX101025,
1:500), PIP3 (GTX119163, 1:500), Akt (GTX128414, 1:500),
phosphorylated (p)-Akt (GTX121937, 1:500), glycogen synthase kinase
(GSK)-3β (GTX111192, 1:500) and p-GSK-3β (GTX132997, 1:500; all
from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) overnight
at 4°C. The membrane was washed with PBS three times to remove
primary antibodies and incubated with horseradish
peroxidase-labeled goat anti-rabbit secondary antibody (GTX77266,
1:500; Beyotime Institute of Biotechnology, Haimen, China) at room
temperature for 1–2 h, washed with PBS three times, and an
electro-chemiluminescence (ECL) color developing kit (Shanghai
Yeasen Biotechnology Co., Ltd., Shanghai, China) for visualization
of the bands, according the manufacturer's instructions. Images
were captured using an in vivo imaging system (LAS-4000; GE
Healthcare Life Sciences) and the relative molecular mass and net
absorption of the target bands were analyzed by a gel image
processing system. The data were normalized with the expression of
β-actin. Using GAPDH as a reference, the relative expression of
target proteins was analyzed using ImageJ version 2×2.1.4.7
software (National Institutes of Health, Bethesda, MD, USA).
Study subjects
The specimens were obtained from 124 patients with
OC who underwent surgical treatment in Linyi Tumor Hospital (Linyi,
China) between February 2013 and February 2015. All the specimens
were confirmed by pathological diagnosis. All patients were females
with childbearing history (the mean age, 48.17±10.32 years; range,
34–73 years), who had received no preoperative chemotherapy.
Surgical-pathological staging (20)
of the patients are as follows: Stage I (n=40); stage II (n=36);
stage III (n=42); and stage IV (n=6). Histological types and
pathological grading (21) of the
patients are as follows: Serous carcinoma (n=74); mucinous
carcinoma (n=29); and endometrioid carcinoma (n=21); and G1
(well-differentiated) (n=47); G2 (moderately-differentiated)
(n=26); and G3 (poorly-differentiated) (n=51).
Lymph-node-metastasis statuses of the patients are as follows:
Positive (n=88) and negative (n=36). The specimens collected from
OC tissues and normal mucosa tissues (5 cm away from the edge of
carcinoma) were treated with liquid nitrogen and stored in a −70°C
freezer. Half of the specimens (n=62) were used for the detection
of miR-214, and the other half (n=62) were fixed with 10% formalin
at room temperature for 30 min, paraffin-embedded and used for
immunohistochemistry. The present study protocol was approved by
the Ethics Committee of Linyi Tumor Hospital, and all patients
signed informed consent.
Analysis of miR-214 expression in OC
tissues and adjacent normal tissues by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Analysis of miR-214 expression in OC tissues and
adjacent normal tissues was performed with RT-qPCR. The total RNA
was extracted from frozen tissues (50 mg) with a miRNeasy Mini kit
(Qiagen, Inc., Valencia, CA, USA) following the manufacturer's
instructions. RNA sample (5 µl) was diluted (20X) with RNase-free
ultra-pure water (Beijing Solarbio Science & Technology Co.,
Ltd., Beijing, China). The adsorptions of UV light at wavelengths
260 and 280 nm (OD260/OD280 ratio) were read from the ultraviolet
spectrophotometer (Mettler Toledo Instruments, Shanghai, China) to
determine purity and contamination of the RNA samples. Only
high-purity RNA samples with an OD260/OD280 ratio of 1.7–2.1 were
used for subsequent analysis. Complementary DNA (cDNA) was obtained
by reverse transcription with a PCR amplification instrument. A
total of 9.0 µl diethyl pyrocarbonate (DEPC)-treated water, 0.6 µl
reverse primers (20 pmol/µl), 0.1 µl enzyme inhibitor (40 U/µl) and
5 µl RNA solution were added to a microcentrifuge tube (0.5 ml,
RNase-free). Following incubation in a water bath at 70°C for 5 min
and in an ice bath for 5 min, the mixture was incubated with 4.0 µl
5xAMV reverse transcription buffer, 1.0 µl dNTP Mixture (10 mol/l),
0.1 µl RNA enzyme inhibitor (40 U/µl) and 0.2 µl AMV Reverse
Transcriptase (5 U/µl) at 42°C for 40 min, followed by
amplification using a PCR instrument. RT-qPCR experiment was
performed using a ABI7500 quantitative PCR instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with the following PCR
conditions: Pre-denaturing (95°C, 10 min); followed by 40 cycles of
denaturing at 95°C for 10 sec and annealing at 60°C for 20 sec; and
extension at 72°C for 34 sec. Real-time detection was performed
using SYBR-Green (Thermo Fisher Scientific, Inc.) with maximum
absorption wavelength 497 nm and emission maximum wavelength 520 nm
for fluorescence signals. The sequences of the primers used are as
follows: miR-214 forward, 5′-CACCTTTCTCCCTTTCCCCTTACTCTCC-3′ and
reverse, 5′-TTTCATAGGCACCACTCACTTTAC-3′; U6 (internal control)
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The 2−ΔΔCq method (22) was used for normalization and was
calculated as follows: 2−ΔΔCq = ΔCqexperiment
group - ΔCqcontrol group. ΔCq = CqmiRNA
- CqU6, where Cq is the number of amplification cycles
when the real-time fluorescence intensity of the reaction reaches
the threshold values. Amplification was performed during a period
of logarithmic growth. Each experiments were repeated 3 times.
Analysis of PTEN expression in OC and
adjacent normal tissues by immunohistochemistry
The 10% formalin fixed and paraffin-embedded
specimens (n=62) were cut into 4 µm-thick sections. The tissue
slices were dried in an oven at 60°C for 1 h, dewaxed with xylene,
dehydrated in graded ethanol (50, 70, 80 and 95%) and incubated
with 3% H2O2 (Sigma-Aldrich; Merck KGaA) at
37°C for 30 min, washed with PBS and placed in 0.01 M citrate
buffer, boiled at 95°C for 20 min, cooled to room temperature and
washed with PBS. The washed tissue sections were incubated at 37°C
for 10 min in blocking medium containing normal goat serum
(Wak-Chemie Medical GmbH, Steinbach, Germany), incubated with the
primary antibody (cat. no. GTX101025; PTEN; 1:100; Santa Cruz
Biotechnology, Inc.) overnight at 4°C, rinsed with PBS, and
additionally incubated with the horseradish peroxidase-labeled
secondary antibody (GTX77266, 1:500, Beijing Biosynthesis
Biotechnology Co., Ltd.) for 30 min at room temperature. The
sections were visualized using 3,3′-diaminobenzidine
tetrahydrochloride (Sigma-Aldrich; Merck KGaA) followed by staining
with hematoxylin (5 mg/ml; Shanghai Bogoo Biotechnology. Co., Ltd.,
Shanghai, China) and mounting. A negative control was prepared by
replacing the primary antibody with PBS, and normal mucosa tissues
were used as a positive control. The expression of PTEN was mainly
observed in the cell nucleus and partially in the cytoplasm,
presenting with yellow or brownish-yellow granules. A total of five
high-power fields (×400, 100 counted cells per field) using a
CX31-LV320 OLYMPUS microscope (Chang Heng Rong technology Co.,
Ltd., Beijing, China) in each tissue section were randomly
selected, and positive cells/all tumor cells >10% was considered
as positive (+) and positive cells ≤10% as negative (−). The
results of immunohistochemistry were evaluated independently by two
experienced observers using a double-blind approach.
Statistical analysis
Continuous variables are presented as the mean ±
standard deviation. The comparison between the two groups was
performed using paired t-test, and the comparison among the groups
was analyzed using one-way analysis of variance. Categorical
variables are presented as frequencies and percentages, and
comparisons were performed with χ2 test. SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA) was used for statistical
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Verification of miR-214 target
gene
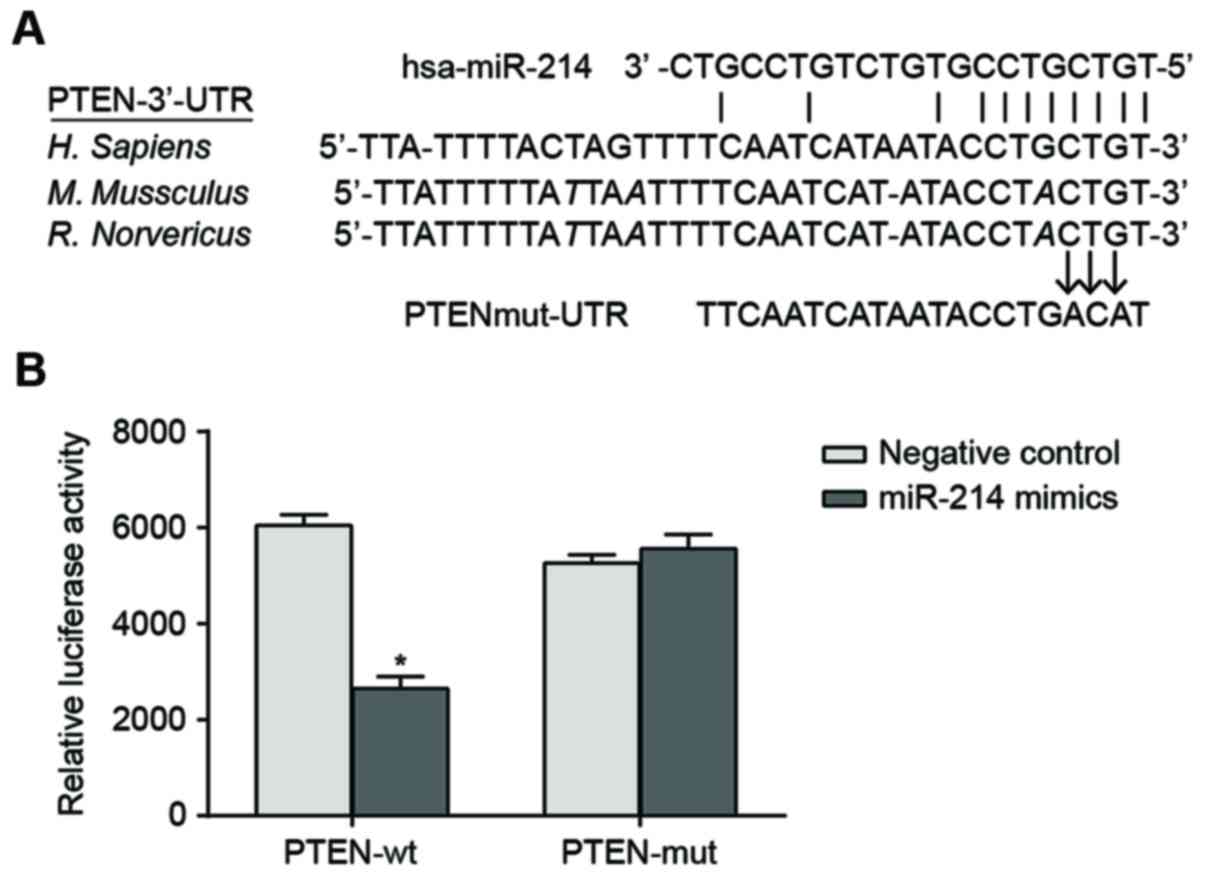
The sequences of miR-214 binding with 3′-UTR region
of PTEN mRNA were illustrated in Fig.
1A. In order to demonstrate that the miR-214 seed region was
able to interact with 3′UTR region of PTEN mRNA and leads to
altered luciferase activity, the designed mutated sequence (PTEN
3′-UTR mRNA which lacks the putative miR-214 binding site) and the
wild sequence were inserted into the reporter gene plasmid. SK-OV-3
cells were co-transfected with miR-214 mimic, wild recombinant
plasmid (Wt-miR-214/PTEN) and mutated recombinant plasmid
(Mut-miR-214/PTEN). The luciferase activity assay indicated that
transfection with miR-214 mimic had no significant effect on the
luciferase activity of the cells in the Mut-miR-214/PTEN plasmid
group compared with cells in the negative control. By contrast,
luciferase activity of the cells in the Wt-miR-214/PTEN plasmid
group decreased by ~62% compared with the negative control
(P<0.05; Fig. 1B).
miR-214 expression
RT-qPCR results indicated that the expression of
miR-214 decreased significantly in the SK-OV-3 cells transfected
with a miR-214 inhibitor compared with the untransfected cells
(blank) and NC groups. By contrast, there was a significant
increase in miR-214 expression in the miR-214 mimic group compared
with the blank (all P<0.05). miR-214 expression was markedly
lower in the miR-214 inhibitor group compared with the miR-214
mimic group (P<0.05). There were no significant differences in
miR-214 expression between the blank and NC P>0.05; Fig. 2).
Effect of miR-214 expression on the
viability of SK-OV-3 cells
MTT assay indicated that, cell viability was
significantly inhibited in the miR-214 inhibitor group compared
with the blank and the NC group (P<0.05). There were significant
differences in OD values between the miR-214 inhibitor group and
the blank group at the time points 24 and 48 h (P<0.05). There
were also significant differences between the miR-214 inhibitor
group and NC group (P<0.05). Cell viability increased
significantly in the miR-214 mimic group compared with the blank
and the NC group. Significant differences in OD values were
observed between the miR-214 mimic group and the blank and the NC
group at the time points 24 and 48 h (both P<0.05).
Significantly increased OD values were observed in the miR-214
mimic group compared with the miR-214 inhibitor group at the time
points of 24 and 48 h (P<0.05). The comparison of OD values
between the blank and the NC group showed no significant difference
at any time points (all P>0.05). With the increase in time,
there were significant differences in OD among different groups
[multivariate analysis of variance (F)=24.544; P<0.001]. There
were significant differences in OD values between the different
time points (F=217.932; P<0.001). Additionally, there was
interaction between the measurement time and group (F=23.839;
P<0.001; Fig. 3).
Cell cycle and apoptotic rate
PI staining results showed that the proportion of
cells in G1 in the blank, NC, miR-214 mimic and miR-214 inhibitor
groups were 56.39±3.82, 57.62±3.34, 60.34±2.38 and 76.40±2.22%,
respectively (Fig. 4). The majority
of cells treated with a miR-214 inhibitor were in G1. By contrast,
the proportion of miR-214 inhibitor-treated cells in the S phase
was decreased, and the proliferation of miR-214 inhibitor-treated
cells was significantly inhibited compared with cells in the blank,
the NC and the miR-214 mimic group. No significant difference in
the proportion of cells in G1, G2 and S existed in the miR-214
mimic group, the blank group and the NC group (all P>0.05).
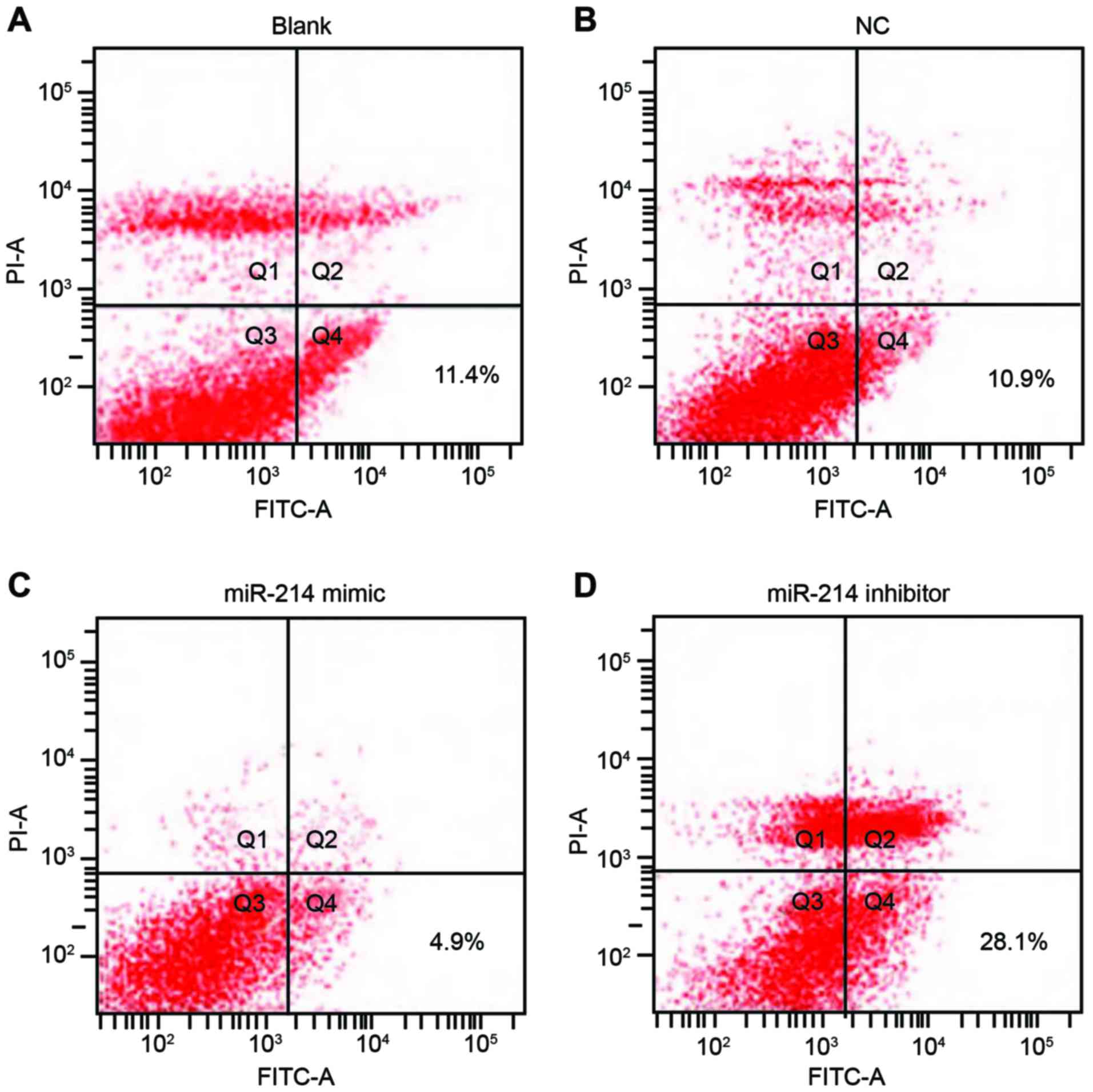
Double staining with Annexin V and PI results
indicated that 48 h following transfection, apoptotic rates in the
blank, the NC, the miR-214 mimic group and the miR-214 inhibitor
group were 11.4, 10.9, 4.9 and 28.1%, respectively. The apoptotic
rate of SK-OV-3 cells in the miR-214 inhibitor group was
significantly increased compared with the other three groups (all
P<0.05). However, the apoptotic rate was not statistically
significant between the blank group and the NC group (P>0.05;
Fig. 5).
Expression of PTEN and PI3K/Akt
pathway-associated proteins
Western blot analysis results revealed significantly
increased expression of PTEN and decreased expression of PIP3,
p-Akt and p-GSK-3β in SK-OV-3 cells transfected with a miR-214
inhibitor (48 h later) compared with the blank group and the NC
group (all P<0.05). SK-OV-3 cells transfected with the miR-214
mimic exhibited significantly downregulated PTEN expression and
significantly upregulated PIP3, p-Akt and p-GSK-3β expression
compared with the blank and the NC group (all P<0.05).
Significant differences in PTEN, PIP3, p-Akt and p-GSK-3β
expression levels were observed between the miR-214 inhibitor group
and the miR-214 mimic group (all P<0.05). There were no
significant differences in Akt and GSK-3β among all groups (all
P>0.05). The expression of each protein in the blank group did
not significantly differ from those in the NC group (all P>0.05;
Fig. 6).
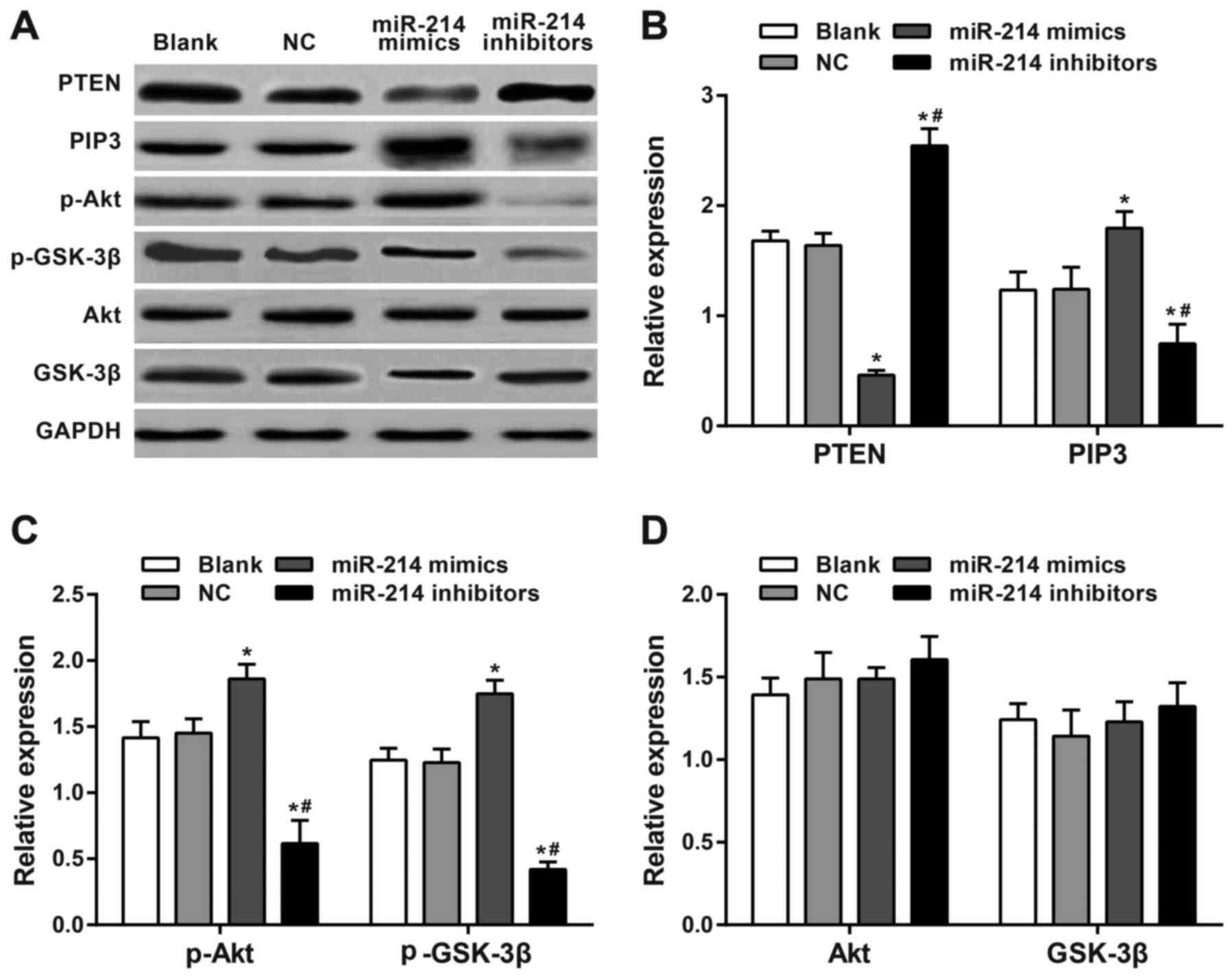 | Figure 6.Western blotting of PTEN and PI3K/Akt
pathway-associated proteins. (A) Expression levels were analyzed by
western blotting following 48-h of transfection. (B) PTEN, PIP3,
(C) p-Akt, GSK-3β, (D) Akt and p-GSK-3β expression levels. There
was a significant increase in expression of PTEN and a significant
decrease in expression of PIP3, p-Akt and p-GSK-3β in
SK-OV-3 cells transfected with a miR-214 inhibitor compared with
the blank, NC and miR-214 inhibitors groups. There was a
significantly downregulated PTEN expression and significantly
upregulated PIP3, p-Akt and p-GSK-3β expression in
miR-214 mimic-transfected cells compared with the blank, NC and
miR-214 mimics groups. There were significant differences in PTEN,
PIP3, p-Akt and p-GSK-3β expression between the miR-214
inhibitor group and the miR-214 mimic group. *P<0.05 compared
with the blank and NC groups; #P<0.05 compared with
miR-214 mimic group; sphatidylinositol (3,4,5)-trisphosphate; GSK, glycogen synthase
kinase; p, phosphorylated; PTEN, phosphatase and tensin homolog
deleted on chromosome 10; PI3K, phosphoinositide 3-kinase; Akt,
protein kinase B; PIP3, phosphatidylinositol (3,4,5)-trisphosphate. |
Expression of miR-214 and PTEN in OC
tissues and adjacent normal tissues
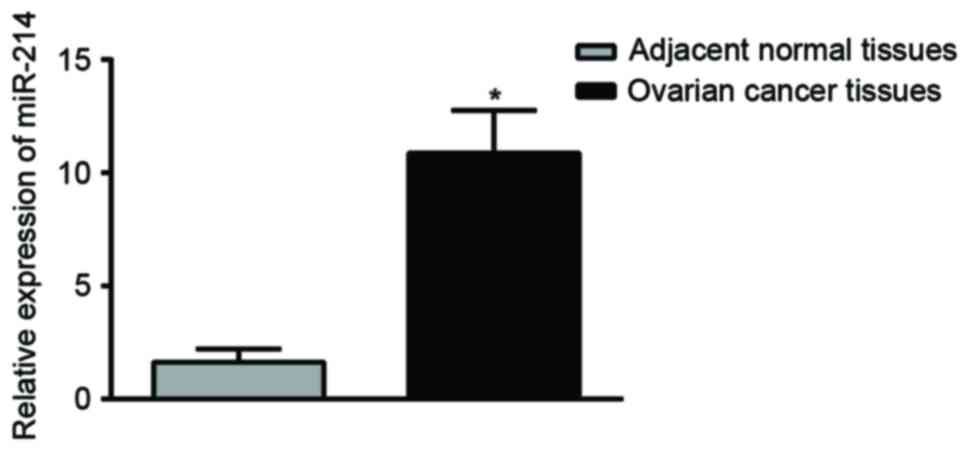
The results from RT-qPCR showed that OC tissues had
a significantly increased expression of miR-214 in comparison with
adjacent normal tissues (P<0.01; Fig.
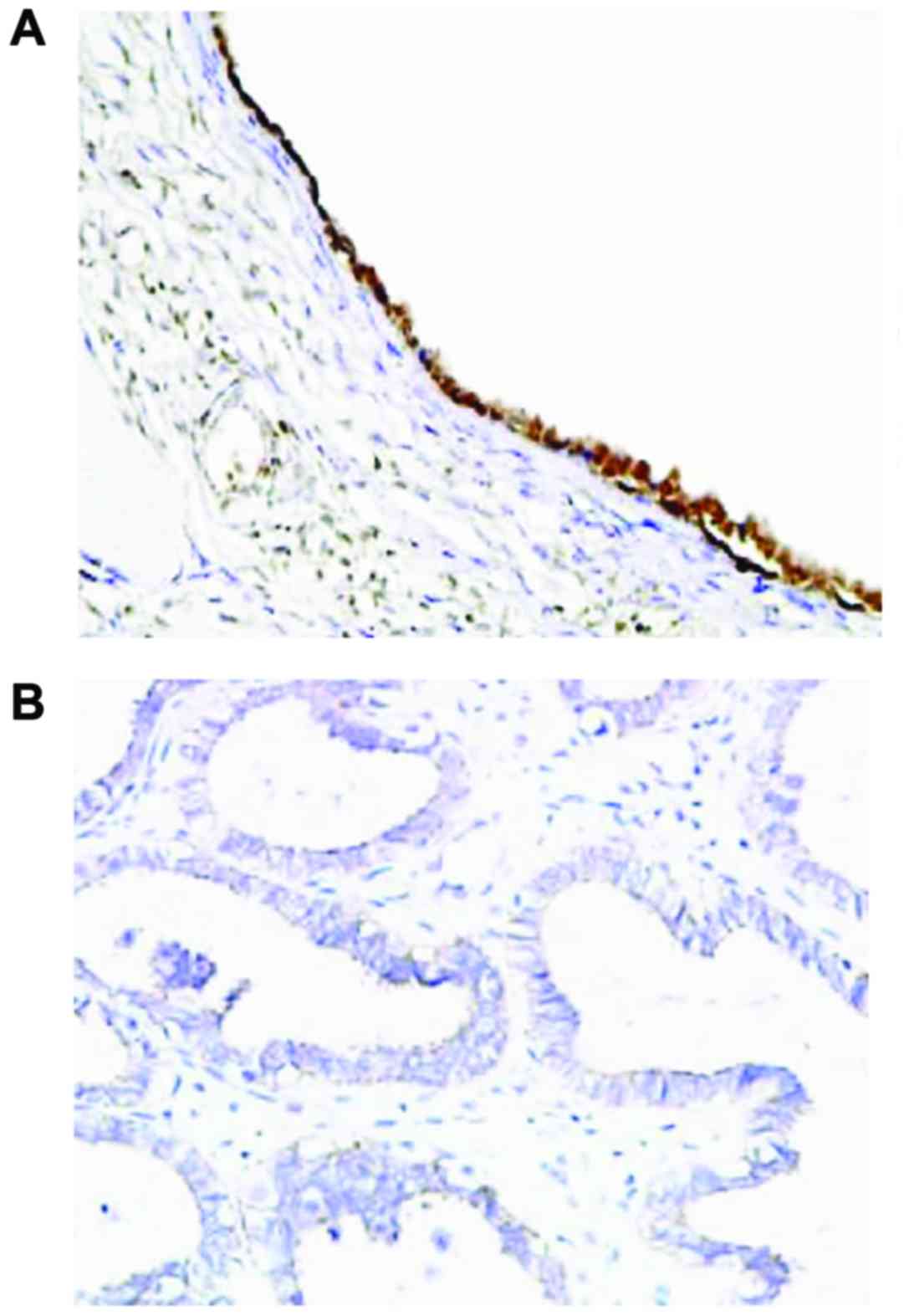
7). Immunohistochemistry results showed that the expression of
PTEN was mainly observed in the cell nucleus, and partially in the
cytoplasm, and presented with yellow or brownish-yellow granules
(Fig. 8). As shown in Table I, adjacent normal tissues exhibited a
significantly increased rate of positive PTEN protein expression
compared with OC tissues [77.4 (96/124) vs. 47.6% (59/124);
P<0.05].
 | Table I.Differences in the positive expression
rate of PTEN protein in ovarian cancer and adjacent normal
tissues. |
Table I.
Differences in the positive expression
rate of PTEN protein in ovarian cancer and adjacent normal
tissues.
|
|
| PTEN expression,
n |
|
|
|---|
|
|
|
|
|
|
|---|
| Type of tissue | Number of cases | Positive | Negative | PTEN positive rate,
% | P-value |
|---|
| Ovarian cancer | 124 | 59 | 65 | 47.6 | <0.001 |
| Normal tissues | 124 | 96 | 28 | 77.4 |
|
Discussion
The functional role of miR-214 in regulating
PTEN-mediated PI3K/Akt signaling pathway was evaluated. It has been
predicted that miR-214, which is located inside the sequence of
long noncoding Dmn3os transcript, targets two activating protein 2
transcription factors, causing downstream effects on genes
regulating vital cell cycle processes, including apoptosis,
proliferation and angiogenesis (11).
Additionally, miR-214 with pleiotropic and tumor-specific functions
may contribute to the formation and progression of OC through its
target genes and primary signaling networks, including PTEN/Akt,
β-catenin, as well as tyrosine kinase receptor (23). The involvement of the PI3K/Akt
signaling pathway in regulating cell survival, cell cycle
progression and cellular growth has been revealed. Components of
the PI3K/Akt signaling pathway are frequently altered in various
types of human cancer, and the PI3K/Akt/mTOR signaling pathway has
been considered as a therapeutic target for OC (24–26). In
the present study, luciferase reporter gene assay indicated PTEN
was as the target gene of miR-214. Important findings in the
present study indicated that OC tissues exhibited increased
expression of miR-214 and increased expression of PTEN protein
compared with the adjacent normal tissues. SK-OV-3 cells
transfected with miR-214 mimic exhibited significantly
downregulated PTEN expression and significantly upregulated
PIP3, p-Akt and p-GSK-3β expression compared with the
blank and the NC group. The cells transfected with miR-214 mimic
exhibited significantly increased viability and proliferation and
markedly decreased apoptotic rate. Combining these significant
results, the present study supported the hypothesis that miR-214
may activate the PI3K/Akt signaling pathway by downregulating PTEN,
which may promote proliferation and inhibit apoptosis of OC
cells.
Extensive roles of miR-214 in promoting tumor cell
proliferation, growth and invasion, as well as tumor progression
and metastasis have been previously reported (12,13,27).
Additionally, miR-214 has been demonstrated to be implicated in the
differentiation of OC stem cells into mature OC cells (28), It has been well documented that
miR-214 targets PTEN and is upregulated in OC, and has oncogenic
and chemoresistant functions (29).
miR-214 may be useful in diagnostic tests for the early detection
of OC. Transient perturbation of miR-31 and miR-214 and miR-155
expression was sufficient to convert normal ovarian fibroblasts
into induced carcinoma-associated fibroblasts that promoted ovarian
tumor growth and increased tumor cell invasiveness and migration
(29,30). Partially consistent with the role of
miR-214 in downregulating PTEN and activating the PI3K/Akt
signaling pathway, a previous study revealed that miR-214 induces
cell survival primarily by targeting the PTEN/Akt pathway,
resulting in downregulation of PTEN protein and activation of Akt
pathway in human OC (15).
In conclusion, OC tissues exhibited increased
expression of miR-214 and decreased expression of PTEN protein
compared with normal tissues. miR-214 may activate the PI3K/Akt
signaling pathway by downregulating the targeted PTEN, which may
promote proliferation and inhibit apoptosis of OC cells and provide
a potential miRNA-based targeted therapy for the treatment of
OC.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chornokur G, Amankwah EK, Schildkraut JM
and Phelan CM: Global ovarian cancer health disparities. Gynecol
Oncol. 129:258–264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lowe KA, Chia VM, Taylor A, O'Malley C,
Kelsh M, Mohamed M, Mowat FS and Goff B: An international
assessment of ovarian cancer incidence and mortality. Gynecol
Oncol. 130:107–114. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chiang YC, Chen CA, Chiang CJ, Hsu TH, Lin
MC, You SL, Cheng WF and Lai MS: Trends in incidence and survival
outcome of epithelial ovarian cancer: 30-year national
population-based registry in Taiwan. J Gynecol Oncol. 24:342–351.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wong KH, Mang OW, Au KH and Law SC:
Incidence, mortality, and survival trends of ovarian cancer in Hong
Kong, 1997 to 2006: A population-based study. Hong Kong Med J.
18:466–474. 2012.PubMed/NCBI
|
|
6
|
Ledermann JA, Raja FA, Fotopoulou C,
Gonzalez-Martin A, Colombo N and Sessa C: ESMO Guidelines Working
Group: Newly diagnosed and relapsed epithelial ovarian carcinoma:
ESMO Clinical Practice Guidelines for diagnosis, treatment and
follow-up. Ann Oncol. 24 Suppl 6:vi24–vi32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Katz B, Trope CG, Reich R and Davidson B:
MicroRNAs in ovarian cancer. Hum Pathol. 46:1245–1256. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Davis-Dusenbery BN and Hata A: MicroRNA in
Cancer: The involvement of aberrant microRNA biogenesis regulatory
pathways. Genes Cancer. 1:1100–1114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tutar L, Tutar E and Tutar Y: MicroRNAs
and cancer; an overview. Curr Pharm Biotechnol. 15:430–437. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xia H, Ooi LL and Hui KM: MiR-214 targets
β-catenin pathway to suppress invasion, stem-like traits and
recurrence of human hepatocellular carcinoma. PLoS One.
7:e442062012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bar-Eli M: Searching for the
‘melano-miRs’: miR-214 drives melanoma metastasis. EMBO J.
30:1880–1881. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Penna E, Orso F, Cimino D, Tenaglia E,
Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De
Pittà C, et al: microRNA-214 contributes to melanoma tumour
progression through suppression of TFAP2C. EMBO J. 30:1990–2007.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Z and Wang T: miR-214 promotes the
proliferation and invasion of osteosarcoma cells through direct
suppression of LZTS1. Biochem Biophys Res Commun. 449:190–195.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Z, Chen S, Luan X, Li Y, Liu M, Li X,
Liu T and Tang H: MicroRNA-214 is aberrantly expressed in cervical
cancers and inhibits the growth of HeLa cells. IUBMB Life.
61:1075–1082. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gadducci A, Sergiampietri C, Lanfredini N
and Guiggi I: Micro-RNAs and ovarian cancer: The state of art and
perspectives of clinical research. Gynecol Endocrinol. 30:266–271.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mabuchi S, Kuroda H, Takahashi R and
Sasano T: The PI3K/AKT/mTOR pathway as a therapeutic target in
ovarian cancer. Gynecol Oncol. 137:173–179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bai H, Li H, Li W, Gui T, Yang J, Cao D
and Shen K: The PI3K/AKT/mTOR pathway is a potential predictor of
distinct invasive and migratory capacities in human ovarian cancer
cell lines. Oncotarget. 6:25520–25532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang X, Cheng Y, Li P, Tao J, Deng X,
Zhang X, Gu M, Lu Q and Yin C: A lentiviral sponge for miRNA-21
diminishes aerobic glycolysis in bladder cancer T24 cells via the
PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 36:383–391. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
FIGO Oncology Committee, . FIGO staging
for gestational trophoblastic neoplasia 2000. FIGO Oncology
Committee. Int J Gynaecol Obstet. 77:285–287. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scully RE and Sobin LH: Histological
Typing of Ovarian Tumours. 2nd. Springer Verlag; New York, NY:
1999, View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Penna E, Orso F and Taverna D: miR-214 as
a key hub that controls cancer networks: Small player, multiple
functions. J Invest Dermatol. 135:960–969. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vara Fresno JA, Casado E, de Castro J,
Cejas P, Belda-Iniesta C and González-Barón M: PI3K/Akt signalling
pathway and cancer. Cancer Treat Rev. 30:193–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Zeng J and Shen K: PI3K/AKT/mTOR
signaling pathway as a therapeutic target for ovarian cancer. Arch
Gynecol Obstet. 290:1067–1078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Faes S and Dormond O: PI3K and AKT:
Unfaithful partners in cancer. Int J Mol Sci. 16:21138–21152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiang R, Wang F, Shi LY, Liu M, Chen S,
Wan HY, Li YX, Li X, Gao SY, Sun BC and Tang H: Plexin-B1 is a
target of miR-214 in cervical cancer and promotes the growth and
invasion of HeLa cells. Int J Biochem Cell Biol. 43:632–641. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yin G, Chen R, Alvero AB, Fu HH, Holmberg
J, Glackin C, Rutherford T and Mor G: TWISTing stemness,
inflammation and proliferation of epithelial ovarian cancer cells
through MIR199A2/214. Oncogene. 29:3545–3553. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chou J and Werb Z: MicroRNAs play a big
role in regulating ovarian cancer-associated fibroblasts and the
tumor microenvironment. Cancer Discov. 2:1078–1080. 2012.
View Article : Google Scholar : PubMed/NCBI
|