Introduction
Lung cancer is a malignant tumor that originates
from lung tissues with uncontrolled cell growth. Lung cancer is the
leading cause of cancer-associated death worldwide (1). This led to 1.56 million mortalities in
2012 (2). Lung cancer is mainly
composed of two types of cancer, consisting of small cell lung
cancer (SCLC) and non-small cell lung cancer (NSCLC). The majority
of lung cancer cases are induced by long-term exposure to tobacco.
The symptoms of lung cancer are mostly severe coughing, chest pain
and weight loss. The current therapeutic methods for lung cancer
are primarily surgery, chemotherapy and radiotherapy (3). Treatment and outcome depends on the
cancer type and the health status of the individual (3). Although there are several types of
therapeutic approaches for lung cancer, the efficacy of the
treatments remains unsatisfactory. Investigating a novel target for
lung cancer therapy is of critical importance.
Insulin-like growth factor binding protein-4
(IGFBP-4) is a member of the IGBP family of proteins. It consists
of an IGF binding domain and a thyroglobulin type-I domain
(4). IGFBP-4 binds both IGF I and IGF
II. Binding of these ligands prolongs the half-life of IGFs and
extends their function. IGFBP-4 alters the interactions between IGF
ligands and cell surface receptors, which in turn promotes cell
proliferation (5). IGFBP-4 expression
is correlated with several types of cancers (6–8). In 2009,
Gupta et al (9) found an
effective anti-cancer stem cell small molecule, salinomycin,
through high-throughput screening of cancer stem cells. Salinomycin
strongly inhibited breast cancer stem cells and downregulated
(LRP6, survivin and histone H3 and H4) or upregulated (p21) several
genes (10,11). It was found that IGFBP-4 gene
expression was downregulated by salinomycin treatment (9). As IGFBP-4 is a cancer cell
growth-associated gene that has, to the best of our knowledge,
never been reported in lung cancer, the present study investigated
whether the gene is associated with the prognosis of patients with
lung cancer.
Materials and methods
Tumor sample collection
In total, 102 patients were selected between January
2014 and May 2016. All tissues were collected with informed
consent. The majority (90 out of 102) of lung cancer patients did
not have metastasis. Overall, 52.0% (53/102) of the patients were
female and 48.0% (49/102) were male. Patients at late stage or
early stages were all included. All experiments were approved by
the Institutional Review Board of Central Hospital of Wuhan (Wuhan,
China).
Immunohistochemistry
All tissues were selected by an experienced
pathologist and embedded in paraffin. The immunohistochemistry
assay was performed following standard protocols (12,13).
Briefly, all the embedded tissues were cut into 3-µm slices. The
slices were deparaffinized with xylene and graded ethanol. The
tissues were treated with 0.3% hydrogen peroxidase, incubated with
a primary rabbit anti-IGFBP-4 antibody (dilution, 1:100; cat. no.
ab4253; Abcam, Cambridge, UK) overnight at 4°C followed by
incubation with anti-rabbit IgG-horseradish peroxidase (HRP)
secondary antibody (dilution, 1:1,000; cat. no. MBS435036;
MyBioSource, San Diego, CA, USA) at room temperature for 1 h. The
tissues were washed three times with PBS between antibody
incubations. The tissues were observed under microscopes (BX4;
Olympus Corporation, Tokyo, Japan; magnification, ×40) and the
expression levels of IGFBP-4 were considered to be extremely
strong, strong, median, weak and none according to the density of
signals.
Cell culture
The lung cancer A549 and lentivirus packaging 293T
cell lines were purchased from Cell Bank of Chinese Academy of
Sciences (Shanghai, China). The cells were maintained in Dulbecco's
modified Eagle's medium (DMEM, Thermo Fisher Scientific, Waltham,
MA, USA) supplemented with 10% fetal bovine serum (FBS,
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and passaged at 1:3
when cells had reached confluence. The cells were incubated in
humid atmosphere at 37°C with 5% CO2.
Short hairpin (sh)RNA
construction
The shRNA construct to target IGFBP-4 was cloned
into lentivirus plasmid pLKO1 (Addgene, Inc., Cambridge, MA, USA).
The targeting sequence of IGFBP-4 was 5′-GCCACTTGCGCCCTGGGCTTG-3′.
The oligo 1
(5′-AATTCGCCACTTGCGCCCTGGGCTTGATGCCAAGCCCAGGGCGCAAGTGGCG-3′) and
oligo 2
(5′-AATTCGCCACTTGCGCCCTGGGCTTGCATCAAGCCCAGGGCGCAAGTGGCGG-3′)
sequences were diluted to 100 µM for annealing. They were mixed
together with T4 DNA ligase buffer, heated at 95°C for 10 min and
cooled slowly at a rate of 1°C/min. The pLKO1 plasmid was cut using
the EcoRI restriction enzyme and ligated using annealed oligos to
generate the pLKO-IGFBP-4 shRNA-1 plasmid. Another IGFBP-4 shRNA
was generated using the same method and its targeting sequence was
5′-GACAAGGACGAGGGTGACCA-3′.
Lentivirus package and stable cell
lines
pLKO-IGFBP-4 shRNA-1 and pLKO-IGFBP-4 shRNA-2 were
co-transfected with vesicular stomatitis virus-G and delta 8.2
(Addgene, Inc.) into 293T cells to package lentivirus. The medium
was changed 12 h post-transfection. The 293T medium was collected
48, 72 and 96 h post transfection. The media were centrifuged at
8,000 × g for 20 min to remove the cell debris. The medium
contained the lenti-IGFBP-4 shRNA-1 or lenti-IGFBP-4 shRNA-2
viruses. The A549 cells were infected with viruses and selected
with 2 µg/ml puromycin 48 h later. The stable cell lines were
passaged and maintained in DMEM 10% FBS with 2 µg/ml puromycin.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted from A549 cells and A549 cells
stably transfected with IGFBP-4 shRNA-1 and IGFBP-4 shRNA-2 with
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. In total, 1 µg RNA
was reverse transcribed into cDNA by Superscript II Reverse
Transcription kit (Invitrogen; Thermo Fisher Scientific, Inc.). The
RT-qPCR were performed with SYBR-Green master mix (Thermo Fisher
Scientific, Inc.), as follows: Preheating at 94°C for 5 min, then
40 cycles of 94°C denaturation for 30 sec, 55°C annealing for 30
sec and 72°C elongation and data collection for 1 min. The primers
for IGFBP-4 were as follows: Forward, 5′-AGGGTGACCACCCCAACAAC-3′;
and reverse, 5′-TCCAGCGCCCGGTGCAGCTC-3′. The primers for the
internal control gene GAPDH were as follows: Forward,
5′-GCGAGATCCCTCCAAAATCAA-3′; and reverse
5′-GTTCACACCCATGACGAACAT-3′. Cq values were processed using
2−ΔΔCq method to calculate the relative expression level
(14).
Western blot analysis
Total protein was extracted from A549 cells, A549
IGFBP-4 shRNA-1 cells and A549 IGFBP-4 shRNA-2 cells using NP40
lysis buffer containing protease inhibitors and phosphatase
inhibitors cocktail (Thermo Fisher Scientific, Inc.). The protein
concentration was measured with bicinchoninic acid assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Denatured protein (40 µg)
was loaded and separated 10% SDS-PAGE. The proteins were
transferred onto polyvinylidene fluoride membranes (Merck KGaA,
Darmstadt, Germany), blocked with 5% non-fat milk, primary rabbit
anti-IGFBP-4 antibody (dilution, 1:100; cat. no. ab4253; Abcam) and
anti-rabbit IgG-HRP secondary antibody (dilution, 1:1,000; cat. no.
MBS435036; MyBioSource). The membranes were exposed to the film for
band development. GAPDH blotted by primary rabbit anti-IGFBP-4
antibody (dilution, 1:100; cat. no. ab8245; Abcam). GAPDH (Cell
Signaling Technology, Inc., Danvers, MA, USA) was used as an
endogenous control in this assay.
Cell proliferation detection
The A549 cells, A549 IGFBP-4 shRNA-1 cells and A549
IGFBP-4 shRNA-2 cells were plated onto 96-well dishes at 2,000
cells per well. The cell viability was measured using an MTT assay
(Thermo Fisher Scientific, Inc.) at the indicated days, according
to the manufacturer's protocol.
Online database
The Kaplan-Meier Plotter online database (8) was used to analyze the association
between IGFBP-4 and lung cancer.
Statistical analysis
The data were expressed as the mean + standard
deviation (SD) or mean ± SD. The differences between groups were
analyzed by one-way analysis of variance and least significant
difference analysis. The survival curve was generated using the
Kaplan-Meier method. The median survival comparison between groups
was calculated using the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Lung cancer tissues expressed
increased levels of IGFBP-4
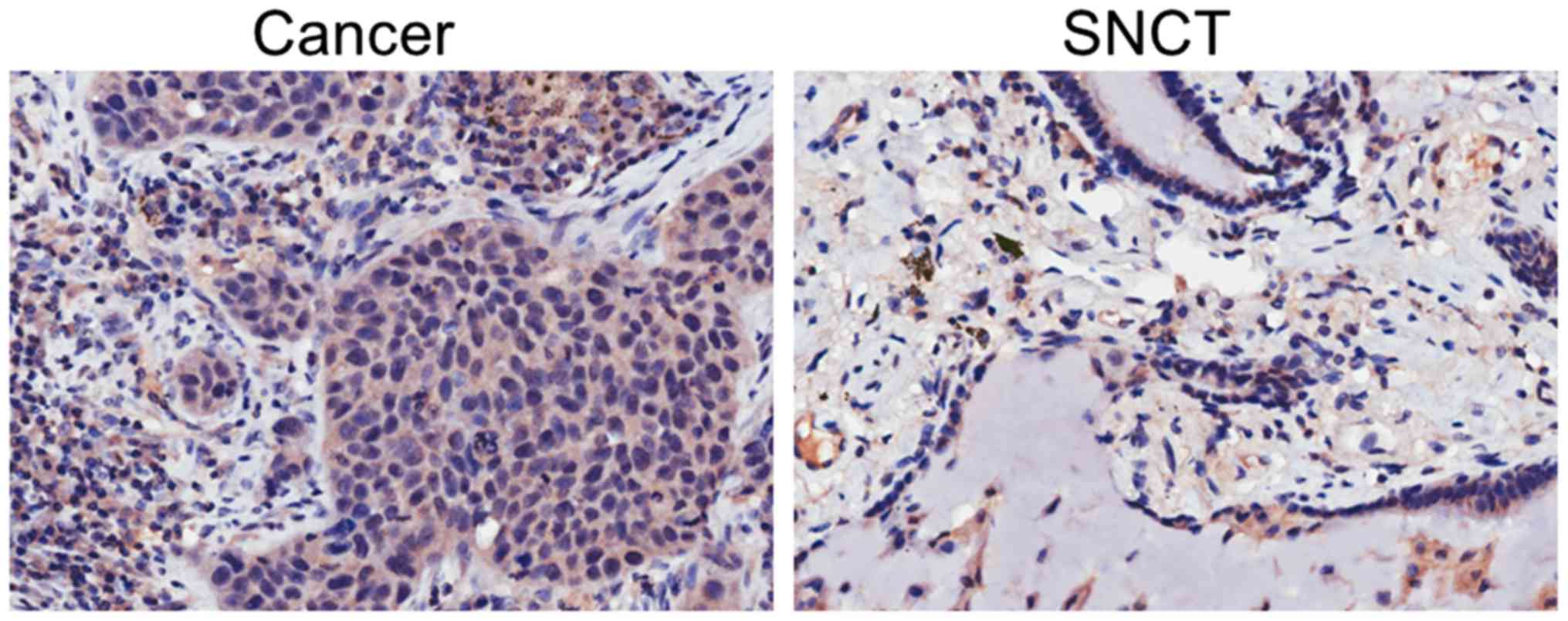
To assess the expression level of IGFBP-4 in lung
cancer tissues and SNCTs, the tissues were stained with IGFBP-4 by
immunohistochemistry. Lung cancer tissues showed increased IGFBP-4
expression compared with SNCT (Fig.
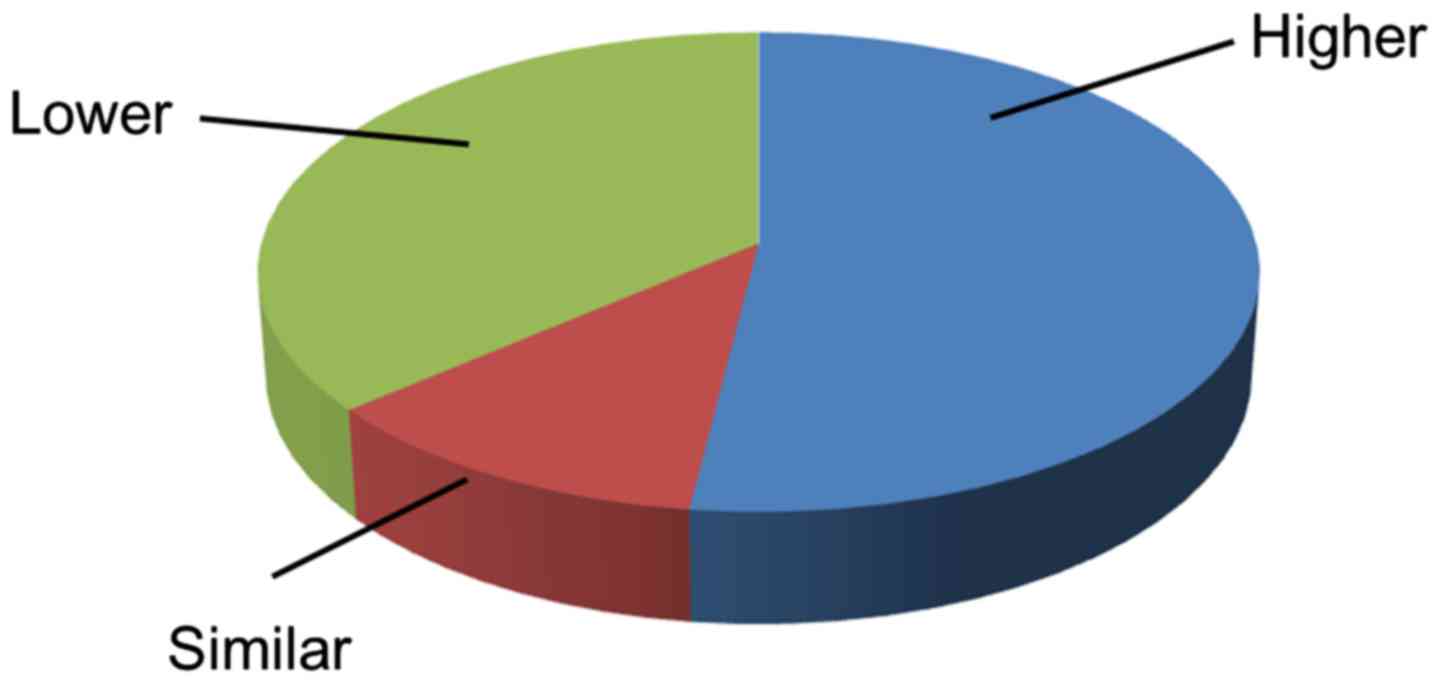
1). In 53 of the 102 paired tissues, cancer tissues expressed
an increased level of IGFBP-4 compared with SNCTs. This percentage
(51.96%) is larger than the proportion of patients with similar
(11.76%) or decreased (36.27%) levels of IGFBP-4 expression in
cancer tissues (Fig. 2).
IGFBP-4 is adversely associated with
lung cancer prognosis
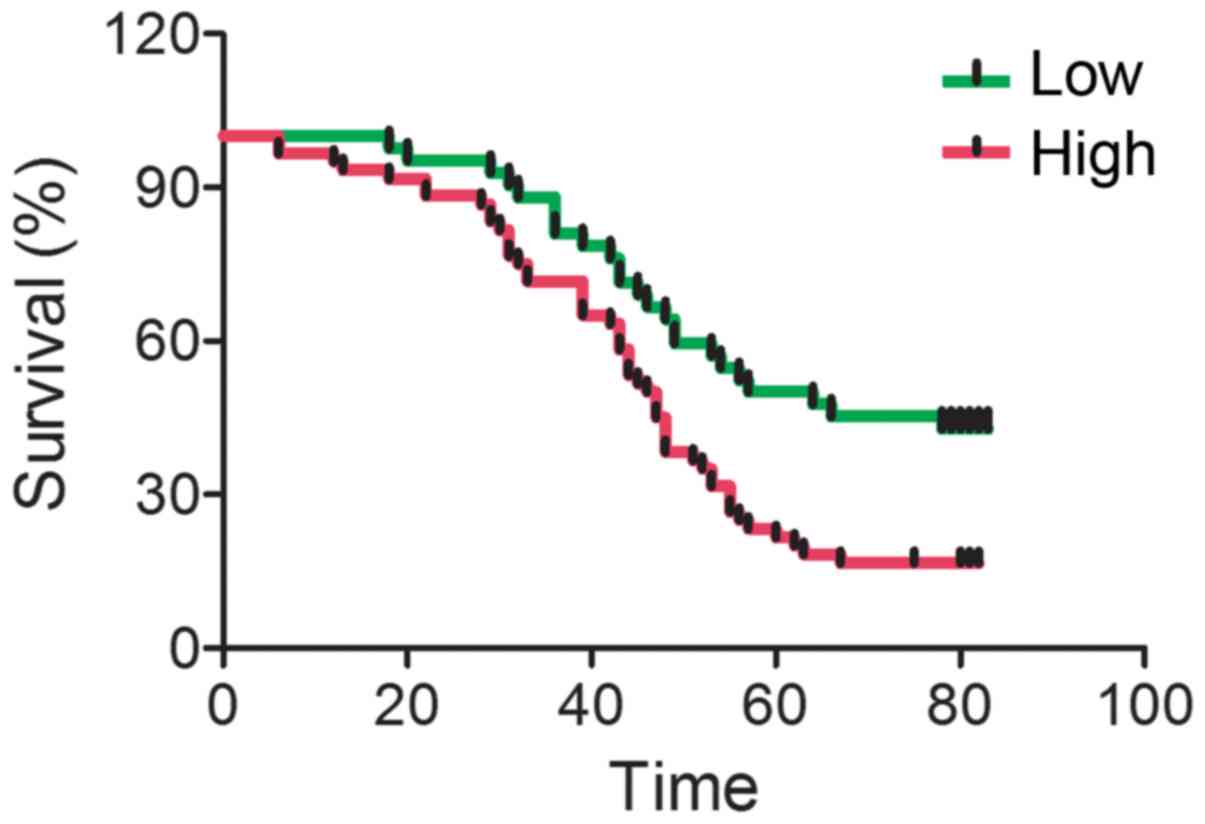
To investigate whether IGFBP-4 expression is
associated with lung cancer prognosis, the present study compared
IGFBP-4 expression with the survival of lung cancer patients. It
was shown that patients with increased IGFBP-4 expression have a
shorter median survival rate (Fig.
3). It was also found that IGFBP-4 expression was associated
with metastasis, TNM stages and tumor malignancy, but not with age,
sex or tumor size (Table I).
 | Table I.Association between IGFBP-4 expression
and pathological parameters in lung cancer tissue specimens. |
Table I.
Association between IGFBP-4 expression
and pathological parameters in lung cancer tissue specimens.
|
| IGFBP-4 expression,
n |
|
|---|
|
|
|
|
|---|
| Parameter | Low | High | P-value |
|---|
| Age |
|
| 0.388 |
| ≥60
years | 21 | 31 |
|
| <60
years | 21 | 29 |
|
| Sex |
|
| 0.286 |
| Male | 19 | 30 |
|
|
Female | 23 | 30 |
|
| Metastasis |
|
| 0.016 |
| Yes | 1 | 5 |
|
| No | 41 | 55 |
|
| TNM stage |
|
| 0.038 |
| I–II | 21 | 26 |
|
|
III–IV | 10 | 38 |
|
| Malignance |
|
| 0.043 |
| High | 20 | 18 |
|
| Low | 22 | 42 |
|
| Tumor size |
|
| 0.216 |
| Big | 25 | 32 |
|
|
Small | 18 | 27 |
|
IGFBP-4 knockdown promoted lung cancer
cell growth
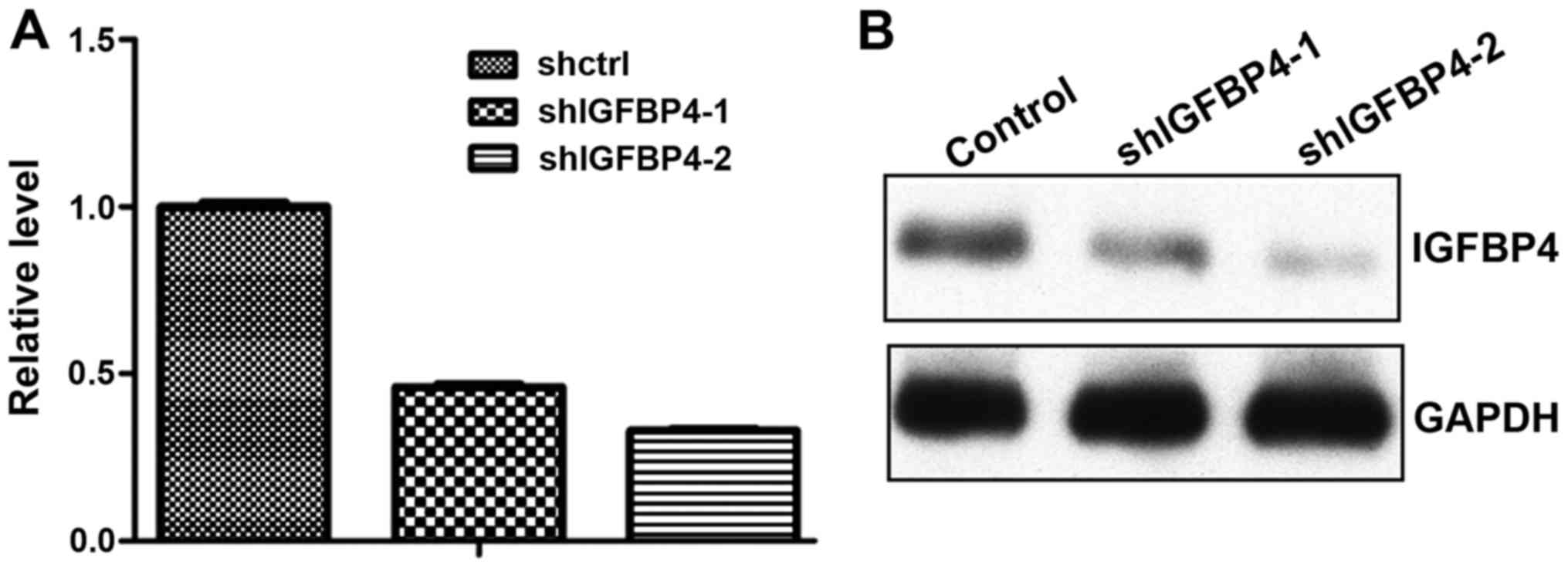
To assess whether blocking the function of IGFBP-4
could inhibit lung cancer progression, the present study knocked
down IGFBP-4 expression by shRNAs. Lentivirus-mediated shRNA
sufficiently knocked down IGFBP-4 expression at the mRNA (Fig. 4A) and protein level (Fig. 4B). With IGFBP-4 knockdown, A549 cells
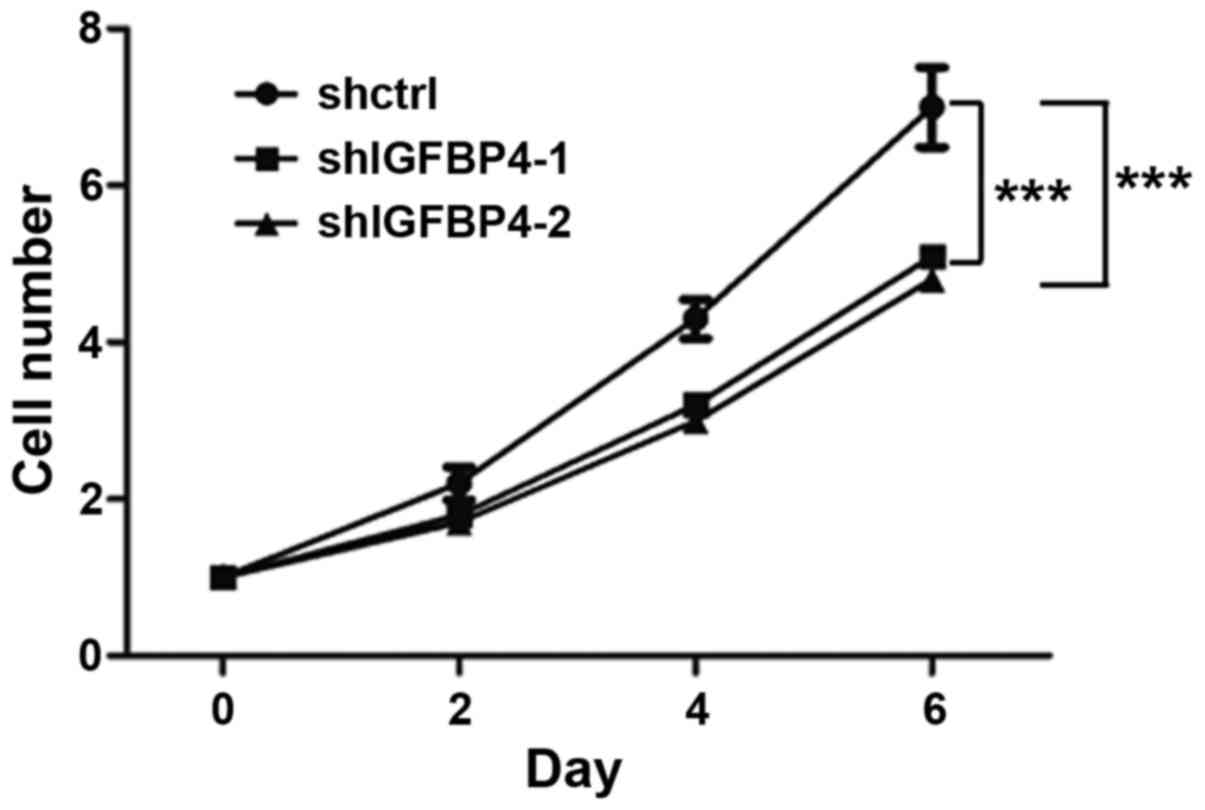
showed a slower proliferation rate (Fig.
5). This indicated that IGFBP-4 promoted lung cancer cell
proliferation.
Discussion
IGFBP-4 is an IGF binding protein that may affect
the half-life of IGFs and promote or inhibit tumor growth. It was
reported in inhibiting colon cancer growth (7,15). In
2009, researchers at the Massachusetts Institute of Technology
investigated an effective drug for the targeting of breast cancer
stem cells. Salinomycin was found to markedly inhibit breast cancer
stem cells. This study compared the effect of salinomycin and the
chemotherapy drug Paclitaxel on cancer stem cells. Salinomycin
could markedly inhibit cancer stem cell properties (9). Salinomycin has been evaluated by several
studies in various types of cancer stem cells (16–20).
Salinomycin downregulated the expression of IGFBP-4 and certain
other genes compared with Paclitaxel treatment. Breast cancer
spheres, namely cancer stem cells, had IGFBP-4 expression that was
~3 folds higher than the expression in adherent cells (9,21). This
suggested that IGFBP-4 may be relevant to cancer stem cells. As
cancer stem cells are the most important subpopulation in various
types of cancers, IGFBP-4 may promote cancer progression.
Lung cancer, which is the most common cancer in the
world, causes 2 million cancer-associated mortalities worldwide
every year (1). Developing an
effective therapeutic method for lung cancer therapy is of marked
importance. To identify whether lung cancer is associated with
IGFBP-4 expression, the Kaplan-Meier Plotter database (22) was used to analyze the association.
Patients with increased IGFBP-4 expression had a decreased survival
rate. IGFBP-4 was adversely associated with lung cancer
progression. This online analysis result is consistent with the
present hypothesis. In the current study, 102 pairs of cancer
tissues and surrounding non-cancerous tissues were collected. The
expression of IGFBP-4 was detected by immunohistochemistry in these
tissues. It was found that IGFBP-4 is highly expressed in lung
cancer tissues (Figs. 1 and 2). Patients with increased IGFBP-4
expression in cancer tissues showed a decreased median survival
time (Fig. 3).
To identify whether IGFBP-4 expression affects lung
cancer progression, its function in the lung cancer A549 cell line
was detected in vitro. The expression of IGFBP-4 was
identified in several different cancer cell lines using the The
Human Protein Atlas (23,24). Lung cancer cell lines were found to
highly express IGFBP-4. The A549 cells particularly expressed a
high level of IGFBP-4. IGFBP-4 was knocked down in A549 cells and
the expression was markedly decreased at the mRNA and protein
levels (Fig. 4). The effect of the
knockdown effect on cell proliferation was detected, and it was
found that IGFBP-4 knockdown could inhibit the proliferation of
A549 cells (Fig. 5).
In the present study, IGFBP-4 was associated with
lung cancer prognosis. Knockdown of IGFBP-4 leads to the inhibition
of proliferation in lung cancer cells. As IGFBP-4 is associated
with breast cancer stem cells, it was likely to also be associated
with lung cancer stem cells. Targeting IGFBP-4 may lead to the
inhibition of lung cancer stem cell growth. As lung cancer stem
cells are associated with chemoresistance, progression and
metastasis of lung cancer (25–27), this
may lead to the long-term inhibition of lung cancer development.
This may be investigated in a future study. In conclusion, the
current study identified the adverse association between IGFBP-4
and lung cancer prognosis. This provides a potential target for
lung cancer therapy.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rabasseda X: A report from the world
conference on lung cancer (September 6–9, 2015-Denver, Colorado,
USA). Drugs Today (Barc). 51:559–567. 2015.PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shimasaki S, Shimonaka M, Zhang HP and
Ling N: Identification of five different insulin-like growth factor
binding proteins (IGFBPs) from adult rat serum and molecular
cloning of a novel IGFBP-5 in rat and human. J Biol Chem.
266:10646–10653. 1991.PubMed/NCBI
|
|
5
|
Mohan S, Nakao Y, Honda Y, Landale E,
Leser U, Dony C, Lang K and Baylink DJ: Studies on the mechanisms
by which insulin-like growth factor (IGF) binding protein-4
(IGFBP-4) and IGFBP-5 modulate IGF actions in bone cells. J Biol
Chem. 270:20424–20431. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mosig RA, Lobl M, Senturk E, Shah H, Cohen
S, Chudin E, Fruscio R, Marchini S, D'Incalci M, Sachidanandam R,
et al: IGFBP-4 is a candidate serum biomarker for detection and
surveillance of early stage epithelial ovarian cancer. Research.
2:13422015. View Article : Google Scholar
|
|
7
|
Mosig RA, Lobl M, Senturk E, Shah H, Cohen
S, Chudin E, Fruscio R, Marchini S, D'Incalci M, Sachidanandam R,
et al: IGFBP-4 tumor and serum levels are increased across all
stages of epithelial ovarian cancer. J Ovarian Res. 5:32012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Drivdahl RH, Sprenger C, Trimm K and
Plymate SR: Inhibition of growth and increased expression of
insulin-like growth factor-binding protein-3 (IGFBP-3) and-6 in
prostate cancer cells stably transfected with antisense IGFBP-4
complementary deoxyribonucleic acid. Endocrinology. 142:1990–1998.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta PB, Onder TT, Jiang G, Tao K,
Kuperwasser C, Weinberg RA and Lander ES: Identification of
selective inhibitors of cancer stem cells by high-throughput
screening. Cell. 138:645–659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu W and Li Y: Salinomycin suppresses LRP6
expression and inhibits both Wnt/β-catenin and mTORC1 signaling in
breast and prostate cancer cells. J Cell Biochem. 115:1799–1807.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Al Dhaheri Y, Attoub S, Arafat K, Abuqamar
S, Eid A, Al Faresi N and Iratni R: Salinomycin induces apoptosis
and senescence in breast cancer: Upregulation of p21,
downregulation of survivin and histone H3 and H4 hyperacetylation.
Biochim Biophys Acta. 1830:3121–3135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fitzgibbons PL, Bradley LA, Fatheree LA,
Alsabeh R, Fulton RS, Goldsmith JD, Haas TS, Karabakhtsian RG,
Loykasek PA, Marolt MJ, et al: Principles of analytic validation of
immunohistochemical assays: Guideline from the College of American
Pathologists Pathology and Laboratory Quality Center. Arch Pathol
Lab Med. 138:1432–1443. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mellors RC: The application of labeled
antibody technics in studying cell antigens. Cancer Res.
28:1372–1381. 1968.PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Durai R, Davies M, Yang W, Yang SY,
Seifalian A, Goldspink G and Winslet M: Biology of insulin-like
growth factor binding protein-4 and its role in cancer (review).
Int J Oncol. 28:1317–1325. 2006.PubMed/NCBI
|
|
16
|
Diehl D, Lahm H, Wolf E and Bauersachs S:
Transcriptome analysis of a human colorectal cancer cell line shows
molecular targets of insulin-like growth factor-binding protein-4
overexpression. Int J Cancer. 113:588–599. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Resham K, Patel PN, Thummuri D, Guntuku L,
Shah V, Bambal RB and Naidu VG: Preclinical drug metabolism and
pharmacokinetics of salinomycin, a potential candidate for
targeting human cancer stem cells. Chem Biol Interact. 240:146–152.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kopp F, Hermawan A, Oak PS, Herrmann A,
Wagner E and Roidl A: Salinomycin treatment reduces metastatic
tumor burden by hampering cancer cell migration. Mol Cancer.
13:162014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naujokat C and Steinhart R: Salinomycin as
a drug for targeting human cancer stem cells. J Biomed Biotechnol.
2012:9506582012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang QL, Zhao ZQ, Li JC, Liang Y, Yin JQ,
Zou CY, Xie XB, Zeng YX, Shen JN, Kang T and Wang J: Salinomycin
inhibits osteosarcoma by targeting its tumor stem cells. Cancer
Lett. 311:113–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Patel N, Baranwal S and Patel BB: A
strategic approach to identification of selective inhibitors of
cancer stem cells. Methods Mol Biol. 1229:529–541. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Győrffy B, Surowiak P, Budczies J and
Lánczky A: Online survival analysis software to assess the
prognostic value of biomarkers using transcriptomic data in
non-small-cell lung cancer. PLoS One. 8:e822412013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Uhlen M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ponten F, Jirström K and Uhlen M: The
human protein Atlas-a tool for pathology. J Pathol. 216:387–393.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Y, Xu H, Huang W, Ding M, Xiao J,
Yang D, Li H, Liu XY and Chu L: Targeting lung cancer stem-like
cells with TRAIL gene armed oncolytic adenovirus. J Cell Mol Med.
19:915–923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen K, Huang YH and Chen JL:
Understanding and targeting cancer stem cells: Therapeutic
implications and challenges. Acta Pharmacol Sin. 34:732–740. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pine SR, Marshall B and Varticovski L:
Lung cancer stem cells. Dis Markers. 24:257–266. 2008. View Article : Google Scholar : PubMed/NCBI
|